Abstract
This review presents an overview of literature that describes the applications of microfluidics to assay individual cells. We quantify the content of an individual mammalian cell, so that we can understand what criteria a single-cell assay must satisfy to be successful. We put in context the justification for single-cell assays and identify the characteristics that are relevant to single-cell assays. We review the literature from the past 24 months that describe the methods that use microfabrication—conventional or otherwise—and microfluidics in particular to study individual cells, and we present our views on how an increasing emphasis on three-dimensional cell culture and the demonstration of the first chemically defined cell might impact single-cell assays.
INTRODUCTION
A mammalian cell contains water, inorganic ions, and a large number of small organic molecules, for example, vitamins, sugars, and fatty acids; these contents represent about 80% of living matter by weight and water is the most abundant in that category.1 The remaining 20% includes proteins and DNA.2 Together, these components represent the contents of a living cell, which is described for our purposes as a cube 10 μm on a side. The corresponding volume is ∼1×10−9 mL and the corresponding weight is ∼1×10−9 g. That 20% of protein and DNA material represents ∼2×10−10 g of the organic material of an individual cell. Given that a cell contains several thousand different proteins (and we assign a molecular weight of 50 000 g∕mol to each protein) each cell contains about a million molecules of each type of protein on average. This number, of course, varies with the type of protein: actin is an abundant protein (∼5×108 molecules∕cell), while the transmembrane insulin receptor protein is much less abundant (∼2×104 molecules∕cell).
A mammalian cell contains several compartments that are sometimes referred to as organelles. These compartments originate from the endosymbiosis of microsomes that contained DNA.3 Several major organelles exist in each cell and they include the mitochondrion, the endoplasmic reticulum, the Golgi apparatus, vacuoles, and one or more nuclei. Minor organelles include autophagosomes, centrioles, cilia, lysosomes, melanosomes, myofibrils, peroxisomes, ribosomes, and vesicles. These organelles define the locations of several types of biomolecules, for example, proteins can bind to membranes, diffuse in the cytoplasm, or be synthesized in the endoplasmic reticulum.
The description presented in the previous paragraphs is complicated by the reality of random variation in the expression of individual genes. One of the important contributions of single-cell assays to understanding the stochastic variation of gene expression was the use of a glucocorticoid-responsive transgene that encoded β-galactosidase;4 the use of this expression reporter revealed large differences between cells in the amount of transgene that was expressed. The assay further revealed that increasing the dose of glucocorticoid increased the probability that an individual cell would express the gene at a high level and not—as might be expected—result in a linear increase in expression in each cell. This and similar experiments have focused the interest of several groups on the problem of understanding variation between individual cells and, consequently, to develop useful assays for understanding the origin of such variations. These experiments are relevant to single-cell assays in two ways. First, they justify the need for single-cell assays in understanding intercellular biochemical kinetics; and second, they clarify the challenge for assays that seek to obtain conclusive data about a particular protein from the study of its location in an individual cell or about the role of a single cell: it is likely that a single-cell assay will require a large population to sample in order to describe intercellular variation that is not a result of variation in gene expression. A better understanding of the role of noise in cellular behavior will be an important consequence of research and development of single-cell assays; however, that same noise will likely challenge the application of single-cell assays to understanding variations in behavior between cells in a given population.
This introduction presents an elementary overview of the challenge for single-cell assays: an individual cell contains a large number of molecules, where each category of molecules, e.g., proteins, can be divided into subcategories, located in several different organelles, and are produced and degraded dynamically during the cell cycle. An individual cell is a moving target for subcellular quantitative analysis and, even if analyzed appropriately, is characterized by stochastic gene expression, which complicates the elucidation of significant differences between cells in a given population.
JUSTIFICATION
For the purposes of this review, we define a single-cell assay as an experiment that quantifies a function or property of an individual cell when the interactions of that cell with its environment can be controlled precisely or can be isolated from the function or property under examination. The practical purpose of a single-cell assay is to identify the function and behavior of individual cells, so that we can understand what function(s) an individual cell plays in a population of cells. We can restate this purpose quantitatively to mean the identification of the function of a family of biomolecules that might constitute one molecule for every million other molecules in a cell as those biomolecules are simultaneously and dynamically produced or degraded at several locations within that cell. Knowing the function of biomolecules and how these functions vary from cell to cell when the cells are of the same or of different type is promoted as information that is critical to understanding normal and pathogenic conditions at the level of an organism.
The driving force for developing tools that assay individual cells is that there are no easy methods available to distinguish the contributions of individual cells from a background signal of millions of cells. It is worthwhile to consider, however, that the information that describes an individual cell lacks context if the information that describes the average of other similar cells is unavailable. We alluded to this need in Sec. 1: random variation in gene expression increases the complexity in quantifying the contribution of a given family of biomolecules to the behavior of an individual cell and requires a large population sample to determine what is actually unique about the cell and whether that uniqueness is due to a specific group of biomolecules. This statement influences our design choices when fabricating single-cell assays, for example, we would prefer assays that make it easy to obtain information about individual cells and large numbers of cells simultaneously. This type of design is best represented by an ordered array of individual cells that is confined to a small, planar substrate: the order in the array makes it easy to observe single cells and the size of the substrate allows, say, a microscope to observe all the cells on the substrate.5 Such a design eliminates the need to perform two experiments: the single-cell assay and the multiple-cell assay are performed together; however, single-cell assays that use microfluidics are often unable to describe the average behavior of multiple cells as readily as a static array.
TRENDS
The research and development of single-cell assays is largely predicated on the notion that genetic variation causes disease and that small subpopulations of cells represent the origin of the disease. This perspective derives from a way of thinking about disease as being caused primarily by one or more malfunctioning genes; however, cells in tissues exhibit plasticity in their differentiated state, a fact that has been used to define a role for the extracellular matrix in normal and pathological cell function.6, 7, 8, 9 Which perspective dominates is unclear; however, the future development of single-cell assays should emphasize the use of external stimuli to simulate the cellular microenvironment.
A consequence of realizing that the extracellular matrix is a signaling element has been the introduction of new methods that culture cells in natural or synthetic hydrogels or that grow spheroid cultures of cells.10, 11 These methods provide one way—using materials to encapsulate cells—to stimulate individual cells; however, to be truly useful single-cell assays that use these materials must define the means to pattern, lyse, and collect (the contents of) individual cells from three-dimensional matrices.
One of the most intriguing developments in recent months was the replacement of a bacterial genome with a chemically synthesized genome.12 The notion that it might be possible to synthesize a cell or, at least, provide the DNA that controls what types of proteins can be produced by the cell invites us to consider the purpose of single-cell assays when the components of the cell can be defined a priori. Our thoughts lead us to realize that even in this circumstance single-cell assays that determine the fate of the products from a synthetic genome might be important in the iteration of the design of a synthetic cell.
This review will provide a description of what we consider important developments in the application of single-cell assays to study biology during the past 24 months.13 We focus on methods that use microfabrication—conventional or otherwise—and microfluidics in particular to assay individual cells and we categorize research in the area of single-cell assays according to the type of information obtained from the assay; these categories are (i) assays that identify or quantify compounds that are secreted from cells; (ii) assays that identify or quantify subcellular components; and (iii) assays that identify or quantify cell-cell interactions. We will also review briefly the methods that organize individual cells.
ASSAYS OF COMPOUNDS SECRETED FROM CELLS
Eukaryotic cells secrete enzymes, surfactants, sebum, and a range of signaling molecules. The ability to detect these molecules, to quantify their number, and even to analyze their structure confirms their presence during a particular phase of the cell cycle and reveals the process that each molecule undergoes inside a cell prior to delivery to the outside, e.g., glycosylation.
Schmid et al. trapped individual Schizosaccharomyces pombe yeast cells in a microfluidic channel using negative dielectrophoresis (nDEP);14 nDEP is a noncontact method that can trap cells in an electric field. In this instance, the cells were engineered to secrete an enhanced Green Fluorescent Protein or eGFP protein that was detected in real-time using confocal microscopy. This method detected 1.2 molecules of eGFP each minute; however, it is unclear if false positives contributed to this result. Presently, the method can differentiate cells that secrete compounds from those that do not when at least four cells are trapped.
Love et al. invented a method that uses microfabrication to fabricate an array of wells (∼50 μm on a side);15 the method requires the dilution of a suspension of cells, so that when the suspension fills the wells in the array the occupancy of each well is, at most, an individual cell (each assay contains 104–105 cells). Their choice of hybridomas was appropriate, because the selection of cells that produce high-activity antibodies is time-consuming (several days). In this method, the use of a glass slide that is placed over the wells patterns an array of physisorbed antibodies on the slide that is indistinguishable from the pattern of wells. Subsequent analysis of the patterned antibodies identifies the location of the well that contains the cell(s) that produces the largest titer of active antibody. These cells can be isolated using an automated system.16
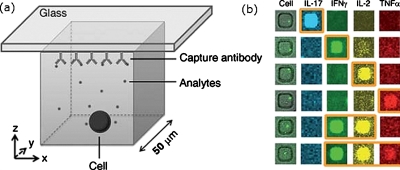
The method can be applied generally: the frequencies and rates of secretion of cytokines from peripheral blood cells were quantified and might be used to identify the subpopulations of cells within a larger set of supposedly homogeneous cells (Fig. 1).17 Kim et al. used a similar approach to quantify the amount of sialylated oligosaccharide on secreted proteins.18 The selection of glycoproteins with a specific glycosylation profile and the cells that produce them avoids the need for high-throughput analysis of large numbers of proteins.
Figure 1.
Method for detecting multiple cytokines that were secreted from T cells. (a) An individual cell is patterned in a well and a glass substrate that contains capture antibodies forms the roof of the well. The T cell secretes cytokines that bind to the capture sites. (b) An image of single viable cells in each microwell (left) and each subsequent column shows the fluorescence from antibodies that bind to each type of cytokine. The brighter spots indicate the location of the well in the array of T cells that produced the most of a given cytokine. Reprinted with permission from Q. Han et al., Lab Chip, 10, 1391 (2010). Copyright 2010 The Royal Society of Chemistry.
Kennedy et al. monitored the secretion of insulin from single islets of Langerhans (these are micro-organs that contain thousands of individual cells, some of which are β-cells that secrete insulin).19 They used a microfluidic device that permitted continuous perfusion of buffer and reagents, which appear to maintain healthy islets for at least 24 h. The device was capable of recording levels of secreted insulin down to the nanomolar level and over periods as long as 39 h. The device is a viable alternative to conventional insulin monitoring tools (e.g., radioimmunoassays or enzyme-linked immnosorbent assays) when continual monitoring and high temporal resolution are required.
Gillis et al. simplified the trapping of single chromaffin cells over electrodes and measured exocytosis20 and Taboryski et al. detected less than 10 000 molecules of dopamine during exocytosis from individual pheochromocytoma cells.21
In this section, we described several methods that isolate individual cells for analysis of secreted compounds. The use of negative dielectrophoresis presents a new way to isolate small numbers of cells; however, this approach is unlikely to scale as efficiently as serial dilution applied to the isolation of high-titer hybridomas. The versatility of serial dilution is revealed in its application to detect several types of secreted compounds and its future is promising with the method applied in later examples in this review. The extension of microfluidic devices to include analytical capability—in this section, amperometric and capillary electrophoresis (CE) analysis were referenced—is another trend that will be repeated in the examples that follow.
ASSAYS OF SUBCELLULAR COMPONENTS
Subcellular analysis can trace the location of biomolecules during the cell cycle and the effect they have on cell function. The use of fluorescent reporters and sensitive detection tools, e.g., CE, can reveal with sub-100 ms temporal precision this type of information.
Allbritton et al. fabricated an array of wells in photoresist on a substrate that contained electrodes;22 each row of wells was located between two electrodes, so that the application of an electric field lyses cells (∼60 ms). Each well could be addressed using a capillary and the contents of the lysed cells were captured by the capillary using electro-osmotic flow; subsequent analysis of the contents of the lysed cells was performed using CE. The CE analysis resolved Oregon Green and fluorescein dyes in less than 2 min. This approach provides a way to isolate individual cells, observe their behavior, and extract components rapidly from them; however, and despite the fast lysis of individual cells, the approach is serial and cultures cells using unconventional materials. The same research group modified this approach, so that a laser was used to lyse cells;23 this modified approach analyzed 1.8 cells each minute, which, when compared to conventional CE (5–35 cells∕day), is a remarkable achievement.
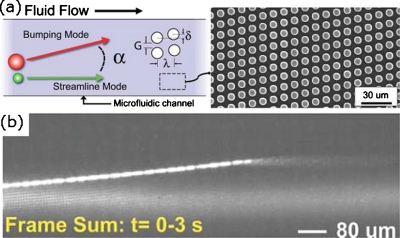
Austin et al. fabricated an array of posts in a microfluidic channel that were not aligned with the direction of flow in the microchannel (Fig. 2).24 Using three independent flow streams (buffer, tracer, and lysis), an individual E. coli cell traversed the channel entering initially the buffer stream and finally being lysed in the lysis stream. The asymmetric alignment of posts with respect to the microchannel caused the cells to move across the channel as liquid flowed along the channel. The chromosome of an individual bacterial cell that was lysed was detected using this device.
Figure 2.
An individual cell can traverse a microfluidic channel that contains an array of posts that are not aligned with the direction of flow along the channel. (a) Left: scheme of the motion of a cell that moves from one side of a microchannel to the other. A tracer stream is added to observe the direction of flow. Only objects above a certain size move across the channel. Right: SEM image of a microchannel that contains an array of posts that are not parallel with the walls of the microchannel. (c) The motion of an individual E. coli cell in a microchannel that contains an array of posts; the tracer stream in the channel is GFP. The cell contains GFP and, as it enters the region of the microfluidic channel that contains a solution that lyses cells, releases the GFP, which travels along the microchannel. Reprinted with permission from K. J. Morton et al., Lab Chip, 8, 1448 (2008). Copyright 2008 The Royal Society of Chemistry.
Lu et al. fabricated a microfluidic device that incorporated a valve over a microchannel that pushed an individual cell against the lower surface of the microchannel.25 The lower surface was a glass substrate, which was used as a waveguide for an evanescent field; this method of imaging, known as total internal reflection fluorescence microscopy, is capable of imaging molecules at the periphery of cells and excludes significant amounts of background interference from the image obtained. The device flowed cells along the microchannel, restricting the passage of cells to one at a time when the valve was actuated, and imaged part of the membrane of each cell that passed into the evanescent field.
Single-molecule imaging of biomolecules in living cells is an exponentially growing field of research.26 Xie et al. quantified the expression of protein and mRNA in individual E. coli cells.27 Cells were attached to a polylysine coated coverslip and imaged; the protein and mRNA copy numbers of an individual cell were not correlated. Chen et al. showed that the serine∕threonine kinase receptors of transforming growth factor-β (TGF-β) dimerize when stimulated by TGF-β1.28 These results provide exquisite information that describes the behavior of individual molecules; however, the development of high-throughput single-molecule imaging remains a challenge. Moreover, the design of assays that exploit single-molecule imaging might be an even greater challenge.
The results presented in this section highlight the creative use of microfabrication, in particular, the use of an array of posts to move a single cell across two fluid boundaries, causing cell lysis. It is, however, unclear if these approaches to subcellular analysis can be scaled to analyze thousands of cells in a short time, and we regard a rapid analysis of subcellular components to remain a challenge for the field.
ASSAYS OF CELL-CELL OR CELL-DRUG INTERACTIONS
The ability to observe an individual cell and its role in signaling to neighboring cells, its response to external stimulants, and to do so in a massively parallel way could reveal precise information that describes the interactions that compose signaling networks and the limits of a particular drug therapy (e.g., identifying drug-resistant phenotypes). Such information would inform new strategies in therapeutics and provide systems biologists with data to infer signaling networks in cellular microenvironments.
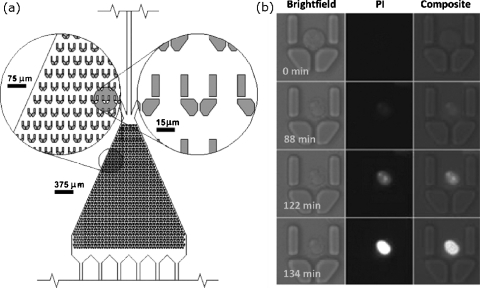
Cooper et al. used an array of microfabricated corrals in a microfluidic channel to pattern several thousand individual and nonadherent cells (Fig. 3).29 The advantages of using microfluidics to pattern cells include the stimulation or labeling of the organized cells with several drugs or fluorophores, which allow for the observation of the effects of a sequence of stimulants to be observed; the shape of the corral minimized the effects of shear flow on the cells. The results of the effects of apoptotic reagents on individual cells were determined and the time-dependent relationship between exposure to these reagents and apoptosis was identified. This approach, however, demonstrates the need for assays that enable the observation of individual as well as average cell behavior.
Figure 3.
Method for organizing individual cells in a microfluidic channel. (a) The microfluidic channel contains barriers that trap individual, nonadherent cells. These traps reduce the effect of shear forces that cells experience in a microfluidic channel. (b) An individual HL60 cell undergoing apoptosis over the course of 134 min. The cell was exposed to staurosporine, a cytotoxic reagent, and propidium iodide, which is impermeable to membranes and therefore reports on membrane integrity. Reprinted with permission from D. Wlodkowic et al., Anal. Chem., 81, 5517 (2009). Copyright 2009American Chemical Society.
Wikswo et al. modified this approach, so that the smaller and more deformable T cells could be organized, and connected two separate devices in sequence, so that the chemicals secreted in one array could be used to activate processes in cells in the second array.30 These cells were cultured for more than 24 h and their calcium signaling pathways were stimulated using chemicals, antibody-coated beads, and dendritic cells. The chaining of arrays maintained secreted chemicals at physiological concentrations and paracrine signaling between cells in one array and cells in the second array was observed. This approach avoids the dilution of secreted chemicals that occurs in conventional tissue culture dishes.
Samuels et al. created a collection of drops that contained different concentrations of a cytotoxic agent, which were fused with another collection of drops that contained individual U937 mammalian cells.31 The cells in drops were incubated and tested for viability using the same chip, so that several functions were performed using one device. The device can sample cells at 100 Hz and the viability of cells encapsulated in drops prior to the addition of the cytotoxic reagent was 80%. This device is a step toward creating multiparameter assays using a drop-based microfluidic device.
Hansen et al. extended the throughput of Quake’s pneumatically controlled microfluidic device32 to perform 256 experiments in parallel that stimulated the pheromone response pathway of Saccharomyces cerevisiae yeast cells over a range of concentrations, time intervals, and frequencies.33
This group of research results is best characterized by the trend to incorporate several functions on a single microfluidic device. For example, the ability to stimulate cells, to change the conditions of stimulation, and to observe their behavior on a single microfluidic chip is representative of the “lab-on-a-chip” concept and the application of this concept to single-cell assays is welcome.
METHODS THAT ORGANIZE INDIVIDUAL CELLS
There are several tens of methods available that pattern individual cells; most methods use random allotment to trap individual or small numbers of cells in a given volume; the statistics of successfully patterning an individual cell in each volume is described using Poisson statistics. The probability of a given volume containing k cells is described by
where λ is the average number of cells per unit volume.
Toner et al. developed a method that overcomes the Poisson nature of encapsulating individual cells in small drops.34 The method flows an ordered sequence of cells along a microchannel to a junction (which flows two streams of oil perpendicular to the direction of flow of cells) that produces drops at a rate equal to or slightly faster than the rate of arrival of cells at the junction. For a given ratio of drops that contain single cells to drops that contain multiple cells, the authors show that their method produces 20 times the ratio predicted using Poisson statistics.
Ismagilov et al. described the random allotment of cells in an array of wells (or drops) as “stochastic confinement.”35 They demonstrated the use of an air bubble to confine individual P. aeruginosa cells in wells (10 μm diameter, ∼1 pL confined volume) and observed the onset of quorum sensing in the presence of very small numbers of cells.36 This observation is important because the onset of quorum sensing was dependent only on an inherent variability in P. aeruginosa: cells in different wells as well as several cells from the same wells did not initiate quorum sensing at the same time. Moreover, quorum sensing was shown to be possibly a function of the amount of biomass per unit volume and might not, or not just, be a characteristic of large numbers of cells.
Individual S. aureus cells were confined to drops in a microfluidic device and assayed for the minimum inhibitory concentration of several antiobiotics.37 The time required to detect changes in fluorescence intensity to a viability indicator was less than 2 h, which was less than the time required for a similar analysis using larger volumes. The microfluidic device was designed so that several antibiotics could be screened in a single experiment and the use of blood plasma to perform the same assays demonstrated that the device was compatible with biological liquids.
Griffiths et al. fabricated a microfluidic device that sorted individual E. coli cells by their fluorescent activity (Fig. 4).38 The device produced aqueous drops (in an oil phase) that contained individual cells; the drops flowed along a microchannel that contained a pair electrodes; when an AC field was applied using the electrodes the drops were directed into one of two exit channels. The rate of sorting was 2000 drops (cells)∕s. This device combines the concept of a mobile microwell—a drop that contains an individual cell—with high-throughput cell sorting; however, the applications of the device are best suited to single-parameter assays. Static arrays of microwells are more appropriate for formulating multiparameter assays.
Figure 4.
Producing and sorting drops that contained cells (image refers to drops that contain fluorescein only; see Ref. 35 for more details). (a) A microfluidic flow-focusing device is used to produce 12 pL aqueous monodisperse drops in oil. (b) The drops could be removed to a Pasteur pipette for incubation. (c) The drops could be reinjected into a microfluidic channel and space could be created between drops using surfactant-free oil. (d) An ac electric field can be applied between two electrodes located near the microfluidic channel. The field caused the drops to flow into the upper channel rather than the lower channel (inset: in the absence of an electric field the drops flow preferentially into the lower channel, because that channel has less hydraulic resistance). The scale bars are 100 μm, except in (b) where it is 1 mm. Reprinted with permission from J.-C. Baret et al., Lab Chip, 9, 1850 (2009). Copyright 2009 The Royal Society of Chemistry.
Straightforward methods that array cells using dilution and that organize individual cells in wells are most appealing for prototyping devices;15, 16 they are also the methods that can be integrated easily with existing technologies. Further research might realize more methods that combine microfluidics with arraying technology; so far, this combination defined strategies that overcome the statistical limitations of patterning individual cells in individual wells and that avoid the dilution of secreted compounds, so that the effect of high concentrations of signaling compounds secreted from cells in wells can be studied. We regard microfluidic single-cell assays as having the greatest potential to revolutionize how we view assays: the integration of several microfluidic modules that flow, fuse, delay, and probe drops that contain individual cells31 provides a compelling argument that several assay steps can be integrated onto a single device in the near future.
CONCLUSION
Several trends are apparent in our review of the past two years of research in the application of microfabrication and microfluidics to single-cell assays. The successful combination of patterning, stimulation, and analysis of a large number of cells on individual chips is exemplified by several reports and is fast becoming a standard for consideration as a useful contribution. The persistent challenges are also evident, namely, a method that can identify and analyze rapidly subcellular components at scale. It is possible that CE and∕or mass spectrometry could be applied usefully to microfluidic chips to address this challenge.
The rapid development of single-cell assays provides several opportunities for new research in biology; however, the needs of biologists are also changing rapidly. We regard the invention of robust methods that assay individual cells in their natural microenvironment as critical to providing the best information about the role of cells in tissues. The challenges in organizing cells in three dimensions as well as detecting responses from individual cells within a matrix will have to be overcome to obtain this information. Finally, the insertion of a synthetic genome into a bacterial cell is evidence that cell-based assays might mean something entirely different when we can program the cell using synthetic DNA versus inferring what is happening inside the cell from external observations.
References
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., and Walter P., Molecular Biology of the Cell (Garland Science, New York, 2002). [Google Scholar]
- This number also includes polysaccharides, which, together with proteins, are referred to as macromolecules.
- A distinction is made between DNA-containing organelles, membrane-bound organelles, and what some scientists call generic organelles; however, in this review we use the term “organelle” generally.
- Ko M. S., Nakauchi H., and Takahashi N., EMBO J. 9, 2835 (1990) [DOI] [PMC free article] [PubMed] [Google Scholar]; For an excellent review on variation in gene expression and noise in biology, see Raj A. and van Oudenaarden A., Cell 135, 216 (2008). 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- It is interesting to note that cells cultured on a Petri dish represent a disordered array of cells and can provide similar information in certain types of experiments, although obtaining data from a disordered array consumes more time and is prone to error. See Ref. for an example of single-molecule imaging of biomolecules in live cells that is, however, compatible with a disordered collection of cells.
- Coulombre J. L. and Coulombre A. J., Dev. Biol. 25, 464 (1971). 10.1016/0012-1606(71)90042-X [DOI] [PubMed] [Google Scholar]
- Ferraris C., Chaloin-Dufau C., and Dhouailly D., Differentiation 57, 89 (1994). 10.1046/j.1432-0436.1994.5720089.x [DOI] [PubMed] [Google Scholar]
- Liu H., Radisky D. C., Wang F., and Bissell M. J., J. Cell Biol. 164, 603 (2004). 10.1083/jcb.200306090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. C., Zhang H., Pallavicini M., Gray J. W., Baehner F., Park C. J., and Bissell M. J., Cancer Res. 66, 1526 (2006). 10.1158/0008-5472.CAN-05-3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon R. B. and Gooden M. D., In Vitro Cell. Dev. Biol.: Anim. 38, 97 (2002). [DOI] [PubMed] [Google Scholar]
- Boudreau N., Myers C., and Bissell M. J., Trends Cell Biol. 5, 1 (1995). 10.1016/S0962-8924(00)88924-2 [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Glass J. I., Lartigue C., Noskov V. N., Chuang R. -Y., Algire M. A., Benders G. A., Montague M. G., Ma L., Moodie M. M., Merryman C., Vashee S., Krishnakumar R., Assad-Garcia N., Andrews-Pfannkoch C., Denisova E. A., Young L., Qi Z. -Q., Segall-Shapiro T. H., Calvey C. H., Parmar P. P., C. A.HutchisonIII, Smith H. O., and Craig Venter J., Science 329, 52 (2010). 10.1126/science.1190719 [DOI] [PubMed] [Google Scholar]
- For reviews that describe earlier research see Chemical Cytometry: Ultrasensitive Analysis of Single Cells, edited by Lu C. (Wiley, Weinheim, 2010) [Google Scholar]; Borland L. M., Kottegoda S., Phillips K. S., and Allbritton N. L., Annu. Rev. Anal. Chem. 1, 191 (2008) 10.1146/annurev.anchem.1.031207.113100 [DOI] [PubMed] [Google Scholar]; Yan H., Zhang B. Y., and Wu H. K., Electrophoresis 29, 1775 (2008) 10.1002/elps.200700561 [DOI] [PubMed] [Google Scholar]; Yi C. Q., Li C. W., Ji S. L., and Yang M. S., Anal. Chim. Acta 560, 1 (2006). 10.1016/j.aca.2005.12.037 [DOI] [Google Scholar]
- Kortmann H., Kurth F., Blank L. M., Dittrich P. S., and Schmid A., Lab Chip 9, 3047 (2009) 10.1039/b908679j [DOI] [PubMed] [Google Scholar]; Kortmann H., Chasanis P., Blank L. M., Franzke J., Kenig E. Y., and Schmid A., Lab Chip 9, 576 (2009). 10.1039/b809150a [DOI] [PubMed] [Google Scholar]
- Love J. C., Ronan J. L., Grotenberg G. M., van der Veen A. G., and Ploegh H. L., Nat. Biotechnol. 24, 703 (2006). 10.1038/nbt1210 [DOI] [PubMed] [Google Scholar]
- Choi J. H., Ogunniyi A. O., Du M., Du M., Kretschmann M., Eberhardt J., and Love J. C., Biotechnol. Prog. 26, 888 (2010). 10.1002/btpr.374 [DOI] [PubMed] [Google Scholar]
- Han Q., Bradshaw E. M., Nilsson B., Hafler D. A., and Love J. C., Lab Chip 10, 1391 (2010). 10.1039/b926849a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim W., Kim Y., Son Y. D., Lee S. -C., Kim E., Kim S. H., Kim J. H., and Kim H. -S., Anal. Chem. 82, 5830 (2010). 10.1021/ac100992n [DOI] [PubMed] [Google Scholar]
- Reid K. R. and Kennedy R. T., Anal. Chem. 81, 6837 (2009). 10.1021/ac901114k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Bhattacharya S., Chen X., Barizuddin S., Gangopadhyay S., and Gillis K. D., Lab Chip 9, 3442 (2009). 10.1039/b913216c [DOI] [PubMed] [Google Scholar]
- Spégel C., Heiskanen A., Pedersen S., Emnéus J., Ruzgas T., and Taboryski R., Lab Chip 8, 323 (2008). 10.1039/b715107a [DOI] [PubMed] [Google Scholar]
- Marc P. J., Sims C. E., Bachman M., Li G. P., and Allbritton N. L., Lab Chip 8, 710 (2008). 10.1039/b719301g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Sims C. E., and Allbritton N. L., Electrophoresis 31, 2558 (2010). 10.1002/elps.201000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton K. J., Loutherback K., Inglis D. W., Tsui O. K., Sturm J. C., Chou S. Y., and Austin R. H., Lab Chip 8, 1448 (2008). 10.1039/b805614e [DOI] [PubMed] [Google Scholar]
- Wang J., Bao N., Paris L. L., Geahlen R. L., and Lu C., Anal. Chem. 80, 9840 (2008). 10.1021/ac801940w [DOI] [PMC free article] [PubMed] [Google Scholar]
- For a recent review see Lord S. J., Lee H. -L. D., and Moerner W. E., Anal. Chem. 82, 2192 (2010). 10.1021/ac9024889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y., Choi P. J., Li G. -W., Chen H., Babu M., Hearn J., Emili A., and Xie X. S., Science 329, 533 (2010). 10.1126/science.1188308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Jiang Y., Wang Q., Ma X., Xiao Z., Zuo W., Fang X., and Chen Y. -G., Proc. Natl. Acad. Sci. U.S.A. 106, 15679 (2009). 10.1073/pnas.0908279106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodkowic D., Faley S., Zagnoni M., Wikswo J. P., and Cooper J. M., Anal. Chem. 81, 5517 (2009). 10.1021/ac9008463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faley S., Seale K., Hughey J., Schaffer D. K., VanCampernolle S., McKinney B., Baudenbacher F., Unutmaz D., and Wikswo J. P., Lab Chip 8, 1700 (2008). 10.1039/b719799c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouzes E., Medkova M., Savenelli N., Marran D., Twardowski M., Hutchison J. B., Rothberg J. M., Link D. R., Perrimon N., and Samuels M. L., Proc. Natl. Acad. Sci. U.S.A. 106, 14195 (2009). 10.1073/pnas.0903542106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science 288, 113 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- Taylor R. J., Falconnet D., Niemistö A., Ramsey S. A., Prinz S., Shmulevich I., Galitski T., and Hansen C. L., Proc. Natl. Acad. Sci. U.S.A. 106, 3758 (2009). 10.1073/pnas.0813416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edd J. F., Carlo D. D., Humphrey K. J., Köster S., Irimia D., Weitz D. A., and Toner M., Lab Chip 8, 1262 (2008). 10.1039/b805456h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M. E., Liu W., Haney E. B., and Ismagilov R. F., Chem. Soc. Rev. 39, 974 (2010). 10.1039/b917851a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedicker J. Q., Vincent M. E., and Ismagilov R. F., Angew. Chem., Int. Ed. 48, 5908 (2009). 10.1002/anie.200901550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedicker J. Q., Ti L., Kline T. R., and Ismagilov R. F., Lab Chip 8, 1265 (2008). 10.1039/b804911d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J. -C., Miller O. J., Taly V., Ryckelynck M., El-Harrak A., Frenz L., Rick C., Samuels M. L., Hutchison J. B., Agresti J. J., Link D. R., Weitz D. A., and Griffiths A. D., Lab Chip 9, 1850 (2009). 10.1039/b902504a [DOI] [PubMed] [Google Scholar]