Abstract
Human infertility is common and frequently linked to poor germ cell development. Yet, human germ cell development is poorly understood, at least in part due to the inaccessibility of germ cells to study especially during fetal development. Here, we explored the function of a highly conserved family of genes, the NANOS genes, in the differentiation of human germ cells from human embryonic stem cells. We observed that NANOS-1, -2 and -3 mRNAs and proteins were expressed in human gonads. We also noted that NANOS3 was expressed in germ cells throughout spermatogenesis and oogenesis and thus, focused further efforts on this family member. NANOS3 expression was highest in human germ cell nuclei where the protein co-localized with chromosomal DNA during mitosis/meiosis. Reduced expression of NANOS3 (via morpholinos or short hairpin RNA) resulted in a reduction in germ cell numbers and decreased expression of germ cell-intrinsic genes required for the maintenance of pluripotency and meiotic initiation and progression. These data provide the first direct experimental evidence that NANOS3 functions in human germ cell development; indeed, NANOS3 is now one of just two genes that has been directly shown to function in germ cell development across diverse species from flies, worms, frogs and mice to humans [the other is BOULE, a member of the Deleted in Azoospermia (DAZ) gene family]. Findings may contribute to our understanding of the basic biology of human germ cell development and may provide clinical insights regarding infertility.
INTRODUCTION
The specification of germ cell versus somatic cell fate is of primary importance to all species and occurs early in embryo development (1). In model organisms, two divergent methods of germ cell specification and maintenance are apparent (1–3). In non-mammalian species, germ cell fate is determined by the inheritance of germ plasm, microscopically distinct oocyte cytoplasm, enriched in RNAs and RNA-binding proteins, that segregates with cells destined to become germ cells (1–3). In contrast, in mammalian species, germ cells are specified independently of germ plasm via inductive signaling (4–10). Fate mapping studies of the pre-implantation mouse epiblast have revealed that germ cells are specified in the proximal epiblast in response to signals such as bone morphogenetic protein 4 (Bmp4) from the neighboring extra-embryonic ectoderm (7,11). Yet it is clear that the proximal epiblast is not predestined to the fate of a germ cell since transplantation of distal epiblast to contact extra-embryonic ectoderm also results in germ cell formation (7). Germ cells are definitively recognized at ∼7.2 days post-coitum as an extra-embryonic cluster of cells that express tissue non-specific alkaline phosphatase, Oct4 and Stella, at the base of the allantois (12–15).
The NANOS gene family is required for germ cell development in diverse model organisms, although the processes that are regulated vary among species and between different homologs. In Drosophila, the single nanos homolog is implicated in germ cell migration, suppression of somatic cell fate in the germ line and maintenance of germ stem cell self-renewal (16–18), in addition to involvement in somatic patterning (19). In general, Nanos is recruited to Nanos-response elements of target mRNAs by its co-factor Pumilio, where the Nanos protein functions to repress translation (20,21). In Caenorhabditis elegans, three homologs exist, with Nos-1 and Nos-2 functioning primarily in germ cell development in maintaining germ cell viability and incorporation into the gonad (22).
Mouse models of Nanos1 did not reveal a discernible germ cell function for this homolog (23). Contrarily, male Nanos2−/− mice have decreased testis size and are infertile due to a loss of germ cells following primordial germ cell (PGC) incorporation into the gonad, while female mice appear developmentally normal and retain fertility (24). Characterization of Nanos3 knockout mice revealed decreased gonad size and infertility in both male and female mice. Similar to Nanos2−/− mice, PGC specification occurred in Nanos3−/− mice, although the PGCs were not maintained during migration (24). Later studies implicated Nanos3 in the maintenance of PGCs during migration via suppression of apoptosis (25). Notably, Nanos2 and Nanos3 phenotypes indicate independent, non-redundant roles for these homologs (26).
While recent years have witnessed remarkable progress in the understanding of germ cell development in model systems, it is difficult to translate information directly to the human system. This difficulty is due, in large part, to several factors. First, many genes required for reproduction have evolved so rapidly that even the same homolog in the closest evolutionary neighbors differ significantly in either sequence or function (27,28). Secondly, genes that reside on sex chromosomes can be expressed at various dosages, dependent on the species of the organism, or in more extreme cases, as in the Deleted in Azoospermia (DAZ) gene, may be missing from the genome altogether, even in closely related mammals (29). Thirdly, in contrast to other species, humans are remarkably imprecise in aspects of germ cell development commonly considered to be highly conserved. For example, meiotic chromosome mis-segregation occurs in yeast in ∼1/10 000 cells. In flies, mis-segregation occurs in 1/1000 to 1/2000 cells and in mice in <1/100 cells. In humans, meiotic mis-segregation occurs in 5–20% of cells, dependent on sex and age (30).
Here, we examined the function of NANOS homologs in the formation and/or differentiation of human germ cells, focusing on NANOS3. Using a previously described differentiation system, in which human embryonic stem cells (hESCs) are differentiated in an adherent monolayer in the presence of BMP4, 7 and 8b (31), we introduced various gene knockdown technologies to specifically focus on the function of one homolog, NANOS3, that demonstrated intriguing expression of mRNA and localization of the protein throughout germ cell development.
RESULTS
mRNA and protein expression of human NANOS homologs
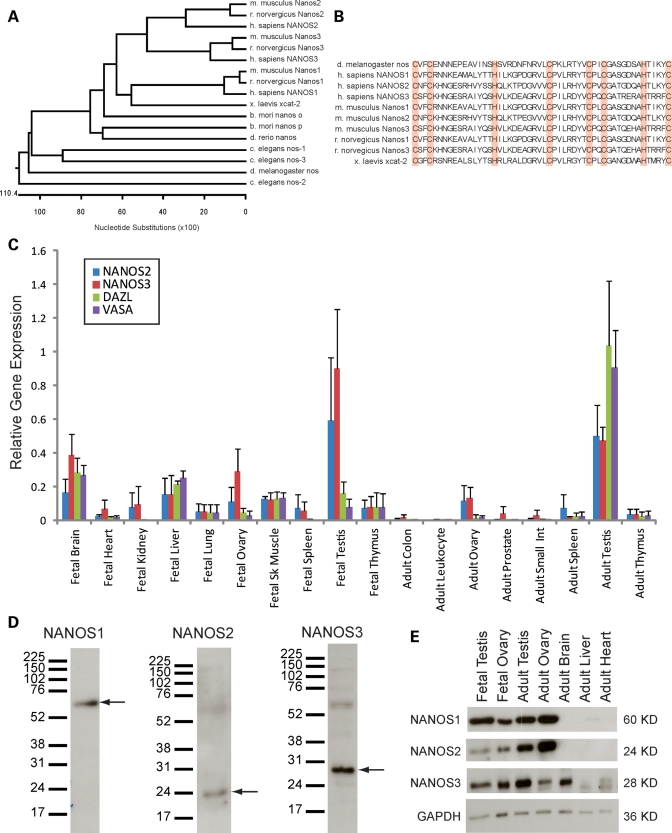
The human genome contains three genes that share extensive sequence similarity with the Nanos genes of other mammalian species (Fig. 1A). Comparison of the Nanos sequences in several model organisms reveals that there is a highly conserved region with 8 invariant cysteine and histidine residues in a CCHC CCHC configuration characteristic of zinc finger motifs (Fig. 1B). Zinc fingers are generally thought to facilitate the binding of DNA and RNA; previous studies determined that these 8 conserved amino acids form the two zinc fingers which confer translational repression properties to the Nanos family (32).
Figure 1.
NANOS is conserved in humans, with enriched expression in human gonadal tissue. (A) Alignment of the protein sequences of NANOS homologs in multiple organisms reveals conservation of the NANOS genes from invertebrates to vertebrates. Mammalian species (M. musculus/mouse, R. norvegicus/rat, H. sapiens/human) each have three NANOS homologs and share the highest sequence similarity, relative to more primitive species (X. laevis/frog, B. mori/silkworm, D. rerio/zebrafish, C. elegans/worm, D. melanogaster/fly). (B) The NANOS protein sequence is mostly divergent outside of the eight amino acids which comprise two zinc finger motifs (CCHC CCHC), shown in red. (C) Quantitative PCR of NANOS2, NANOS3, DAZL and VASA in fetal and adult human tissue cDNA. NANOS2 and 3 expression are highest in the fetal and adult testis, and are likewise enriched in the fetal and adult ovary. Gene expression is normalized to GAPDH, RPLPO and EEF1A1. (D) Characterization of NANOS expression in adult human testis lysates via western blot analysis with custom NANOS antibodies. NANOS1 is detected at ∼60 kDa. NANOS2 is detected at ∼24kDa. NANOS3 is detected at ∼28kDa. (E) NANOS expression in human gonadal tissue is determined via western blot analysis of human tissue lysates. NANOS1, 2 and 3 are expressed in the fetal and adult ovaries and testes; NANOS3 is also expressed in the adult brain. GAPDH is included as a loading control.
Analysis of NANOS mRNA expression in human fetal and adult tissues indicated that expression of NANOS-2 and -3 was enriched in the fetal ovary, testis and brain, as well as in the adult ovary and testis (Fig. 1C). This pattern was similar to that of the germ cell-specific markers, VASA and DAZL and was confirmed by polymerase chain reaction (PCR) (Supplementary Material, Fig. S1A). In contrast, NANOS1 was expressed ubiquitously (Supplementary Material, Fig. S1B).
To examine protein expression and localization, we generated antibodies against each of the 3 homologs. One antibody was designed to detect NANOS1, and two antibodies were designed against NANOS2 and NANOS3, although one of the NANOS2 peptides failed to initiate an immune response. Of the two NANOS3 antibodies designed, NANOS3-1 more effectively detected the NANOS3 protein via western blot analysis, whereas NANOS3-2 better detected the NANOS3 protein within tissue sections and in fixed cells. Preliminary experiments revealed that both NANOS3 antibodies were able to detect exogenous NANOS3–green fluorescent protein (GFP). The NANOS1, NANOS2 and NANOS3 antibodies detected proteins of ∼60, 24 and 28kDa, respectively (Fig. 1D). Notably, NANOS2 and NANOS3 antibodies also appear to detect NANOS1 as bands of ∼60 kDa were detected when blotting with each of these antibodies. This cross-reactivity could indicate the presence of NANOS1–NANOS3 or NANOS1–NANOS2 complexes, or may be attributed to sequence similarity between homologs. Western blot analysis revealed that NANOS-1, -2 and -3 proteins are expressed in fetal and adult gonadal tissue, with NANOS3 expression also detected in the human brain (Fig. 1E). While western blot analysis appears to reveal slight expression of NANOS3 in the adult heart, further characterization via immunohistochemistry indicates that NANOS3 is not expressed in any heart-specific cell types (Supplementary Material, Fig. S2B). Western blot analysis of the adult liver demonstrated only a non-specific band corresponding to a protein smaller than NANOS3; this lack of NANOS3 expression in liver was confirmed via immunohistochemistry (Supplementary Material, Fig. S2D).
NANOS3 localizes to germ cells during human oogenesis and spermatogenesis
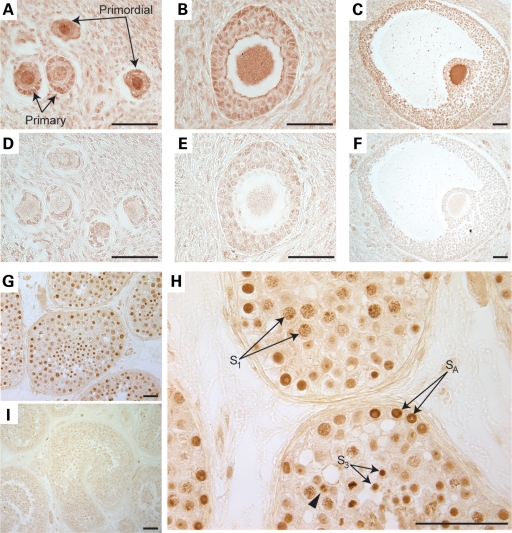
We focused further experiments on human NANOS3 and examined expression in both the human ovary and testis. For this purpose, biopsies of adult human ovaries were analyzed via immunohistochemistry and assessed for NANOS3 expression and localization. We observed that the NANOS3 protein was expressed during multiple stages of human oogenesis, including primordial, primary, secondary and antral follicles (Fig. 2A–C) with the highest expression in the oocytes; negative controls did not demonstrate specific staining (Fig. 2D–F). Parallel analysis of human testis sections also revealed that NANOS3 was expressed in germ cells of the testis (Fig. 2G). Cells positive for NANOS3 include those at multiple stages of spermatogenesis, including type A spermatogonia (SA), primary spermatocytes (S1), round spermatids (S3) (Fig. 3H) and elongated spermatids (S4) (Supplementary Material, Fig. S3). However, NANOS3 expression was not detected in immature sperm (Supplementary Material, Fig. S3). Most striking in these studies, and in contrast to the cytoplasmic expression of Nanos homologs in model organisms (22,33), human NANOS3 localized to the nuclei of human germ cells and co-localized with DNA during cellular division (arrowhead, Fig. 2H). NANOS3 protein expression in the human fetal testis (as determined via western blot analysis) was confirmed upon staining of human fetal testis, although no specific cell types were identified in this assay (Supplementary Material, Fig. S2). Negative controls were non-reactive in both the fetal and adult testis (Supplementary Material, Fig. S2; Fig. 2I).
Figure 2.
NANOS3 is expressed during human oogenesis and spermatogenesis. (A–C) Immunohistochemical analysis of cross-sections of the adult human ovary reveals expression of NANOS3 in human follicles. NANOS3 is expressed in the oocyte proper as well as the granulosa cells of primordial and primary follicles (A) and secondary follicles (B). (C) NANOS3 is highly expressed in antral follicles, with highest expression in the oocyte. (D–F) Serial sections treated with pre-immune anti-serum as a negative control. (G–I) Adult testis sections analyzed via immunohistochemistry. (G) NANOS3 is expressed in human testis cross-sections, in a highly specific staining pattern. (H) Increased magnification of testis sections probed with the NANOS3 antibody. NANOS3 is expressed specifically by germ cells at varying stages of spermatogenesis. Type A spermatogonia (SA), primary spermatocytes (S1) (pre-meiotic) and round spermatids (S3) stain positively for NANOS3. NANOS3 localization is predominantly nuclear in all positive cell types with the protein remaining bound to the DNA during cellular division (arrowhead). (I) No positive signal is detected using the pre-immune anti-serum to stain the adult human testis. The scale bar is 50 μm.
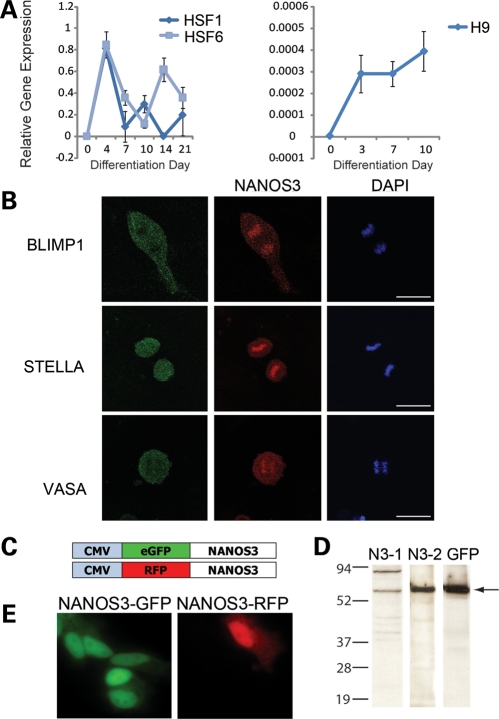
Figure 3.
Characterization of NANOS3 expression in hESC-derived germ cells. (A) Expression of NANOS3 in undifferentiated hESCs and cells differentiated as embryoid bodies. NANOS3 is not expressed in undifferentiated hESCs in HSF1, HSF6 or H9, although expression increases upon differentiation and persists throughout differentiation. (B) H9 hESCs, differentiated adherently for 7 days in medium supplemented with BMP4, 7 and 8b, were stained for the known germ cell markers BLIMP1, STELLA and VASA and imaged via confocal microscopy. NANOS3 expression is detected in dividing cells which co-express these germ cell markers. NANOS3 expression is predominantly nuclear, although cytoplasmic expression is also detected. BLIMP1 and VASA expression are largely restricted to the cytoplasm; STELLA is both cytoplasmic and nuclear. Cellular division was scored via DNA morphology (DAPI). (C) Constructs were created in which the NANOS3 ORF was labeled with either GFP or RFP, under the control of the CMV promoter. (D) NANOS3-GFP was generated via the pET expression system (Novagen) and then detected via western blot analysis using NANOS3-1 (N3-1), NANOS3-2 (N3-2) and GFP antibodies; arrow indicates NANOS3-GFP. (E) mSSCs were transfected with constructs over-expressing NANOS3-GFP or NANOS3-RFP. NANOS3 localization is predominantly nuclear, although the NANOS3 protein is also detected in the cytoplasm.
NANOS3 mRNA and protein expression in undifferentiated and differentiated hESCs
We next explored NANOS3 function during differentiation of hESCs to germ cells. We differentiated three hESC lines [HSF1 (XY-bearing), HSF6 and H9 (both XX-bearing)] spontaneously as embryoid bodies. HSF1 and HSF6 were differentiated in parallel experiments for 21 days, with sample time points taken at 0, 4, 7, 10, 14 and 21 days. We observed that NANOS3 mRNA expression increased upon spontaneous differentiation of hESCs, peaking at 4 days of differentiation in both cell lines assessed (Fig. 3A). In contrast, NANOS1 was expressed throughout differentiation, while NANOS2 expression was not detected throughout the period of hESC differentiation, in either line (Supplementary Material, Fig. S4A and B). Subsequently, the H9 hESC line was also differentiated for a total of 10 days. NANOS3 was not expressed in undifferentiated H9 cells, but increased at day 3 and remained expressed through day 10 (Fig. 3A). This later observation may reflect differences in derivation and/or genetic background but suggests potential inherent differences in germ cell development. Similar to HSF1 and HSF6, NANOS1 was expressed in H9 hESCs throughout differentiation, while NANOS2 was not detected at any of the time points sampled (Supplementary Material, Fig. S4C and D). Thus, in hESCs, NANOS3 is not detected in undifferentiated cells but later increases upon differentiation.
NANOS3 co-localizes with BLIMP1, STELLA and VASA in dividing hESC-derived germ cells
We examined the localization of NANOS3 during hESC differentiation via a 7-day protocol in which cells were differentiated adherently in media supplemented with BMPs and then analyzed via immunofluorescence. We observed that a subpopulation of differentiated hESCs expressed the NANOS3 protein in: (i) both the cytoplasm and nucleus, with the protein co-localized with the chromosomes and (ii) in actively dividing cells (Fig. 3B). We then assayed expression of other proteins required for germ cell development in diverse organisms, namely BLIMP1, STELLA and VASA. We observed that all three of these germ cell-specific proteins were also co-expressed in mitotically dividing cells, where, in contrast to NANOS3, each protein localized to the cytoplasm, with STELLA expression also identified in the nucleus (Fig. 3B). Thus, within the differentiating hESC population, we observed cells that express the NANOS3 protein in the nucleus, and also are positive for the germ cell proteins BLIMP1, STELLA and VASA. The co-localization of these four proteins is diagnostic of germ cells in vivo, and suggests that these cells are in vitro-derived germ cells and that NANOS3 localizes to the cytoplasm and nucleus where it co-localizes with the chromatin (Fig. 3B). To further verify antibody specificity, we generated GFP-tagged NANOS3 constructs (NANOS3-GFP) for over-expression in a bacterial expression system (Fig. 3C). Detection of the GFP-tagged NANOS3 protein was achieved by probing with NANOS3-1, NANOS3-2 or GFP (JL-8) antibodies (Fig. 3D). With each antibody, NANOS3-GFP was detected, although a non-specific band was also detected when probing with NANOS3-1. This band, however, is likely an artifact of the bacterial expression system, as it is not detected when probing human testicular lysates with the same antibody. NANOS3-GFP [or red fluorescent protein (RFP)] constructs were transfected into mouse spermatogonial stem cells (mSSCs), with high levels of fluorescence expression observed in the germ cell nuclei, and lower levels of expression in the cytoplasm (Fig. 3E). These data provide further corroborating evidence that NANOS3 predominantly localizes to the nuclei of mammalian germ cells.
Decreased expression of NANOS3 results in reduced germ cell numbers and aberrant expression of germ cell markers
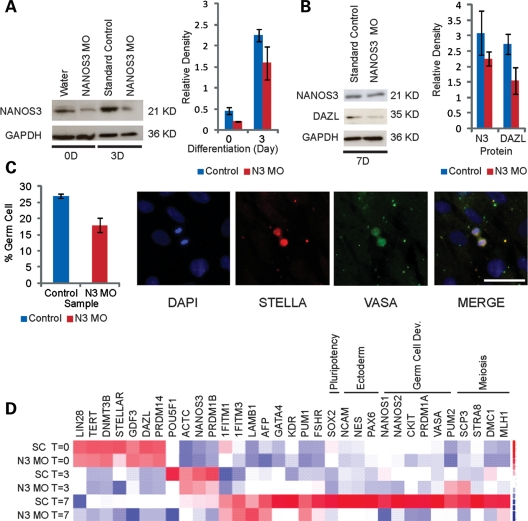
To examine whether NANOS3 is required for the differentiation of germ cells from hESCs, we developed a knockdown strategy that relied on morpholino oligonucleotides (MOs) (typically ∼25bp in length) to inhibit protein synthesis (34). MOs block protein synthesis by altering the DNA backbone linkages, thereby conferring resistance to digestion by nucleases. The oligonucleotides are generally targeted to the 5′ untranslated region (UTR) or start codon of the genes of interest to avoid displacement by the translation machinery. We designed MOs to target the NANOS3 start site (NANOS3 MO/N3 MO) and introduced them into undifferentiated hESCs via nucleofection. Following nucleofection, hESCs were maintained as undifferentiated hESCs or differentiated in the presence of BMPs. Cells were harvested at days 0, 3, and 7 and then analyzed via western blot to assess the efficiency of NANOS3 protein reduction. As shown, we detected an ∼50% reduction in the NANOS3 protein levels upon introduction of the NANOS3 MO at day 0 (Fig. 4A). Following 3 days of differentiation, NANOS3 protein levels were still reduced, relative to the control (Fig. 4A). By day 7, NANOS3 expression returned to levels comparable with those found in cells nucleofected with the standard control (SC) (Fig. 4B). Further analysis of the cells collected after 7 days of NANOS3 MO-knockdown and differentiation revealed, however, that DAZL expression was reduced by ∼50% (Fig. 4B).
Figure 4.
Morpholino knockdown of NANOS3 in hESCs. A morpholino targeting NANOS3 (NANOS3 MO/N3 MO), a control morpholino (standard control/SC) or water was nucleofected into H9 hESCs. (A) NANOS3 is expressed at lower levels in the NANOS3 MO lines relative to either water or the SC in undifferentiated hESCs (0D) or in hESCs differentiated for 3D in the presence of BMPs. (B) NANOS3 expression is restored to SC levels by day 7. DAZL expression is decreased in the NANOS3 MO line relative to the SC. (C) The SC MOs or the NANOS3 MOs were independently introduced into hESCs and then differentiated for 7 days in the medium supplemented with BMPs. Germ cell number is decreased in the NANOS3 MO line, relative to the SC. hESC-derived germs cells were quantified by screening for cells undergoing cellular division and which positively express both VASA and STELLA. The scale bar represents 50 μm. (D) Gene expression analysis was performed on undifferentiated hESCs and hESCs differentiated for 3 or 7 days in the presence of BMPs. Gene expression profiles from cells transduced with either the SC or the NANOS3 MO were compared by clustering both samples and genes. Control and MO profiles from undifferentiated hESCs (T = 0) and cells differentiated for 3 days (T = 3) are virtually identical. The SC sample, after 7 days of differentiation (SC T = 7), expresses genes required for germ cell development (CKIT, PRDM1A, VASA), meiosis (MLH1, STRA8, SCP3, DMC1) and ectodermal differentiation (NCAM, NES, PAX6). While the NANOS3 MO line (N3 MO T = 7) upregulates expression of some genes required for germ cell development (1FITM1, 1FITM3, FSHR), most of the genes activated in the SC sample are not expressed in the NANOS3-knockdown line.
Germ cell numbers in both the control and NANOS3-knockdown populations were quantified by counting the actively dividing cells that expressed both STELLA and VASA. We found that 27% of the dividing cells in the control population were positive for both germ cell markers, relative to 18% in cells nucleofected with the NANOS3 MO (Fig. 4C). Taken together, these findings suggest that NANOS3 either directly or indirectly regulates expression of the DAZL gene and subsequent development, in particular cellular division of differentiating human germ cells.
We then examined gene expression in undifferentiated and differentiated cells that were nucleofected with the NANOS3 MO. For this purpose, cells were cultured in the presence of BMPs for 0, 3 and 7 days of differentiation and then harvested for gene expression analysis. Data clustering revealed that gene expression at days 0 and 3 was similar between NANOS3 MO and SC samples (Fig. 4D). Analysis of gene expression data from cells differentiated for 7 days, however, indicated significant differences between the NANOS3 MO and SC MO (Fig. 4D). In the control population, several genes required for germ cell development (PUM2, cKIT, PRDM1A, VASA), meiosis (MLH1, STRA8, SCP3, DMC1) and also, notably, ectodermal development (NCAM, NES, PAX6) were highly expressed, as were NANOS1 and NANOS2. In the cells with the reduced NANOS3 protein, however, expression of each of these genes was significantly lower than in the control population (Fig. 4D). This suggests that wild-type levels of the NANOS3 protein are required for proper regulation and expression of key regulators of germ cell development, meiotic progression and ectodermal differentiation.
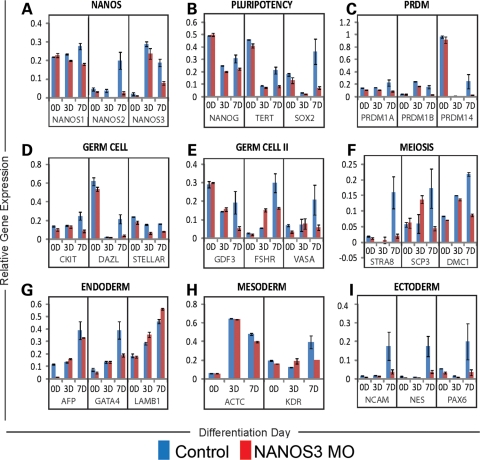
Quantitation of the gene expression in cells nucleofected with the NANOS3 MO and SC MO was also carried out. We detected decreased NANOS-1, -2 and -3 expression at 3 and 7 days (Fig. 5A). We also observed that decreased levels of NANOS3 resulted in reduced expression of three key regulators of pluripotency: NANOG, TERT and SOX2 (Fig. 5B). In addition, decreased NANOS3 expression resulted in lower expression levels of the early germ cell markers PRDM1A, PRDM1B, PRDM14, CKIT, DAZL, STELLAR and GDF3 (Fig. 5C–E), as well as decreased levels of the late germ cell genes FSHR and VASA (Fig. 5E). Moreover, a comparison of expression levels of the meiotic regulators STRA8, SCP3 and DMC1 indicated that all showed decreased expression upon reduction in NANOS3 expression (Fig. 5F). In contrast, key markers of endodermal and mesodermal differentiation did not reveal any lineage-specific differences between cells transduced with either the NANOS3 MO or the control MO (Fig. 5G and H). On the contrary, however, expression of all three ectodermal markers assessed showed significantly decreased levels of expression, relative to control cells: NCAM, NES and PAX6 (Fig. 5I).
Figure 5.
Gene expression analysis of NANOS3-knockdown cells. H9 hESCs, transduced with either the SC MO or with the NANOS3 MO, were differentiated for 0, 3 or 7 days in the medium supplemented with BMPs. (A) NANOS1, 2 and 3 expression are decreased in the NANOS3-knockdown line at 3 and 7 days. (B) NANOG, TERT and SOX2, regulators of pluripotency, are decreased in the 7D population in the NANOS3 MO line. (C–E) Expression of early regulators of germ cell development (PRDM1A, PRDM1B, PRDM14, CKIT, DAZL, STELLAR, GDF3) as well as late markers (FSHR, VASA) decrease after 7 days of differentiation, in the NANOS3 MO line relative to the SC. (F) Three genes required for meiosis (STRA8, SCP3, DMC1) are decreased in the NANOS3-knockdown cells at day 7. (G–H) No change was detected in the overall expression profile of either the endodermal or mesodermal lineages. (I) Markers of ectodermal development (NCAM, NES and PAX6) show decreased expression at 7D in the NANOS3 MO population.
To confirm our data generated with the morpholino system, we likewise reduced NANOS3 expression via introduction of short hairpin RNAs (shRNAs) that targeted the NANOS3 open reading frame (ORF) or 3′ UTR. We discovered that introduction of each shRNA decreased NANOS3 expression in undifferentiated hESCs relative to the LACZ negative control (Supplementary Material, Fig. S5A). We analyzed gene expression in two independent shRNA lines (825 and 826) and found that expression of both NANOS1 and NANOS2 were also decreased in the NANOS3-knockdown lines (Supplementary Material, Fig. S5A). Moreover, we discovered that decreased NANOS3 expression also resulted in a reduction in the expression of two key regulators of pluripotency, SOX2 and TERT (Supplementary Material, Fig. S5B). Analysis of the expression of genes required for human germ cell development revealed decreased expression of DAZL and ablated STELLAR expression (Supplementary Material, Fig. S5C), although gene expression analysis of three meiotic markers (MLH1, SCP3, DMC1) did not reveal significant changes in expression levels (Supplementary Material, Fig. S5D).
In cells carrying NANOS3 shRNA, we also sought to assess whether the decreased expression levels of NANOS3 resulted in changes in cellular apoptosis. We assayed expression levels of both caspase3 and caspase7, key players in the execution phase of apoptosis, in undifferentiated hESCs. We compared expression levels of caspase3/7 in two negative control lines (H9 hESCs and ES cells with shRNA against LACZ) and in two independent NANOS3 shRNA lines (133 and 826). Upon comparison of caspase3/7 expression, however, we did not detect significant differences in cellular apoptosis (Supplementary Material, Fig. S6).
Taken together, these data provide the first experimental evidence to suggest that NANOS3 functions in human germ cell development, given that decreased NANOS3 levels significantly alter the germ cell number and expression patterns of genes that regulate important germ cell properties, including maintenance of pluripotency, germ cell development and meiotic progression.
DISCUSSION
A curiosity of germ cell formation and differentiation across species is the conservation of genes that encode components of germ plasm in diverse species regardless of the mode of germ cell specification. For example, in Drosophila, the disruption of genes such as Oskar, Vasa, Tudor, Germ cell-less and Aubergine results in the lack of a germ line (35). The function of these genes is to assemble germ plasm, which contains highly conserved, interacting RNA-binding proteins such as Pumilio and Nanos which are thought to act to repress translation and indirectly silence gene transcription in nascent germ cells (35–38). In Pumilio and Nanos mutants, nascent germ cells may divide prematurely, migrate abnormally and subsequently die during early embryonic development (35). Several years ago, homologs of several germ plasm components were identified in mammalian germ cells, including those of humans (24,39–44). In a screen for proteins that interact with DAZ and DAZL, Moore et al. identified a set of genes that included PUMILIO homologs, PUM1 and PUM2 (39). In other studies, it was shown that a human NANOS homolog interacts with PUMILIO homologs and is expressed in germ cells as well (24,44). These interactions are intriguing given that, in humans, deletions and variants of DAZ homologs are associated with the production of very few or no germ cells and, in diverse model organisms, these genes are required solely in the development of the germ cell lineage (24,29,44–51). Thus, it has been hypothesized that the protein complex containing human DAZ, PUMILIO and NANOS homologs likely functions in germ cell formation and/or differentiation by regulating RNA localization, transcription, translation or stability (39,44,52–56). However, previously, it was not possible to directly test this hypothesis in humans. Indeed, human germ cell development remains very poorly understood in spite of the fact that defects in germ cell development contribute to a majority of infertility in the human population (57).
Here, we report our analysis of a highly conserved family of proteins that is implicated in germ cell development in diverse species (22,24,58). We show that, in human ovary and testis tissues, NANOS3 is expressed in human germ cells, with the protein localized to the germ cell nucleus. Nuclear localization of the NANOS3 protein and co-expression with known germ cell proteins BLIMP1, VASA and STELLA were also observed in hESC-derived germ cells. Moreover, we found that, in the absence of NANOS3, the molecular program(s) which drive human germ cell development were altered, thereby altering gene expression and quantifiably reducing the total germ cell number in active cell division. Taken together, these findings suggest that NANOS3 functions during human germ cell development in a cell-autonomous fashion and provides the first direct experimental evidence that NANOS3 functions in human germ cell development. Thus, NANOS3 is one of just two genes that has been shown directly to function in germ cell development from flies and worms to frogs, mice and now, humans (the other is BOULE, a member of the DAZ gene family).
The decreased mRNA levels of NANOS3 following 7 days of differentiation are somewhat surprising, given that MOs should only affect protein levels. It is possible, however, that a negative-feedback loop exists whereby NANOS3 mRNA levels are regulated based on the NANOS3 protein levels. Alternatively, NANOS3 mRNA may have been converted to protein following the decreased effectiveness of the MOs after 7 days. Given that so little is known regarding the mechanism of NANOS3 expression regulation, this is an area of study which warrants further investigation.
Due to the lack of an internal reporter system when using MOs, we were unable to track each cell in which NANOS3 expression levels were decreased. Despite this inability to track individual cells in such a heterogeneous cell population, we were able to quantify that fewer hESCs differentiated to germ cells in culture when NANOS3 expression was decreased.
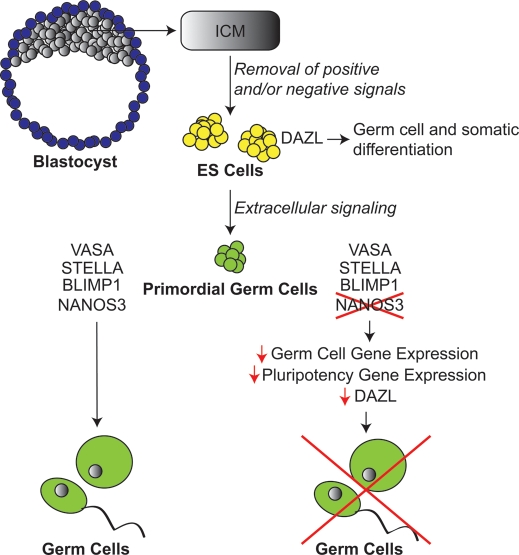
Based on our data, we propose a model for NANOS3 expression in human hESC-derived germ cells (Fig. 6). The inner cell mass (ICM) of a human blastocyst can be extracted, and, in concert with the loss of positive and/or negative signals, gives rise to hESCs which can be maintained in vitro. ES cells, which express DAZL, are capable of differentiating to both germ and somatic cellular lineages. With the addition of extracellular signaling, PGCs can be derived in vitro from hESCs. PGCs express numerous factors required for germ cell development, including (but not limited to) VASA, STELLA and BLIMP1, and give rise to the germ cell lineage exclusively. In this study, we show that PGCs also express the highly conserved germ cell factor NANOS3. Based on our observations, we propose that down-regulation of NANOS3 results in decreased expression of genes required for several processes required for the development of human germ cells including (i) decreased expression of germ cell genes, (ii) decreased expression of genes required for the maintenance of pluripotency and (iii) reduced DAZL protein levels. The resultant down-regulation of the germ cell molecular profile ultimately disrupts the process of human germ cell development.
Figure 6.
Proposed model. The ICM of a human blastocyst can be dissected, and in concert with the removal of positive and/or negative signals yields hESCs which can be maintained in vitro indefinitely. ES cells have previously been shown to express DAZL and can give rise to both germ and somatic cellular lineages. With the addition of extracellular signaling, PGCs can be derived in vitro from hESCs. PGCs express numerous factors required for germ cell development including, but not limited to VASA, STELLA and BLIMP1, and give rise to the germ cell lineage exclusively. In this study, we have shown that PGCs also express a member of the highly conserved Nanos family: NANOS3. Based on our observations, we propose that down-regulation of NANOS3 results in decreased expression of genes required for germ cell development and pluripotency as well as decreased levels of the DAZL protein; this down-regulation of the germ cell molecular profile ultimately disrupts the process of human germ cell development.
While the mechanism of germ cell regulation is as yet unknown, based on the results described here, it is likely that NANOS3 modulates essential aspects of human germ cell development perhaps during the cell cycle. Furthermore, we present the novel discovery that NANOS3 localizes to the nuclei of human germ cells and may regulate human germ cell development at both mRNA and DNA levels in contrast to the exclusive regulation at the mRNA level that has been previously reported (32,59–61). Further investigation of the specific mechanism and potential targets of NANOS3 would undoubtedly increase our knowledge of human germ cell development and could potentially provide information for the purpose of advancing human fertility studies as well as treatments.
MATERIALS AND METHODS
hESC culture and differentiation
Three federally approved (as of the time of this study) hESC lines were used in this study: HSF1 (XY), HSF6 (XX) and H9 (XX). Human ES cell cultures were maintained on a uniform layer of cesium-irradiated mouse embryonic fibroblasts (MEFs). Cultures were grown at 37°C with 5% CO2 in KSR + recombinant human basic fibroblast growth factor (bFGF) medium [knockout Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% knockout serum replacer, 1 mm l-glutamine, 0.1 mm non-essential amino acids, 0.1 mm β-mercaptoethanol and 4 ng/ml bFGF (R&D systems)]. To differentiate spontaneously, hESCs were treated with type IV collagenase (1 mg/ml) for 10 min, scraped with a 5 ml sterile pipette, then transferred to a low-adhesion culture dish in differentiation media (knockout DMEM supplemented with 20% fetal bovine serum (FBS), 1 mm l-glutamine, 0.1 mm non-essential amino acids, 0.1 mm β-mercaptoethanol and 50 ng/ml recombinant human BMP4, BMP7 and BMP8b (R&D Systems)].
For adherent differentiation, hESCs were either: (i) treated with type IV collagenase (1 mg/ml) for 10 min and then scraped with a 5 ml pipette or (ii) manually passaged under a dissection microscope with a sterile cell lifter. Approximately 5 × 104 hESCs (to achieve <50% confluency in a six-well tissue-culture plate) were transferred onto Matrigel-coated six-well plates (1:30, BD Biosciences). Cells were grown on Matrigel for 1–2 days in MEF-conditioned media (KSR + bFGF media collected following overnight (ON) incubation on irradiated MEFs), rinsed once in phosphate buffered saline (PBS) without Ca2+ and Mg2+, and then cultured in differentiation media. Media were changed after 7 days of culture, if further differentiation was required.
Western blot analysis
To collect undifferentiated and adherently differentiated hESCs for western blot analysis, cells were washed with 3 ml of PBS without Ca2+ and Mg2+, and then treated with 0.25% trypsin–ethylenediaminetetraacetic for 5 min at 37°C. Trypsin was inactivated with media and then harvested. The cell suspension was centrifuged at 1000rpm for 5 min, media were aspirated and the cell pellet was either frozen at −80°C or lysed in RIPA buffer (Sigma) supplemented with Protease Inhibitors (Complete Mini, Roche). The lysates were vortexed vigorously and then incubated at 4°C with agitation for 30 min. Cell debris was centrifuged out at 15 000rpm for 30 min, and the supernatant was harvested. Protein concentration was determined via BCA Assay (Pierce).
Cells were denatured in 3× sample buffer at 95°C for 5 min, and then loaded onto 8, 10 or 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gels. The SDS–PAGE gels were run at 225 V for 45 min and then transferred to polyvinylidene fluoride membranes for 60 min at 100 V. Blots were then blocked in 5% non-fat milk with 10% FBS for 1 h at room temperature (RT). Primary antibody in 5% blocking solution was added and the blots were incubated ON at 4°C [1:500 NANOS1, 1:1000 NANOS2-1, 1:1000 NANOS3-1, 1:10 000 GAPDH (Abcam), 1:1000 DAZL, 1:500 VASA (Abcam)]. Blots were washed three times in PBST (1× PBS, 0.1% Tween 20) for 15 min each, and then incubated in secondary antibody (1:10 000) for 1 h at RT (NANOS1: anti-chicken horseradish peroxidase (HRP); NANOS2: anti-mouse HRP; NANOS3, VASA, DAZL, GAPDH: anti-rabbit HRP). Following 3 × 15 min washes in PBST, blots were visualized using ECL (GE) on film. Human tissue lysate sources are as follows: Fetal Testis, Fetal Ovary: ABR; Adult Testis, Adult Ovary: Novus Biologicals; Adult Brain, Liver and Heart: Pro-Sci.
The relative band intensity was quantified as described in: http://www.lukemiller.org/journal/2007/08/quantifying-western-blots-without.html. Briefly, the mean intensity of each band was multiplied by the number of pixels to calculate the absolute intensity. The absolute intensity of each experimental band was normalized against the absolute intensity of the standard to determine the relative intensity (reported value).
Quantitative PCR
Cells were scraped from tissue-culture dishes in cold PBS and then harvested. Total RNA was collected using RNeasy (Qiagen) or Picopure (Arcturus) RNA extraction kits and cDNA was prepared using Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer's protocols.
Quantitative PCR was performed using either the ABI 7300 (Applied Biosystems) or Biomark Dynamic Arrays (Fluidigm) using gene-specific Taqman Probes and Taqman Master Mix (Applied Biosystems). Gene expression was normalized in one of the two ways: (i) against GAPDH using the calculation 2−(CT, Sample − CT, Reference) or (ii) using the program GeNorm, with gene expression normalized to at least three of the following housekeeping genes (GAPDH, RPLPO, EEF1A1, CTNNB1, TBP or ACTB) as previously described (62,63). Briefly, a normalization factor was calculated (via GeNorm) based on the geometric mean of multiple housekeeping genes and was used to normalize the expression levels of the genes of interest within each experiment.
shRNA vectors
shRNA vectors targeting NANOS3 were constructed using the Block-It inducible H1 lentiviral system (Invitrogen). The shRNAs were cloned into the pENTR/H1/T0 vector and then transferred into pLenti4/BLOCK-It-DEST destination vectors. Viral supernatants were prepared as described for pLVGV (31). hESCs were transduced with NANOS3 shRNA lentiviral supernatant as described for pLVGV, although selection was performed using 2 mg/ml of zeocin in conditioned media for 3 days. NANOS3 shRNA primer sequences are available upon request.
GFP and RFP fusion proteins
Human testis cDNA was amplified via PCR using primers to amplify NANOS2 and NANOS3 ORFs (available upon request). ORFs were cloned into the peGFP-C2 (Clontech) or pDSRed2-C1 (BD Biosciences) vectors. Eight clones were mini-prepped according to the manufacturer's instructions (Qiagen), digested, run on 1.5% tris-borate-EDTA gels, gel extracted (QIAquick gel extraction kit, Qiagen) and sequenced to confirm that no mutations were introduced. Using the pET Expression System (Novagen), the GFP–NANOS3 protein was produced in bacteria, and then assessed via western blot analysis using antibodies against NANOS3 (NANOS3-1 and NANOS3-2) and GFP (JL-8). Constructs were also introduced into mSSCs via Lipofectamine 2000 (Invitrogen) transfection for use in immunofluorescence experiments.
Custom antibodies
Custom antibodies were designed against NANOS1, NANOS2 and NANOS3 using Custom Polyclonal Antibody Production (Invitrogen). The NANOS1 antibody (NANOS1 RSARDGPPGKKLR) was raised in chickens, harvested after 10 weeks, and then purified via IgY Purification (Invitrogen). Two NANOS2 peptides (NANOS2-1 GQRLETQEIEEPS; NANOS2-2 RRSGRNSAGRRVKR) were introduced into mice, with the antibody harvested after 10 weeks, although NANOS2-2 peptide did not generate an immunological response. Two NANOS3 antibodies were raised in rabbits (NANOS3-1 GKEGPETRLSPQPE; NANOS3-2 KAKTQDTGHRRGG). NANOS3-1 antibody was used for all western blot protocols and NANOS3-2 antibody was used for all immunohistochemistry and immunofluorescence protocols. Each western blot, immunohistochemistry and immunofluorescence experiment was performed in parallel with the corresponding pre-immune serum to confirm antibody specificity.
Immunofluorescence
hESCs were grown on Lab-Tek II chambered coverglass (Nunc) coated with Matrigel (BD Biosciences) according to the manufacturer's protocol. Media were aspirated, the cells were washed once with 1× PBS and then fixed in 4% paraformaldehyde for 10 min at RT. Cells were washed three times in 0.1% Tween 20 in PBS (PBST) for 5 min each at RT, followed by permeabilization with 1% Triton X in PBST for 10 min (for cytoplasmic proteins) or 1 h and 30 min (for nuclear proteins) at RT. Cells were blocked ON in 4% goat serum (NANOS, STELLA, SSEA1) or 4% chicken serum (VASA) in PBST at 4°C. Primary antibody was added in 1% serum for 4 h at RT [NANOS3 1:500 (Custom, Invitrogen), VASA 1:200 (R&D), STELLA 1:200, SSEA1 1:200 (Millipore), BLIMP1 1:200 (Novus Biologicals)]. Cells were washed three times in PBST, 5 min each and then secondary antibody in 1% serum was applied for 30 min at RT (NANOS3: goat anti-rabbit IgG 1:500, VASA: chicken anti-goat IgG 1:500, STELLA, BLIMP1: goat anti-mouse IgG 1:500, SSEA1: goat anti-mouse IgM 1:500) (Molecular Probes). If cells were stained using two independent antibodies, the immunofluorescence protocol (block, primary antibody and secondary antibody) was performed sequentially. Cells were washed three times with PBST for 5 min each, counterstained with DAPI/PBS and imaged using either a Zeiss LSM510 Meta inverted confocal microscope with an environmental control chamber or a Leica DMI6000B.
Immunohistochemistry
Tissue sections on glass slides were de-paraffinized in 2 × 10 min xylene washes. Sections were rehydrated in descending concentrations of ethanol washes for 3–5 min each (2 × 100%, 95%, 70%, 50%), incubated in Coplin jars filled with Target Retrieval Solution (Dako) in 95–99°C water bath for 30 min, cooled at RT for 20 min and then washed in PBS, three times at 5 min each. Ten percent hydrogen peroxide in methanol was used to block endogenous peroxidase activity (20 min, RT). Slides were washed 3 × 10 min in PBS + 1% BSA + 0.1% Tween (PBST). Triton X at 0.1% in PBS was used to permeabilize cell membranes (5 min), followed by three 5 min washes in PBST. Tissues were blocked in 4% goat serum in PBST for 1 h at RT. Primary antibody was added (in 1% serum) ON at 4°C in humid chamber (NANOS3-2 or Pre-Immune, 1:1000). Following ON incubation, sections were equilibrated to RT and then washed 3 × 10 min in PBST. Secondary antibody (1:200 anti-rabbit HRP) in 1% goat serum/PBST was applied to slides for 45 min at RT, followed by 3 × 10 min washes in PBST. Protein localization was imaged using Vectastain ABC reagent (Vector Labs) and DAB (Vector Labs) according to the manufacturer's protocols. DAB reactivity was stopped by incubating slides with water. Tissue sections were rehydrated for 3–5 min each in 50, 70, 95 and 2 × 100% ethanol washes. Cells were mounted and cover-slipped and then imaged using a Leica DM4000B upright microscope. Tissue section sources are as follows: Adult Human Testis: Joerg Gromoll Laboratory, Germany; Adult Human Ovary: Zyagen; Adult Thymus and Heart: Imgenex.
Morpholino oligonucleotides
Two anti-sense non-overlapping MOs were designed to target either the 5′ UTR of NANOS3 or the NANOS3 start codon in the ORF (Gene-tools, LLC); both MO constructs yielded similar results. A SC MO (SC: CCTCTTACCTCAGTTACAATTTATA) (Gene-tools, LLC) was used in parallel, as a negative control. MOs were nucleofected using the Human Stem Cell (H9) 96-well Nucleofector Kit (Amaxa/Lonza) according to the manufacturer's specifications. Following nucleofection, cells were replated in 12-well tissue-culture dishes (Falcon) or Lab-Tek II Chambered Coverglass (Nunc) coated with Matrigel (BD Biosciences) in the conditioned medium.
Quantification of dividing germ cells
Cells were prepared for analysis via immunofluorescence, as previously described using the following primary antibodies: [VASA 1:200 (R&D), STELLA 1:200 (Millipore)]. Secondary antibodies are chicken anti-goat 1:500 IgG and goat anti-mouse IgG 1:500 (Molecular Probes). Cells were counterstained using DAPI in PBS. Germ cells were quantified by screening for cells undergoing cellular division (as assessed via DNA morphology), and then assessing the percentage of dividing cells which positively express both STELLA and VASA.
Apoptosis
Apoptosis was assessed using the Caspase-Glo 3/7 Assay (Promega) according to the manufacturer's recommendations. Briefly, Caspase-Glo 3/7 reagents were reconstituted and added to identical cell numbers (as determined via flow cytometry) and volumes for the assays (H9, LACZ, 133, 826), negative control samples and blanks. Following incubation for 30 min–3 h, luminescence of each sample was read using a plate-reading luminometer as directed by the manufacturer.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (#F31HD054017 to V.T.A.J. and U54HD055764 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research to R.A.R.P.) and the California Institute for Regenerative Medicine (CIRM Comprehensive Grant #RC1-00137 to R.A.R.P.). Funding to pay the Open Access publication charges for this article was provided by the National Institute of Child Health and Human Development #U54HD055764 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Drs Shawn Chavez and Sohyun McElroy for assistance with differentiation experiments, Drs Shawn Chavez and Sonya Schuh-Huerta for critical review of this manuscript, Dr Joerg Gromoll for tissue section procurement and members of the RRP lab for valuable discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Saffman E.E., Lasko P. Germline development in vertebrates and invertebrates. Cell Mol. Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houston D.W., King M.L. Germ plasm and molecular determinants of germ cell fate. Curr. Top. Dev. Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- 3.Wylie C. Germ cells. Curr. Opin. Genet. Dev. 2000;10:410–413. doi: 10.1016/s0959-437x(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 4.Ying Y., Qi X., Zhao G.-Q. Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proc. Natl Acad. Sci. 2001;98:7858–7862. doi: 10.1073/pnas.151242798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ying Y., Liu X.M., Marble A., Lawson K.A., Zhao G.Q. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol. Endocrinol. 2000;14:1053–1063. doi: 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimizu T., Obinata M., Matsui Y. Stage-specific tissue and cell interactions play key roles in mouse germ cell specification. Development. 2001;128:481–490. doi: 10.1242/dev.128.4.481. [DOI] [PubMed] [Google Scholar]
- 7.Tam P., Zhou S. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by position of the cells in the gastrulating mouse embryo. Dev. Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 8.McLaren A. Signalling for germ cells. Genes Dev. 1999;13:373–376. doi: 10.1101/gad.13.4.373. [DOI] [PubMed] [Google Scholar]
- 9.Lawson K.A., Dunn N.R., Roelen B.A., Zeinstra L.M., Davis A.M., Wright C.V., Korving J.P., Hogan B.L. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson K.A., Hage W.J. Germline Development: Ciba Foundation Symposium. Vol. 182. West Sussex, UK: John Wiley and Sons; 1994. pp. 68–84. [Google Scholar]
- 11.Fujiwara T., Dunn N.R., Hogan B.L. Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. Proc. Natl Acad. Sci. 2001;98:13739–13744. doi: 10.1073/pnas.241508898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiquoine A. The identification, origin and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- 13.Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 14.Scholer H., Dressler G., Balling R., Rohdewohld H., Gruss P. Oct-4: a germ line specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholer H., Ruppert S., Suzuki N., Chowdhury K., Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 16.Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K., Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi Y., Hayashi M., Kobayashi S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc. Natl Acad. Sci. 2004;101:10338–10342. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z., Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- 19.Gavis E., Lehmann R. Translational regulation of nanos by RNA localization. Nature. 1994;369:315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 20.Sonoda J., Wharton R.P. Drosophila brain tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis D., Apfeld J., Lehmann R. Nanos is an evolutionarily conserved organizer of anterior-posterior polarity. Development. 1995;121:1899–1910. doi: 10.1242/dev.121.6.1899. [DOI] [PubMed] [Google Scholar]
- 22.Subramaniam K., Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- 23.Haraguchi S., Tsuda M., Kitajima S., Sasaoka Y., Nomura-Kitabayashi A., Kurokawa K., Saga Y. nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech. Dev. 2003;120:721–731. doi: 10.1016/s0925-4773(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 24.Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki A., Tsuda M., Saga Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development. 2007;134:77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H., Tsuda M., Kiso M., Saga Y. Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev. Biol. 2008;318:133–142. doi: 10.1016/j.ydbio.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Hendry A.P., Wenburg J.K., Bentzen P., Volk E.C., Quinn T.P. Rapid evolution of reproductive isolation in the wild: evidence from introduced salmon. Science. 2000;290:516–518. doi: 10.1126/science.290.5491.516. [DOI] [PubMed] [Google Scholar]
- 28.Swanson W.J., Vacquier V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 29.Reijo R., Lee T.Y., Salo P., Alagappan R., Brown L.G., Rosenberg M., Rozen S., Jaffe T., Straus D., Hovatta O., et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 30.Hunt P.A., Hassold T.J. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- 31.Kee K., Angeles V., Flores M., Nguyen H., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtis D., Treiber D., Tao F., Zamore P., Williamson J., Lehmann R. A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 1997;16:834–843. doi: 10.1093/emboj/16.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sada A., Suzuki A., Suzuki H., Saga Y. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science. 2009;325:1394–1398. doi: 10.1126/science.1172645. [DOI] [PubMed] [Google Scholar]
- 34.Corey D., Abrams J. Morpholino antisense oligonucleotides: tools for investigating vertebrate development. Genome Biol. 2001;2:Rev1015. doi: 10.1186/gb-2001-2-5-reviews1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos A., Lehmann R. Germ cell specification and migration in Drosophila and beyond. Curr. Biol. 2004;14:R578–R589. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 37.Forbes A., Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 38.Parisi M., Lin H. The Drosophila Pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis, and embryogenesis. Genetics. 1999;153:235–250. doi: 10.1093/genetics/153.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore F.L., Jaruzelska J., Fox M.S., Urano J., Firpo M.T., Turek P.J., Dorfman D.M., Reijo Pera R.A. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (deleted in AZoospermia) and DAZ-Like proteins. Proc. Natl Acad. Sci. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka S.S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 41.Rongo C., Broihier H.T., Moore L., Van Doren M., Forbes A., Lehmann R. Germ plasm assembly and germ cell migration in Drosophila. CSH Symp. Quant. Biol. 1997;62:1–11. [PubMed] [Google Scholar]
- 42.Mochizuki K., Nishimiya-Fujisawa C., Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev. Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- 43.Castrillon D.H., Quade B.J., Wang T.Y., Quigley C., Crum C.P. The human VASA gene is specifically expressed in the germ cell lineage. Proc. Natl Acad. Sci. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaruzelska J., Kotecki M., Kusz K., Spik A., Firpo M., Reijo R.A. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev. Genes Evol. 2003;213:120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- 45.Reijo Pera R., Alagappan R.K., Patrizio P., Page D.C. Severe oligospermia resulting from deletions of the Azoospermia Factor gene on the Y chromosome. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 46.Karashima T., Sugimoto A., Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- 47.Houston D.W., King M.L. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 48.Eberhart C.G., Maines J.Z., Wasserman S.A. Meiotic cell cycle requirement for a fly homologue of human Deleted in AZoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 49.Maegawa S., Yasuda K., Inoue K. Maternal mRNA localization of zebrafish DAZ-like gene. Mech. Dev. 1999;81:223–226. doi: 10.1016/s0925-4773(98)00242-1. [DOI] [PubMed] [Google Scholar]
- 50.Ruggiu M., Speed R., Taggart M., McKay S.J., Kilanowski F., Saunders P., Dorin J., Cooke H. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 51.Haston K., Tung J., Reijo Pera R.A. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS ONE. 2009;4:e5654. doi: 10.1371/journal.pone.0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fox M., Urano J., Reijo Pera R.A. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and Deleted in Azoospermia-Like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Collier B., Gorgoni B., Loveridge C., Cooke H., Gray N. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reynolds N., Collier B., Maratou K., Bingham V., Speed R., Taggart M., Semple C., NK N.G., Cooke H. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum. Mol. Genet. 2005;14:3899–3909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- 55.Ruggiu M., Cooke H. In vivo and in vitro analysis of homodimerisation activity of the mouse Dazl protein. Gene. 2000;252:119–126. doi: 10.1016/s0378-1119(00)00219-5. [DOI] [PubMed] [Google Scholar]
- 56.Tsui S., Dai T., Roettger S., Schempp W., Salido E.C., Yen P.H. Identification of two novel proteins that interact with germ-cell specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 57.Matzuk M., Lamb D. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 2002;4S:s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi S., Yamada M., Asaoka M., Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- 59.Kadyrova L., Habara Y., Lee T., Wharton R. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- 60.Macdonald P. Translational control: a cup half full. Curr. Biol. 2004;14:R282–R283. doi: 10.1016/j.cub.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 61.Wreden C., Verrotti A.C., Schisa J.A., Lieberfarb M.E., Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 62.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.