Abstract
Sexually reproducing metazoans establish a cell lineage during development that is ultimately dedicated to gamete production. Work in a variety of animals suggests that a group of conserved molecular determinants function in this germ line maintenance and function. The most universal of these genes are vasa and vasa-like DEAD box RNA helicase genes. However, recent evidence indicates that vasa genes also function in other cell types, distinct from the germ line. Here we evaluate our current understanding of vasa function and its regulation during development, addressing vasa’s emerging role in multipotent cells. We also explore the evolutionary diversification of the amino-terminal domain of this gene and how this impacts the association of vasa with nuage-like perinuclear structures.
Keywords: vasa, germ line, multipotent cells, zinc-knuckle, primordial germ cell
Introduction
Segregation and maintenance of the germ line is required for all sexually reproducing metazoans. In many animals, a population of primordial germ cells (PGCs) is set aside early in embryogenesis that is dedicated for the germ cell lineage while other cells in the embryo differentiate into soma. PGCs migrate to the somatically-derived gonads and proliferate into germ line stem cells that can self renew and differentiate into gametes. At least 3 general mechanisms are used to specify PGCs within the animal kingdom. The germ line can form (1) early in embryogenesis from an inheritance of maternal factors (maternally derived, also referred to as preformation) used in flies and nematodes, (2) by cell-cell interactions early in embryogenesis (inductive, also referred to as epigenetic) as seen in mice, and (3) any time in the animal’s life, even in adulthood, from a multipotent stem cell precursor (persistent multipotent cell derived germ cells), such as in planaria and hydra.(1–4) Despite these developmental differences, animals employ a group of conserved molecular determinants for PGC specification. The most common of these is the gene vasa.(5–7) While the exact function of Vasa is unclear, its extensive conservation underscores its universal importance in germ line development (Table 1).
Table 1.
Full-length vasa ortholog cDNAs
| Group | Common name | Gene name | Polypeptide length | CCHC Zn-fingers | Genebank accession |
|---|---|---|---|---|---|
| Porifera | Sponges | N/A | N/A | N/A | N/A |
|
| |||||
| Cnidaria | Hydra | ||||
| Hydra magnipapillata | CnVasl | 797 | 7 | BAB13307 | |
| CnVas2 | 890 | 1 | BAB13308 | ||
| Hydractinia echinata | HeVas | 680 | 3 | ABO93350 | |
|
| |||||
| Platyhelminthes | Flatworms | ||||
| Macrostomum lignano | Macvasa splice-form 1 | 929 | 3 | CAL91031 | |
| Macvasa splice-form 2 | 860 | 2 | CAL91030 | ||
| Neobenedenia girellae | Ngvlg3 | 634 | 7 | BAF44661 | |
| Paragonimus westermani | vasa2n | 606 | 0 | ABM30180 | |
| vasa3n | 606 | 0 | ABM30181 | ||
|
| |||||
| Echinoderms | |||||
| Strongylocentrotus purpuratus | Purple urchin | SpVasa splice-form 1 | 766 | 4 | ACM80369 |
| SpVasa splice-form 2 | 732 | 3 | |||
| Lytechinus variegates | Green urchin | LvVasa splice-form 1 | 5 | ||
| LvVasa splice-form 2 | 679 | 0 | ACM80368 | ||
| Eucidaris tribuloides | Pencil urchin | EtVasa | 498 | 1 | ACM80367 |
| Asterias forbesi | Forbes’s Starfish | AfVasa | 715 | 3 | ACM80365 |
| Asterina miniata | Batstar | Am Vasa | 730 | 4 | ACM80366 |
|
| |||||
| Tunicates | Sea Squirts | ||||
| Ciona intestinalis | DEAD1 | 669 | 3 | BAA36710 | |
| Ciona savignyi | CsVHa | 688 | 3 | BAB12216 | |
| CsVHb | 770 | 6 | BAB12217 | ||
| Halocynthia roretzi | Vasa | 691 | 3 | ACJ64200 | |
| Botryllus primigenus | BpVas | 687 | 5 | BAE44472 | |
| Botryllus schlosseri | BS-Vasa | 547 | 1 | ACJ69403 | |
| Polyandrocarpa misakiensis | PmVas | 705 | 5 | BAE94497 | |
| Botrylloides violaceus | Vasa protein | 630 | 4 | ABM74410 | |
|
| |||||
| Lancelets | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Vertebrates | |||||
| Homo sapiens | Human | Ddx4 | 690 | 0 | AAH47455 |
| Mus musculus | Mouse | Ddx4 | 728 | 0 | AAI37602 |
| Gallus gallus | Chicken | Cvh | 662 | 0 | BAB 12337 |
| Danio rerio | Zebrafish | Vasa protein | 688 | 0 | AAI29276 |
| Vasa-like protein | 715 | 0 | AAL89410 | ||
| Sus scrofa | Wild boar | Vasa-like protein | 722 | 0 | AAT46129 |
| Oncorhynchus mykiss | Rainbow trout | Vasa | 647 | 0 | BAA88059 |
| Oryzias latipes | Medaka | Vasa | 617 | 0 | BAB61047 |
| Rattus norvegicus | Rat | Ddx4 | 713 | 0 | Q64060 |
| Xenopus laevis | Clawed frog | Vlgl | 700 | 0 | AAC03114 |
| Rana lessonae | Pond frog | DEAD box protein | 724 | 0 | CAH56439 |
| Carassius auratus | Silver crucian carp | CagVasa | 701 | 0 | AAV70960 |
| Oreochromis niloticus | Nile tilapia | Vasa | 645 | 0 | BAB 19807 |
| Monopterus albus | Swamp eel | Vasa-like protein | 618 | 0 | ABA54551 |
| Silums meridionalis | Southern catfish | Vasa | 662 | 0 | ACD62525 |
| Pan troglodytes | Chimpanzee | Vasa | 703 | 0 | XP 517757 |
| Macaca fascicularis | Crab-eating macaque | Ddx4 | 725 | 0 | Q4R5S7 |
| Thunnus orientalis | Pacific bluefin tuna | Vasa | 644 | 0 | ABY77970 |
| Trachurus japonicus | Japanese jack mackerel | Vasa | 657 | 0 | BAG72093 |
| Salvelinus leucomaenis | Whitespotted char | Vasa | 662 | 0 | ACA33927 |
| Leucopsarion petersii | Ice goby | Vasa homolog | 645 | 0 | BAD04052 |
| Cyprinus carpio | Carp | Vasa | 691 | 0 | AAL87139 |
| Am by stoma mexicanum | Axolotl | AxVH | 724 | 0 | AAT09162 |
|
| |||||
| Molluscs | |||||
| Crassostrea gigas | Pacific oyster | Oyvlg | 758 | 4 | AAR37337 |
| Chlamys farreri | Japanese scallop | Vasa | 801 | 5 | ABE27759 |
| Haliotis asinina | Vetigastropod | Vasa | 763 | 2 | ACT35657 |
|
| |||||
| Pogonophora | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Vestimentifera | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Annelids | |||||
| Platynereis dumerilii | Dumeril’s clam worm | Vasa homolog | 712 | 3 | CAJ15139 |
| Vasa protein isoform | 732 | 4 | CAJ38803 | ||
| Enchytraeus japonensis | Vasa-related | 990 | 8 | BAF767096 | |
|
| |||||
| Rotifers | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Nematodes | |||||
| Caenorhabditis elegans | Germ-line helicase 1 | 763 | 4 | AAB52901 | |
| Germ-line helicase 2 | 974 | 6 | AAB03510 | ||
| Germ-line helicase 3 | 720 | 2 | AAC28388 | ||
| Germ-line helicase 4 | 1156 | 5 | AAC28387 | ||
| Caenorhabditis briggsae | GLH-1 related | 795 | 4 | CAP31774 | |
|
| |||||
| Onychophora | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Tardigrades | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Myriapods | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Insects | |||||
| Drosophila melanogaster | Fruit fly | Vasa | 661 | 0 | CAA31405 |
| Anopheles gambiae | African malaria mosquito | Vasa-like protein | 596 | 0 | AAY41942 |
| Apis mellifera | Honey Bee | Vasa protein | 630 | 0 | NP 001035345 |
| Acyrthosiphon pisum | Pea aphid | Vasa-like protein | 669 | 0 | XP 001948608 |
| Culex quinquefasciatus | Southern house mosquito | Vasa | 641 | 0 | XP 001850751 |
| Tribolium castaneum | Red flour beatle | Vasa | 627 | 0 | NP 001034520 |
| Bombyx mori | Silkworm | BmVLG | 601 | 0 | BAA19572 |
| Drosophila virilis | Fruit fly | Vasa | 625 | 0 | AAM49782 |
| Aedes aegypti | Yellow fever mosquito | Vasa-like protein | 638 | 0 | AAY41941 |
| Melipona scutellaris | Stingless bee | Vasa | 624 | 0 | ABQ96192 |
| Melipona quadrifasciata | Stingless bee | Vasa | 624 | 0 | AC102436 |
| Scaptotrigona postica | Stingless bee | Vasa | 624 | 0 | ABQ96191 |
| Frieseomelitta varia | Stingless bee | Vasa | 624 | 0 | ACI02437 |
| Copidosoma floridanum | Parasitic wasp | Vasa | 708 | 0 | AAT12450 |
| Gryllus bimaculatus | Two-spotted cricket | Vasa | 650 | 0 | BAG65665 |
|
| |||||
| Crustaceans | |||||
| Parhyale hawaiensis | Amphipod crustacean | Vasa | 676 | 5 | ABX76969 |
| Daphnia magna | Water flea | DmaVas | 779 | 6 | BAE00180 |
| Litopenaeus vannamei | Pacific white shrimp | Vasa-like protein | 703 | 3 | AAY89069 |
| Fenneropenaeus chinensis | Chinese white shrimp | Vasa | 712 | 3 | ABQ00071 |
| Macrobrachium rosenbergii | Giant freshwater prawn | MrVLG | 710 | 3 | ABC87271 |
| Moina macrocop | Water flea | MmVas | 843 | 6 | BAD99524 |
| Artemia franciscana | Brine shrimp | AfVas | 726 | 3 | BAD99523 |
|
| |||||
| Horseshoe Crabs | N/A | N/A | N/A | N/A | N/A |
|
| |||||
| Arachnids | N/A | N/A | N/A | N/A | N/A |
Vasa is a member of the DEAD box protein family which functions in a broad range of molecular events involving duplex RNA. Nine conserved sequence motifs typify all DEAD-box genes (Figure 1A.(8) Biochemical analyses show how these motifs, in Vasa and other DEAD-box proteins, confer its ATP-dependant RNA helicase catalytic activity. Structural data also suggest that Vasa unwinds duplex RNA in a non-processive manner.(8–11) DEAD-box proteins may operate as chaperones that unwind local secondary structures to facilitate proper RNA folding and interactions with accessory proteins. (12,13) These features are evident in examples encompassing pre-mRNA splicing, ribosome biogenesis, nuclear export, translational regulation and degradation.(8)
Figure 1. Vasa gene family conservation.
(A) Nine conserved sequence motifs specific to all DEAD-box genes. Residues important for ATP-binding and hydrolysis are indicated in green and residues involved in RNA binding are indicated in purple. The function of the remaining conserved motifs in white is unclear. (B) A schematic representation of the phylogenetic relationship between Vasa genes, PL10 genes and the closest related DEAD box gene, p68 (Adapted from Mochizuki et al., 2001). The star indicates the evolutionary split between Vasa and PL10 genes. (C) CCHC Zn-knuckle domain-containing Vasa genes from Table 1 were used to illustrate the phylogenetic conservation of this motif in the N-terminal portion of Vasa proteins. Animals with Vasa CCHC Zn-knuckles are indicated in red, animals lacking Vasa Zn-knuckles are shown in black and animals with insufficient Vasa sequence information are indicated in grey (Phylogenetic tree adapted from Dunn et al., 2008).
Functional analyses of Vasa
What is Vasa’s role in germ line development? Vasa-null animals have been generated in Drosophila, Caenorhabditis elegans and mice by gene knockout, by reduction of Vasa mRNA by RNA-interference (RNAi) and by Vasa protein reduction by antisense morpholino treatment (Knockdown).(14–20)
Drosophila leads the way in understanding vasa function
Genetic screens for maternal-effect genes in Drosophila first revealed vasa function in oocyte development.(21) Subsequent mutational and gene inactivation studies showed vasa function in posterior patterning and in germ cell specification in the embryo.(15,22,23) Identifying the molecular targets of this DEAD-box helicase though has proven difficult. Vasa produced from bacterial recombinant can bind RNA and has ATP-dependent RNA helicase activity in vitro. This activity is absent with mutations in the conserved DEAD-box sequence motifs.(10,24) Structural analysis of the DEAD-box region in Drosophila Vasa suggests that it unwinds duplex RNA in a nonprocessive manner by binding and bending short stretches of the duplex.(10) Consistent with its RNA-binding ability, biochemical and genetic data suggest Drosophila Vasa acts as a translational regulator. Indeed, its direct binding to the translational initiation factor eIF5B is required for proper translation of maternal gurken transcripts.(15,25–28)
Does Vasa bind RNA in a sequence specific manner?
Recently, Liu et al., (2009) screened for mRNAs that co-purified with Vasa from Drosophila embryos. They identified 221 candidate mRNAs that bound to Vasa, 24 of which were mRNAs in the pole cells – where Vasa is in vivo. Mei-P26 was one of the candidates – its protein product represses microRNA activity and promotes differentiation of the germ line stem cells. Liu et al. found that in vasa mutants, mei-P26 translation is substantially reduced.(29) This is intriguing in that mei-P26 was previously shown to interact with one of the Argonaut proteins of the miRNA pathway (Ago1) to repress the miRNA interference of target mRNAs in the germ line. Thus, the absence of vasa resulted in low mei-P26 synthesis, and therefore miRNA interference was functional in the germ line. Perhaps more important was that this Vasa/mei-P26 mRNA interaction was shown to be sequence specific; Vasa bound specifically to a (U)-rich motif in the mei-P26 39 untranslated region in vitro, and expression of a GFP-mei-P26 transgenes in vivo was dependent on the same (U)-rich 39 UTR domain. Moreover, mei-P26 translation was significantly reduced by a mutation in Vasa that reduced its interaction with the translational initiation factor eIF5B.(29) These results are important for several reasons: 1) it provides an important gene regulatory link to understand miRNA regulation in the germ line, 2) it suggests that Vasa interacts with mRNAs selectively, and 3) Vasa interacts with mRNAs in a sequence selective fashion, perhaps linking the sequences in the 3′UTR target mRNA to the initiation factor important for translation of the sequence. Further identification of other sequences in this Vasa-interactome will be important to understand the consensus mRNA sequences for its interaction and the resulting sequences important for germ line development.
Overlapping functions of multiple vasa genes in C. elegans
Germ line development in the nematode Caenorhabditis elegans is dictated by the inheritance of localized cytoplasmic determinants in the egg and early embryo. Despite some notable differences in germ plasm composition between Drosophila and other organisms, such as the lack of an Oskar homolog outside of diptera, and the presence of the unique PLG-1 gene in C. elegans, the localization of Vasa in the germ plasm remains a conserved feature.(4) C. elegans has four germ line helicases (GLH 1-4), each are Vasa homologs, each present in the germ granules (P-granules) and each are present exclusively in germ line blastomeres during development.(16–18) Loss of function analyses suggests that GLH-1 is most important for germ line development; glh-1 mutants have a dramatic decrease in germ cells and mature gametes.(18,19) Although GLH 2–4 transcripts are present in the germ line and their proteins localize to P-granules, deletion of glh-2, glh-3 or glh-4 genes alone are not sufficient to cause adult sterility.(17,19) Unfortunately, the mechanism of GLH-function is not known, and may require a biochemical approach as recently accomplished in Drosophila.(29)
The picture of vasa function in mammals is dim
Vasa-null mice develop normally and the females are completely fertile. In contrast, male vasa-null mice are infertile due to deficiencies in male germ cell proliferation and differentiation.(14) The Vasa-like gene, DBY in humans, also appears to be required for male fertility.(30) These examples of sex-specific phenotypes in mammals are just the opposite to vasa mutants in Drosophila, where females are sterile and males are fertile. Mutants of the vasa-related gene (PL10, also called Belle) in Drosophila, however, are male infertile.(31) Thus, although vasa is present in both male and female germ lines, it must either have different targets of function or is regulated differently in the two germ line types. The major impediment in our progress here is that we have so few clues as to the functional mRNA targets of vasa. Overcoming this deficiency will likely require the biochemical approaches of Vasa-mRNA co-purification. Such an approach is difficult though in many animals in which only a small amount of relevant tissue is accessible.
Vasa function is required in diverse organisms – studies with loss of function approaches
While work in Drosophila, C. elegans and mice constitutes the most extensive analysis of any vasa gene, a growing body of data from several different animals have contributed to our understanding of Vasa function. Abrogating Vasa expression by utilizing RNAi in embryos and adults has been useful for collecting functional data in animals lacking stable transgenics. For example, the flatworm M. lignano displays Vasa expression in the multipotent neoblast stem cells in addition to germ cells. However, RNAi knockdown of Vasa had no effect on stem cell maintenance, neoblast proliferation, gonad formation or gonad development.(32) This suggests either a nonessential role of Vasa in adults or a functional overlap with other Vasa-like genes in flatworm gonads and neoblasts, similar to that seen in the germ line of the roundworm. So too in colonial ascidians,(33–35) oysters,(36) in teleosts,(20,37) Xenopus,(38) the parasitic wasp,(39) and the crustacean Parhyale hawaiensis,(40) vasa function and or association in germ cell development is widespread.
Vasa function is required in diverse organisms – gain of function in the chicken and the human
Recent Vasa gain-of-function analyses in human and chicken cell lines have provided more insight into Vasa’s role in development. Ectopic Vasa expression has been reported in epithelial ovarian cancer cells where it abrogates the DNA damage-induced G2 checkpoint by down regulating 14-3-3σ expression.(41) However, it is unclear whether the abnormal presence of Vasa reflects a causative role in ovarian tumorigenesis. This could be tested in mice by restoring 14-3-3σ expression in a Vasa-expressing ovarian cancer cell line, reintroducing these cells into a host animal and analyzing whether it abolishes tumorigenicity. Nonetheless, this observation illustrates the importance of correct Vasa expression in development. Ectopic Vasa expression in chicken embryonic stem cells (ESCs) induces expression of specific germ line and meiotic genes.(42) As a consequence, following their injection into chick embryos, these ESCs exhibit improved germ line colonization and adopt a germ cell fate. This supports a fundamental role of Vasa in germ line identity and function.
Together these data suggest Vasa has an essential and evolutionarily conserved role in many aspects of germ line development including germ cell specification, proliferation and maintenance. Identification and analyses of Vasa targets will help resolve the mechanistic details behind this conserved germ line function as well as additional roles outside the germ line. The results also show that no one approach (gene inactivation versus gene over-expression), no one cell-type, no one animal, and no one gene will likely solve this problem. Instead, using diverse experimental approaches in many different animals will hopefully enable the most efficient approach to definitely identify vasa function.
Beyond the germ line: Vasa in multipotent stem cells
Although Vasa has proven a reliable marker for the germ lineage in several animals, these examples mostly consist of insects and vertebrates (Table 1).(5) Emerging data have provided a broader phylogenetic perspective on vasa and demonstrate its function also in multipotent cell types. In Cnidarians, for example, the multipotent interstitial cells (I-cells) contribute not only to the germ line, but also to somatic cell types such as nematocytes (nerve cells) and gland cells.(7,43,44) Flatworms possess remarkable regenerative capabilities due to their pool of multipotent stem cells (neoblasts).(45) Vasa is expressed in the ovaries, the testes and the neoblast stem cells.(32) The function(s) of Vasa in these persistent multipotent cells of cnidarian and flatworm is yet unknown, but with recent technological advances in the experimentation of both of these animals, vasa’s function can be tested by RNAi or morpholino knock-down of Vasa followed by testing the ability of I-cells or neoblasts not only to proliferate and differentiate into somatic cell types, but also into a germ line.
While no examples of Vasa expression are reported outside of germ line cells in vertebrates and insects, data from other deuterostomes and from various arthropods prove otherwise. For example, the crustacean Polyascus polygenea (a colonial barnacle) contains Vasa in a cluster of multipotent stem cells in its buds and stolon during the parasitic stage of its life cycle.(46) Vasa is present in the auxiliary cells of the oyster ovary, in the somatic cells of the reconstructing gonadal tissues during its development for the next spawning cycle,(47) and in the snail, is present in non-germ-line lineages.(48) In the polychaete annelid, Vasa is enriched in the progenitor mesodermal posterior growth zone (MPGZ). The MPGZ cells are multipotent somatic stem cells, highly proliferative and contribute to both mesodermal tissue and PGCs.(49) In the oligochaete as well, Vasa is present in non-genital segments during its development.(50)
Tunicates are close relatives to vertebrates and analyses in the colonial ascidians (Botryllus primigenus and Polyandrocarpa misakiensis) show vasa mRNA expression in their germ lines along with cell aggregates containing somatic-derived multipotent hemoblasts.(51,52) Regenerating buds induce vasa expression de novo at every budding cycle suggesting that vasa may have functions in regeneration activities independent of the germ line.(2,35,53) Indeed, many of the animals with Vasa in their multipotent cells are capable of tissue regeneration in the adult to varying degrees.(54,55) This is also true in the more limited regenerative context for oyster gonad regeneration as well as asexual reproductive budding in colonial ascidians and the parasitic barnacle. However, Vasa expression in multipotent cells outside of a regenerative context in the polychaete annelid may be indicative of a more general germ cell developmental phenomenon where localized cytoplasmic factors may help set aside a multipotent stem cell population of which a subset are later designated for the germ line. Overall, the presence of Vasa in non-germ line cells in various taxa is becoming standard, but still lacking is an experimental test of vasa function during the regeneration, the development, or in the maintenance of potency of non-germ line cells.
Regulation of Vasa expression
Transcriptional and epigenetic control of vasa expression
Localized Vasa expression is a common feature throughout phylogeny and in order to control this, animals employ a variety of mechanisms to regulate both vasa mRNA and protein accumulation. Several studies suggest vasa transcriptional regulation contributes to cell and tissue-specific Vasa expression in developing embryos and adults. (56) In Drosophila embryonic development, zygotic transcription of vasa occurs specifically in the pole cells immediately after gastrulation and remains germ line-specific into adulthood.(57) DNA methylation is one form of epigenetic regulation directing differential gene expression, where hypermethylation is associated with gene silencing.(58) A recent study in humans suggests that the methylation state of the vasa gene promoter controls its specific transcription in the testes. The vasa promoter is hypomethylated in the testes and methylated in all other tissues, which do not express vasa.(59) A clinical study showed that spermatogenesis defects such as idiopathic azoospermia or severe oligospermia were also associated with a hypermethylated vasa promoter in some individuals.(60) Transcriptional regulation during gonad development and germ cell maturation likely also involves hormonal signaling.(61,62)
Post-transcriptional regulation of vasa
The presence of different vasa splice forms in several animals such as M. lignano, S. purpuratus, L. variegatus and P. dumerilii, suggests RNA processing contributes to vasa regulation (Table 1).(32,49,63) Differential vasa expression can also result from regulation of vasa mRNA stability. For example, the vasa transcript in the amphipod crustacean Parhyale hawaiensis is maternally provided and uniformly distributed during early cleavage stages before localizing in 32-cell stage embryos. These Vasa-positive cells were determined to be PCGs by lineage tracing analysis and vasa transcript localization is dependent on its 3′UTR to preferentially stabilize it in the germ line.(40,64) Differential vasa transcript stability has been seen in several other animals, such as zebrafish and other teleosts, and thus appears to be an important general process for post-transcriptional regulation.(37,65–67)
Translational repression is another regulatory mechanism that allows localized protein production from a ubiquitous transcript during embryonic development.(68) Sequences within a transcript 5′ and 3′UTRs may contain cis-regulatory elements which form secondary RNA structures or which bind trans-acting factors that inhibit its translation.(69) Relief of this translational repression can direct localized vasa expression. For instance, in mice, Dazl protein binds the 3′UTR of mouse vasa mRNA in vivo, stimulates its translation in Xenopus oocyte extracts and Dazl knockout mice have reduced levels of Vasa protein, suggesting this regulation is crucial for Vasa translation and spermatogenesis.(70)
Post-translational regulation of Vasa
Work in Drosophila has detailed multiple mechanisms that regulate Vasa protein localization to the pole plasm. For example, Vasa directly interacts with Oskar protein in vitro and this interaction may facilitate Vasa anchoring to polar granules in the posterior pole of the oocyte.(71) Indeed, all mutant oskar alleles are defective in Vasa protein localization.(72,73)
Evidence also suggests that proteolysis may play a regulatory role in Vasa localization. In Drosophila, Vasa is ubiquitinated in the oocyte and its pole plasm accumulation is dependent on the deubiquitinating enzyme Fat facets.(74) Since ubiquitylation can target a protein for degradation, Fat facets may stabilize Vasa in the pole plasm. Normal Vasa pole plasm localization also depends on Gustavus and Fsn, two paralogous B30.2/SPRY domain proteins.(75,76) Both Gustavus and Fsn directly interact with Vasa through their B30.2/SPRY domain and share several features indicative of an E3 ubiquitin ligase function.(76–78) Gustavus contains a SOCS-box domain that complexes with Elongin B/C in vitro and Cullin-5 in vivo.(75–77)Fsn has an F-box domain that interacts with Cullin-1 in vivo.(76) While Gustavus and Fsn both appear to target Vasa for ubiqutination, mutational and overexpression analyses suggest their functions are not identical and may contribute to a delicate regulatory balance of Vasa ubiquitination required for its proper localization.(76) These results raise several important questions concerning this regulation. What is the nature of Vasa ubiquitination by Gustavus and Fsn? Are there mono-ubiquitination, K48-ubiquitin chains or K63-ubiquitin chains species of Vasa and do these impart different stability and localization properties?
Regulation of proteolysis may also direct Vasa localization during embryonic development in the sea urchin Strongylocentrotus purpuratus. Vasa transcript is present uniformly during early embryogenesis through blastula stage, but Vasa protein is strongly enriched in the 16-cell stage micromeres and subsequent small micromeres.(63,79) The Vasa coding region is sufficient for its small micromere accumulation and proteasome inhibition increases Vasa protein levels throughout the embryo (Gustafson et al., in preparation). Vasa expression is also regulated by proteolysis in C. elegans.(19) The Jun N-terminal kinase member KGB-1 and COP9 signalosome subunit 5 (CSN-5) both bind GLH-1.(80) Phosphorylation of GLH-1 by KGB-1 targets it for proteasomal degradation, whereas CSN-5 association in GLH-1 enhances its stability in the germ line.(80) In addition, recent data from Vasa orthologs in mice, Xenopus and Drosophila suggest arginine methylation is a conserved aspect of Vasa regulation. Furthermore, in Drosophila, the arginine methyltransferase Capsuleen (dPRMT5/csul/dart5) is required for symmetric dimethyl-ariginine modifications of Vasa in vivo.(81) However, additional work is required to elucidate the functional consequences of Vasa arginine methylation. Does arginine methylation influence Vasa’s binding specificity to target mRNAs and other proteins or are they important for Vasa’s localization and protein stability?
Sub-cellular Vasa localization suggests a dynamic role in mRNA association - the Nuage-like structures
In addition to its cell type and tissue-specific expression, another universal feature of Vasa expression is its subcellular localization. Germ cells in all metazoan animals contain perinuclear electron-dense ribonucleoprotein (RNP) structures.(82) These RNP rich structures are often called nuage, the mitochondrial cloud, polar granules, P-granules, chromatoid bodies and in some somatic cells, P bodies. (82,83) While the various names of these structures correspond to differences in morphology, composition and animals in which they were first identified, it is believed they are related entities.(83,84) Several of the proteins identified as nuage components are known to function in mRNA regulation.(4) Although it is still unclear whether these structures are centers for translation, polysomes have been reported adjacent to nuage in Drosophila and rats.(83,85) Ultrastructural analyses in C. elegans show nuage structures are frequently associated with nuclear pores.(86) One of the most common nuage components is Vasa (Figure 2).(17,86–90) Examples in several animals suggest that at some point in development, either Vasa mRNA or protein displays a nuage association or nuage-like localization (Table 2).
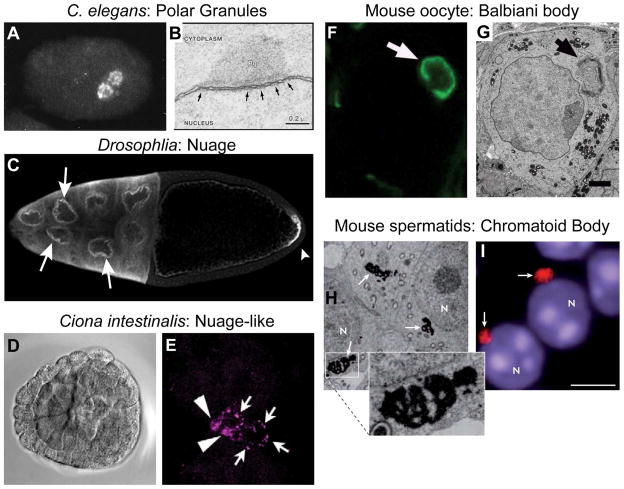
Figure 2. Vasa protein association with nuage-like structures.
Vasa protein localizes to perinuclear structures in a wide variety of metazoans. C. elegans immunofluorescent imaging of Vasa protein (A) and TEM image of polar granules (B) (black arrows indicate nuclear pores). (C) In Drosophila melanogaster developing egg chamber, Vasa protein localizes to the pole plasm in the oocyte (white arrowhead) and the pernuclear nuage in nurse cells (red arrows). DIC (D) and Vasa immunoflourescence (E) imaging of a Ciona intestinalis gastrula embryo with both punctate (arrows) and diffuse (arrowheads). Vasa protein (F, immunofluorescence) localizes to the Balbiani body (G, TEM) in mouse oocytes. In mouse spermatids, chromatoid bodies (black arrows) (H, TEM) contain Vasa protein (white arrows) (I, immunoflourescence).
Table 2.
Subcellular localization of Vasa during development
| Species | Nuage-localization |
|---|---|
| Cnidaria | |
| Hydractinia echinata (Hydrazoa) | Protein: Perinuclear nuage material in oocytes and early embryos(44) |
| Platyhelminthes | |
| Macrostomum lignano (Flatworm) | Protein: Perinuclear localization in developing germ cells in ovaries and testes as well as multipotent neoblasts(32) |
| Echinoderms | |
| Strongylocentrotus purpuratus (Purple sea urchin) | VasaGFP Localizes to perinuclear granular structures resembling nuage in small micromere cells (Gustafson et al., unpublished) |
| Annelids | |
| Platynereis dumerilii (Dumerili’s clam worm) | Protein: Localizes to a perinuclear ring in oocytes corresponding to previously described nuage material(49,111) |
| Tunicates | |
| Ciona intestinalis (Ascidian) | Protein: Perinuclear nuage-like particles during the late tailbud stage of embryonic development(90) |
| Vertebrates | |
| Homo sapiens (Human) | Protein in Gonocytes and oogonia: Not expressed in first trimester fetuses, but progressively increases in developing female germ cells between weeks 12–20 and coincides with a change from a uniform cytoplasmic distribution to a punctuated perinuclear ring localization(112–114) |
| Protein in Oocytes: Localizes to a single compact perinuclear body in developing embryos(113) | |
| Mus musculus (Mouse) | Protein in Spermatocytes: Localizes to the cytoplasm in a perinuclear fashion resembling the chromatoid body(115) |
| Gallus gallus (Chicken) | Protein: Localizes to spherical mitochondrial cloud structures in developing oocytes(116) |
| Sparus aurata (Gilt head sea bream) | mRNA: Localizes to the perinuclear cytoplasm in early midvitellogenic and vitellogenic oocytes(62) |
| Oryzias latipes (Medaka) | mRNA: Localizes to patches with the early oocyte cytoplasm(117) |
| Danio rerio (Zebrafish) | Protein: Granular localization in PGCs and Vasa(20) |
| mRNA: localizes to nuage-like structures in 1000-cell embryos(118) | |
| Xenopus Laevis (African clawed frog) | Protein: Localizes to germ granules and germ plasm during early embryonic development(38,119) |
| Chaetognaths | |
| Sagitta inflata (Arrow worms) | Protein: Localizes to the large germ granule formed in fertilized eggs (120) |
| Insects | |
| Drosophila melanogaster (Fruit Fly) | Protein: Found in perinuclear nuage of nurse cells in developing egg chambers(72,73,121) |
| mRNA: Localizes to a perinuclear granules in the egg(122) | |
| Bombyx mori (Silkworm) | |
| Nematodes | |
| Caenorhabditis elegans | Protein: Localizes to P-granules during embryonic development(16–18) |
| Molluscs | |
| Crassostrea gigas (Oyster) | mRNA: Localizes distinctly to cytoplasmic granules at the vegetal pole in unfertilized oocytes(47) |
It is unclear however whether nuage is the location of Vasa function. Instead, this subcellular localization may be used to direct a lineage-specific segregation of Vasa during early embryonic development. In many animals, Vasa-containing nuage structures are thought to be exclusively germ line-specific. However, the presence of Vasa-positive nuage-like structures in multipotent neoblasts of the flatworm Magnostatum lignano suggests they may be more widespread (Table 2).(32) Now that at least one vasa target, mei-P26, has been identified, its relative association with the nuage will be important to determine in order to resolve the consistent role of vasa in this specialized cellular structure.
Does vasa interact with the RNAi pathway?
Several studies indicate a functional relationship between Vasa and both the small interfering RNA and micro-RNA processing pathways. One essential component in both of these pathways is the RNase III endonuclease Dicer,(91) which, in mice, colocalizes with Vasa in nuage.(89) Furthermore, ectopically expressed Vasa and Dicer protein interact in COS cell lysates and this interaction requires the C-terminal portion of Vasa.(89) The C-terminal RNaseIII region of Dicer is sufficient to interact with Vasa and the remaining N-terminal ATPase/helicase-PAZ domain region appears independent of Vasa.(89)
PIWI proteins represent a subgroup of Argonautes required for germ line stem cell maintenance and fertility in several animals.(92) Drosophila PIWI is a polar granule component that interacts with Vasa and PIWI-mutant flies have normal Vasa protein expression and abdominal patterning of the embryo, but exhibit a severe deficiency in pole cell formation.(93) Over-expressing PIWI results in a dose-dependent increase in Vasa protein levels and pole cell formation. These data suggest that a PIWI-mediated piRNA pathway regulates the levels of Vasa and Oskar proteins and possibly other genes involved in the germ line determination pathway in Drosophila.(93) A similar interaction is found in mouse, where Vasa protein binds to both recombinant and endogenous MIWI and MILI, which are mouse PIWI homologs.(81,94) Indeed, MILI and Vasa knockout mice have similar phenotypes and defects in spermatogenesis indicative of cooperative molecular functions. (14,94) In MIWI knockout mice, Vasa protein does not localize to the nuage structures.(94) However, it is still unknown whether MIWI is required for nuage and ultrastructural studies in MIWI knockout mice are needed. Exactly how these specific interactions influence Vasa, MIWI or MILI function is unclear.
Recent work has identified Maelstrom as a nuage component that interacts with both mouse Vasa and MIWI, is required for spermatogenesis and also is involved in silencing transposable elements.(95,96) In Drosophila, Maelstrom protein localizes to nuage in a Vasa-dependent manner. In maelstrom mutant oocytes, a higher molecular weight Vasa protein species is evident indicating that Maelstrom is required for proper Vasa modification or processing.(87) Although still not definitive, the consistent association in multiple animals of vasa and members of the RNAi pathway argues strongly that they have a functional relationship. This may not be surprising since both are postulated to be involved in translational regulation, and the engagement of each mRNA with the ribosome is likely a continuously evaluated process that responds with incremental increases, decreases, and rates of translation. Researchers often have difficulty in dissecting overlapping pathways by classic genetic means, so an alternative approach is to make use of blossoming in vitro cell free lysate assays both for mRNA translational activity, as well as for mRNA stability as a result of miRNAs.(97,98) In the context of vasa function in the RNAi pathway, it is hard to ignore the intersection here that the small RNAs are 20–30 bases in length, and that vasa is capable of unwinding 20–25 bases of dsRNA.
Vasa evolution resulted in divergent N-termini while retaining the conserved helicase domain
Comparative phylogenetic data suggests that the Vasa gene family originated from a duplication in a PL10-related DEAD-box gene early in metazoan evolution.(7) While animals have both vasa and PL10 genes, plants and fungi have only PL10 genes and lack vasa genes (Figure 1B). At some point after this gene duplication, vasa genes acquired CCHC Zn-knuckle domains in the region N-terminal of the conserved DEAD box. The number of Zn-knuckle domains found Vasa sequences vary from 1 to 8. However, vertebrates and insects may have both independently lost these Zn-knuckle domains (Figure 1C, Table 1).
CCHC Zn-knuckles can be categorized as a “classical zinc-finger” based its zinc chelation topology of a short β-hairpin followed by an α-helix.(99) They can bind single and double-stranded DNA or RNA and may be involved in transcriptional regulation.(100,101) Most examples of CCHC Zn-knuckles come from human retroviruses which all require Zn2+ binding for proper nucleocapsid protein folding.(102–104) The CCHC Zn-knuckle domains of retroviral nucleocapsid proteins interact with specific structures in the viral RNA genome during packaging.(105–109) A non-viral CCHC Zn-knuckle example includes the C. elegans Lin28 protein, which has 2 CCHC Zn knuckles that are crucial for its localization to P-granules and stress granules.(110) While the exact functional significance of these Zn-knuckle domains is unknown, their RNA-binding properties point to a role in Vasa’s RNA target specificity. Zn-fingers are versatile and can also target binding to other proteins.(99)
Do the CCHC Zn-knuckles impart additional functional dimensions to Vasa proteins? While the presence of the 9 conserved DEAD-box sequence motifs in Vasa allow inference into their RNA helicase catalytic activities, the highly divergent N-terminal regions are more cryptic. The presence of Vasa Zn-knuckles correlates, in many animals, with an expanded expression pattern (and possible functional role) outside of the germ line. The presence or absence of Zn-knuckles may reflect differences in RNA or protein target binding. These different target interaction properties may be important to potential expanded functions outside of the germ line. One exception to this notion is the presence of Zn-knuckles in the 4 Vasa homolog Germ-line helicase (GLH 1-4) genes in C. elegans, whose expression is restricted to the germ line.(16–18) Alternatively, it is possible that the loss of Zn-knuckles in insect and vertebrate vasa genes coincided with the emergence of Zn-knuckle containing cofactor proteins which now confer the target specificity. However, no such Vasa cofactors have been identified yet in either insects or vertebrates.
Summary and Future directions
Although vasa has been known to be an important gene in development for almost 30 years, we know remarkably little about how it works. With recent progress and technological breakthroughs recently, however, we anticipate that the next few years will result in a plethora of answers and new understanding. We feel the following are the most important questions to address:
What are the mRNA targets of vasa in translational regulation?
Does vasa function similarly in germ cells as it does in multipotent stem cells?
What role does the highly divergent N-terminus of vasa play in its function?
Is the nuage a site of vasa function or storage?
Is vasa an integrator of RNAi and translational regulation?
Despite the tremendous diversity in embryonic development displayed in metazoan animals, the broad conservation of Vasa underscores its importance for reproduction. Despite numerous functional analyses though, our understanding of Vasa’s specific molecular function in translational regulation is meager. Further Vasa functional analyses will greatly expand our understanding of Vasa’s purpose during development and how it is localized so effectively. Analysis of Vasa expression in less-studied animals outside of vertebrates and insects imply that the role of Vasa is not strictly confined to the germ line, which coincides with the divergence of its N-terminus. Perhaps the animals containing Zn-knuckles on their N-termini have different functional capabilities than those without a defined N-terminal structure. Alternatively, vasa may simply interact with a Zn-knuckle containing protein and thereby have similar functions. Ectopic expression studies and gene swapping experiments will certainly help in this regard. Most importantly, if Vasa utilizes its helicase activity in translation, does it function as a general translation factor or does it interact with specific target transcripts? Although Vasa is capable of binding RNA in vitro, it is currently unknown whether Vasa interacts with mRNAs in vivo.(10,24) Gurken is one of the only examples for which Vasa has been shown to be required for translation.(23) However, we still do not know if Vasa directly binds Gurken transcript. A screen for mRNAs that interact with Vasa proteins will help identify its targets. With the break in this wall by Liu et al., 2009, we may now be able to crack the function of vasa in both the germ line, and multipotent cells.
Abbreviations
- PGC
primordial germ cell
- DEAD-box
single-letter amino acid code (aspartic acid, glutamic acid, alanine, aspartic acid) within the helicase domain of a family of conserved proteins
- MPGZ
mesodermal posterior growth zone
References
- 1.Extavour C. Evolution of the bilaterian germ line: lineage origin and modulation of specification mechanisms. Integrative and Comparative Biology. 2007;47:770–785. doi: 10.1093/icb/icm027. [DOI] [PubMed] [Google Scholar]
- 2.Rosner A, Moiseeva E, Rinkevich Y, Lapidot Z, Rinkevich B. Vasa and the germ line lineage in a colonial urochordate. Dev Biol. 2009;331(2):113–28. doi: 10.1016/j.ydbio.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130(24):5869–84. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 4.Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127(5):891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1(3):REVIEWS1017. doi: 10.1186/gb-2000-1-3-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noce T, Okamoto-Ito S, Tsunekawa N. Vasa homolog genes in mammalian germ cell development. Cell Struct Funct. 2001;26(3):131–6. doi: 10.1247/csf.26.131. [DOI] [PubMed] [Google Scholar]
- 7.Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211(6):299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- 8.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34(15):4168–80. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 11.Linder P, Lasko P. Bent out of shape: RNA unwinding by the DEAD-box helicase Vasa. Cell. 2006;125(2):219–21. doi: 10.1016/j.cell.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109(7):797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 13.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109(6):769–79. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14(7):841–53. [PMC free article] [PubMed] [Google Scholar]
- 15.Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125(9):1569–78. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- 16.Roussell DL, Bennett KL. glh-1, a germ-line putative RNA helicase from Caenorhabditis, has four zinc fingers. Proc Natl Acad Sci U S A. 1993;90(20):9300–4. doi: 10.1073/pnas.90.20.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1996;93(24):13837–42. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans Development. 2000;127(13):2907–16. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- 19.Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, Kuznicki K, Yee C, Bennett K, Strome S. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics. 2008;178(4):1973–87. doi: 10.1534/genetics.107.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braat AK, van de Water S, Korving J, Zivkovic D. A zebrafish vasa morphant abolishes vasa protein but does not affect the establishment of the germline. Genesis. 2001;30(3):183–5. doi: 10.1002/gene.1060. [DOI] [PubMed] [Google Scholar]
- 21.Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113(2):443–8. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- 22.Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335(6191):611–7. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- 23.Tomancak P, Guichet A, Zavorszky P, Ephrussi A. Oocyte polarity depends on regulation of gurken by Vasa. Development. 1998;125(9):1723–32. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- 24.Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120(5):1201–11. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 25.Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5(1):181–7. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- 26.Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131(17):4167–78. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 27.Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- 28.Dahanukar A, Wharton RP. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 1996;10(20):2610–20. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- 29.Liu N, Han H, Lasko P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev. 2009;23(23):2742–52. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foresta C, Ferlin A, Moro E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet. 2000;9(8):1161–9. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- 31.Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev Biol. 2005;277(1):92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Pfister D, De Mulder K, Hartenstein V, Kuales G, Borgonie G, Marx F, Morris J, Ladurner P. Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319(1):146–59. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 33.Mukai H, Watanabe H. Studies on the formation of germ cells in a compound ascidian Botryllus primigenus Oka. J Morphol. 1976;148(3):377–62. doi: 10.1002/jmor.1051480306. [DOI] [PubMed] [Google Scholar]
- 34.Kawamura K, Fujiwara S. Cellular and molecular characterization of transdifferentiation in the process of morphallaxis of budding tunicates. Semin Cell Biol. 1995;6(3):117–26. doi: 10.1006/scel.1995.0017. [DOI] [PubMed] [Google Scholar]
- 35.Sunanaga T, Watanabe A, Kawamura K. Involvement of vasa homolog in germline recruitment from coelomic stem cells in budding tunicates. Dev Genes Evol. 2007;217(1):1–11. doi: 10.1007/s00427-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 36.Fabioux C, Corporeau C, Quillien V, Favrel P, Huvet A. In vivo RNA interference in oyster--vasa silencing inhibits germ cell development. Febs J. 2009;276(9):2566–73. doi: 10.1111/j.1742-4658.2009.06982.x. [DOI] [PubMed] [Google Scholar]
- 37.Miyake A, Saito T, Kashiwagi N, Ando D, Yamamoto A, Suzuki T, Nakatsuji N, Nakatsuji T. Cloning and pattern of expression of the shiro-uo vasa gene during embryogenesis and its roles in PGC development. Int J Dev Biol. 2006;50(7):619–25. doi: 10.1387/ijdb.062172am. [DOI] [PubMed] [Google Scholar]
- 38.Ikenishi K, Tanaka TS. Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev Growth Differ. 1997;39(5):625–33. doi: 10.1046/j.1440-169x.1997.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhurov V, Terzin T, Grbic M. Early blastomere determines embryo proliferation and caste fate in a polyembryonic wasp. Nature. 2004;432(7018):764–9. doi: 10.1038/nature03171. [DOI] [PubMed] [Google Scholar]
- 40.Ozhan-Kizil G, Havemann J, Gerberding M. Germ cells in the crustacean Parhyale hawaiensis depend on Vasa protein for their maintenance but not for their formation. Dev Biol. 2009;327(1):230–9. doi: 10.1016/j.ydbio.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto H, Sudo T, Mikami Y, Otani M, Takano M, Tsuda H, Itamochi H, Katabuchi H, Ito M, Nishimura R. Germ cell specific protein VASA is over-expressed in epithelial ovarian cancer and disrupts DNA damage-induced G2 checkpoint. Gynecol Oncol. 2008;111(2):312–9. doi: 10.1016/j.ygyno.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Lavial F, Acloque H, Bachelard E, Nieto MA, Samarut J, Pain B. Ectopic expression of Cvh (Chicken Vasa homologue) mediates the reprogramming of chicken embryonic stem cells to a germ cell fate. Dev Biol. 2009;330(1):73–82. doi: 10.1016/j.ydbio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Bosch TC, David C. Stem Cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol. 1987;121(1):182–191. [Google Scholar]
- 44.Rebscher N, Volk C, Teo R, Plickert G. The germ plasm component Vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia echinata. Dev Dyn. 2008;237(6):1736–45. doi: 10.1002/dvdy.21562. [DOI] [PubMed] [Google Scholar]
- 45.Baguna J, Salo E, Romero R. Effects of activators and antagonists of the neuropeptides substance P and substance K on cell proliferation in planarians. Int J Dev Biol. 1989;33(2):261–6. [PubMed] [Google Scholar]
- 46.Shukalyuk AI, Golovnina KA, Baiborodin SI, Gunbin KV, Blinov AG, Isaeva VV. vasa-related genes and their expression in stem cells of colonial parasitic rhizocephalan barnacle Polyascus polygenea (Arthropoda: Crustacea: Cirripedia: Rhizocephala) Cell Biol Int. 2007;31(2):97–108. doi: 10.1016/j.cellbi.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Fabioux C, Pouvreau S, Le Roux F, Huvet A. The oyster vasa-like gene: a specific marker of the germline in Crassostrea gigas. Biochem Biophys Res Commun. 2004;315(4):897–904. doi: 10.1016/j.bbrc.2004.01.145. [DOI] [PubMed] [Google Scholar]
- 48.Swartz SZ, Chan XY, Lambert JD. Localization of Vasa mRNA during early cleavage of the snail Ilyanassa. Dev Genes Evol. 2008;218(2):107–13. doi: 10.1007/s00427-008-0203-6. [DOI] [PubMed] [Google Scholar]
- 49.Rebscher N, Zelada-Gonzalez F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306(2):599–611. doi: 10.1016/j.ydbio.2007.03.521. [DOI] [PubMed] [Google Scholar]
- 50.Oyama A, Shimizu T. Transient occurrence of vasa-expressing cells in nongenital segments during embryonic development in the oligochaete annelid Tubifex tubifex. Dev Genes Evol. 2007;217(10):675–90. doi: 10.1007/s00427-007-0180-1. [DOI] [PubMed] [Google Scholar]
- 51.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439(7079):965–8. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 52.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–9. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 53.Sunanaga T, Saito Y, Kawamura K. Postembryonic epigenesis of Vasa-positive germ cells from aggregated hemoblasts in the colonial ascidian, Botryllus primigenus. Dev Growth Differ. 2006;48(2):87–100. doi: 10.1111/j.1440-169X.2006.00849.x. [DOI] [PubMed] [Google Scholar]
- 54.Galliot B, Schmid V. Cnidarians as a model system for understanding evolution and regeneration. Int J Dev Biol. 2002;46(1):39–48. [PubMed] [Google Scholar]
- 55.Newmark PA, Wang Y, Chong T. Germ cell specification and regeneration in planarians. Cold Spring Harb Symp Quant Biol. 2008;73:573–81. doi: 10.1101/sqb.2008.73.022. [DOI] [PubMed] [Google Scholar]
- 56.Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol. 2006;18(3):291–8. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8(4):243–6. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 59.Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, Nemoto N, Song F, Ghosh S, Held WA, Yoshida-Noro C, et al. Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics. 2007;89(3):326–37. doi: 10.1016/j.ygeno.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugimoto K, Koh E, Sin HS, Maeda Y, Narimoto K, Izumi K, Kobori Y, Kitamura E, Nagase H, Yoshida A, et al. Tissue-specific differentially methylated regions of the human VASA gene are potentially associated with maturation arrest phenotype in the testis. J Hum Genet. 2009;54(8):450–6. doi: 10.1038/jhg.2009.59. [DOI] [PubMed] [Google Scholar]
- 61.Park SY, Jameson JL. Minireview: transcriptional regulation of gonadal development and differentiation. Endocrinology. 2005;146(3):1035–42. doi: 10.1210/en.2004-1454. [DOI] [PubMed] [Google Scholar]
- 62.Cardinali M, Gioacchini G, Candiani S, Pestarino M, Yoshizaki G, Carnevali O. Hormonal regulation of vasa-like messenger RNA expression in the ovary of the marine teleost Sparus aurata. Biol Reprod. 2004;70(3):737–43. doi: 10.1095/biolreprod.103.021428. [DOI] [PubMed] [Google Scholar]
- 63.Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314(2):276–86. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerberding M, Browne WE, Patel NH. Cell lineage analysis of the amphipod crustacean Parhyale hawaiensis reveals an early restriction of cell fates. Development. 2002;129(24):5789–801. doi: 10.1242/dev.00155. [DOI] [PubMed] [Google Scholar]
- 65.Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C. An evolutionary conserved region in the vasa 3′UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol. 2002;12(6):454–66. doi: 10.1016/s0960-9822(02)00723-6. [DOI] [PubMed] [Google Scholar]
- 66.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124(16):3157–65. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 67.Pelegri F, Knaut H, Maischein HM, Schulte-Merker S, Nusslein-Volhard C. A mutation in the zebrafish maternal-effect gene nebel affects furrow formation and vasa RNA localization. Curr Biol. 1999;9(24):1431–40. doi: 10.1016/s0960-9822(00)80112-8. [DOI] [PubMed] [Google Scholar]
- 68.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev Genet. 2003;4(8):626–37. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 69.Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101(5):251–62. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 70.Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, Semple CA, Gray NK, Cooke HJ. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet. 2005;14(24):3899–909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- 71.Breitwieser W, Markussen FH, Horstmann H, Ephrussi A. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10(17):2179–88. doi: 10.1101/gad.10.17.2179. [DOI] [PubMed] [Google Scholar]
- 72.Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109(2):425–33. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- 73.Lasko PF, Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990;4(6):905–21. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- 74.Liu N, Dansereau DA, Lasko P. Fat facets interacts with vasa in the Drosophila pole plasm and protects it from degradation. Curr Biol. 2003;13(21):1905–9. doi: 10.1016/j.cub.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 75.Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3(6):865–76. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- 76.Kugler JM, Woo JS, Oh BH, Lasko P. Regulation of Drosophila Vasa in vivo through paralogous Cullin-RING E3 ligase specificity receptors. Mol Cell Biol. doi: 10.1128/MCB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woo JS, Imm JH, Min CK, Kim KJ, Cha SS, Oh BH. Structural and functional insights into the B30.2/SPRY domain. Embo J. 2006;25(6):1353–63. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo JS, Suh HY, Park SY, Oh BH. Structural basis for protein recognition by B30.2/SPRY domains. Mol Cell. 2006;24(6):967–76. doi: 10.1016/j.molcel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 79.Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300(1):406–15. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 80.Orsborn AM, Li W, McEwen TJ, Mizuno T, Kuzmin E, Matsumoto K, Bennett KL. GLH-1, the C. elegans P granule protein, is controlled by the JNK KGB-1 and by the COP9 subunit CSN-5. Development. 2007;134(18):3383–92. doi: 10.1242/dev.005181. [DOI] [PubMed] [Google Scholar]
- 81.Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy EM. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229–80. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 83.Parvinen M. The chromatoid body in spermatogenesis. Int J Androl. 2005;28(4):189–201. doi: 10.1111/j.1365-2605.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- 84.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306(1–2):17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 85.Mahowald AP. Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool. 1968;167(2):237–61. doi: 10.1002/jez.1401670211. [DOI] [PubMed] [Google Scholar]
- 86.Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 2000;219(2):315–33. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- 87.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130(5):859–71. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 88.Pepling ME. From primordial germ cell to primordial follicle: mammalian female germ cell development. Genesis. 2006;44(12):622–32. doi: 10.1002/dvg.20258. [DOI] [PubMed] [Google Scholar]
- 89.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A. 2006;103(8):2647–52. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shirae-Kurabayashi M, Nishikata T, Takamura K, Tanaka KJ, Nakamoto C, Nakamura A. Dynamic redistribution of vasa homolog and exclusion of somatic cell determinants during germ cell specification in Ciona intestinalis. Development. 2006;133(14):2683–93. doi: 10.1242/dev.02446. [DOI] [PubMed] [Google Scholar]
- 91.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6(2):127–38. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 92.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26(5):603–9. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 93.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16(19):1884–94. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 94.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–49. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 95.Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 2006;15(15):2324–34. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- 96.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15(2):285–97. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35(6):868–80. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oulhen N, Boulben S, Bidinosti M, Morales J, Cormier P, Cosson B. A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system. PLoS One. 2009;4(3):e5070. doi: 10.1371/journal.pone.0005070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32(2):63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 100.Rajavashisth TB, Taylor AK, Andalibi A, Svenson KL, Lusis AJ. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989;245(4918):640–3. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- 101.Espinosa JM, Portal D, Lobo GS, Pereira CA, Alonso GD, Gomez EB, Lan GH, Pomar RV, Flawia MM, Torres HN. Trypanosoma cruzi poly-zinc finger protein: a novel DNA/RNA-binding CCHC-zinc finger protein. Mol Biochem Parasitol. 2003;131(1):35–44. doi: 10.1016/s0166-6851(03)00187-7. [DOI] [PubMed] [Google Scholar]
- 102.Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232(4749):485–7. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 103.Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271(5252):1081–5. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 104.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254(4):523–37. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 105.Tisne C, Roques BP, Dardel F. Specific recognition of primer tRNA Lys 3 by HIV-1 nucleocapsid protein: involvement of the zinc fingers and the N-terminal basic extension. Biochimie. 2003;85(5):557–61. doi: 10.1016/s0300-9084(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 106.Ramboarina S, Druillennec S, Morellet N, Bouaziz S, Roques BP. Target specificity of human immunodeficiency virus type 1 NCp7 requires an intact conformation of its CCHC N-terminal zinc finger. J Virol. 2004;78(12):6682–7. doi: 10.1128/JVI.78.12.6682-6687.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.South TL, Kim B, Hare DR, Summers MF. Zinc fingers and molecular recognition. Structure and nucleic acid binding studies of an HIV zinc finger-like domain. Biochem Pharmacol. 1990;40(1):123–9. doi: 10.1016/0006-2952(90)90187-p. [DOI] [PubMed] [Google Scholar]
- 108.Karpel RL, Henderson LE, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262(11):4961–7. [PubMed] [Google Scholar]
- 109.Green LM, Berg JM. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: spectroscopic studies and a proposed three-dimensional structure. Proc Natl Acad Sci U S A. 1989;86(11):4047–51. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4(1):16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 111.Fischer A. Stages and stage distribution in early oogenesis in the Annelid, Platynereis dumerilii. Cell Tissue Res. 1974;156(1):35–45. doi: 10.1007/BF00220100. [DOI] [PubMed] [Google Scholar]
- 112.Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci U S A. 2000;97(17):9585–90. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Albamonte MS, Willis MA, Albamonte MI, Jensen F, Espinosa MB, Vitullo AD. The developing human ovary: immunohistochemical analysis of germ-cell-specific VASA protein, BCL-2/BAX expression balance and apoptosis. Hum Reprod. 2008;23(8):1895–901. doi: 10.1093/humrep/den197. [DOI] [PubMed] [Google Scholar]
- 114.Anderson RA, Fulton N, Cowan G, Coutts S, Saunders PT. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol. 2007;7:136. doi: 10.1186/1471-213X-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93(1–2):139–49. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 116.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127(12):2741–50. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 117.Shinomiya A, Tanaka M, Kobayashi T, Nagahama Y, Hamaguchi S. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev Growth Differ. 2000;42(4):317–26. doi: 10.1046/j.1440-169x.2000.00521.x. [DOI] [PubMed] [Google Scholar]
- 118.Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149(4):875–88. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Komiya T, Itoh K, Ikenishi K, Furusawa M. Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994;162(2):354–63. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- 120.Carre D, Djediat C, Sardet C. Formation of a large Vasa-positive germ granule and its inheritance by germ cells in the enigmatic Chaetognaths. Development. 2002;129(3):661–70. doi: 10.1242/dev.129.3.661. [DOI] [PubMed] [Google Scholar]
- 121.Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55(4):577–87. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- 122.Nakao H. Isolation and characterization of a Bombyx vasa-like gene. Dev Genes Evol. 1999;209(5):312–6. doi: 10.1007/s004270050257. [DOI] [PubMed] [Google Scholar]