Abstract
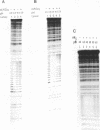
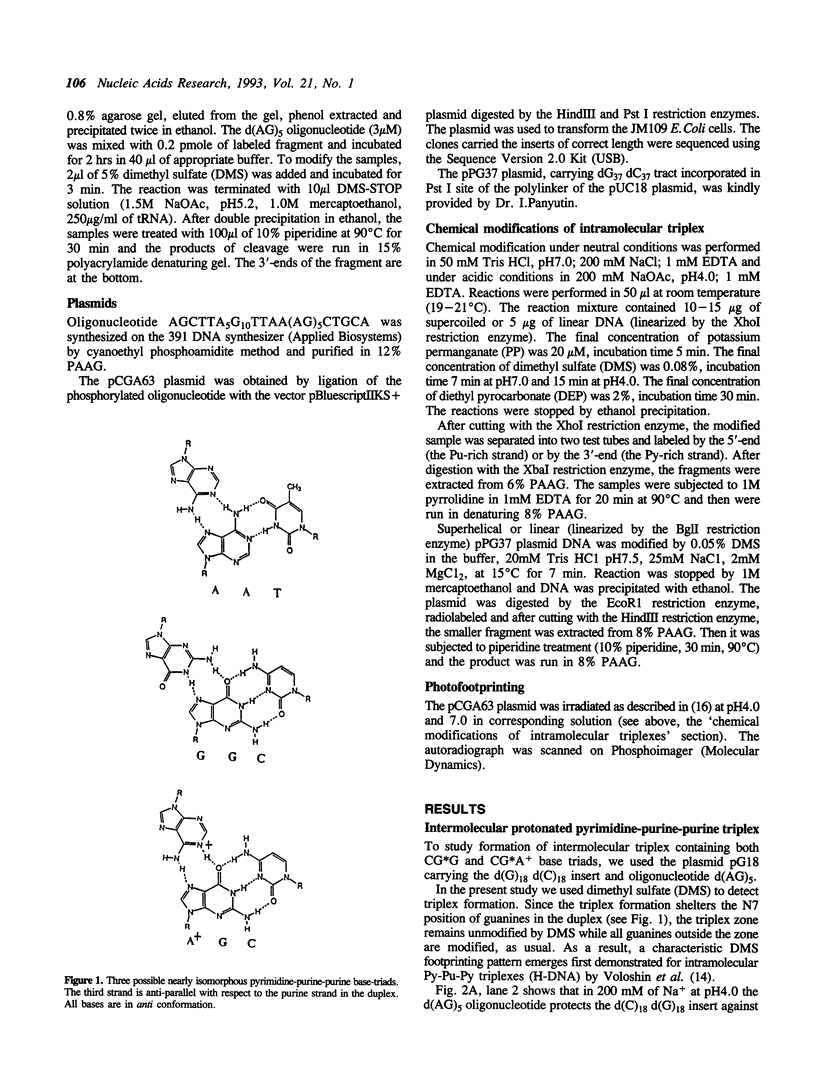
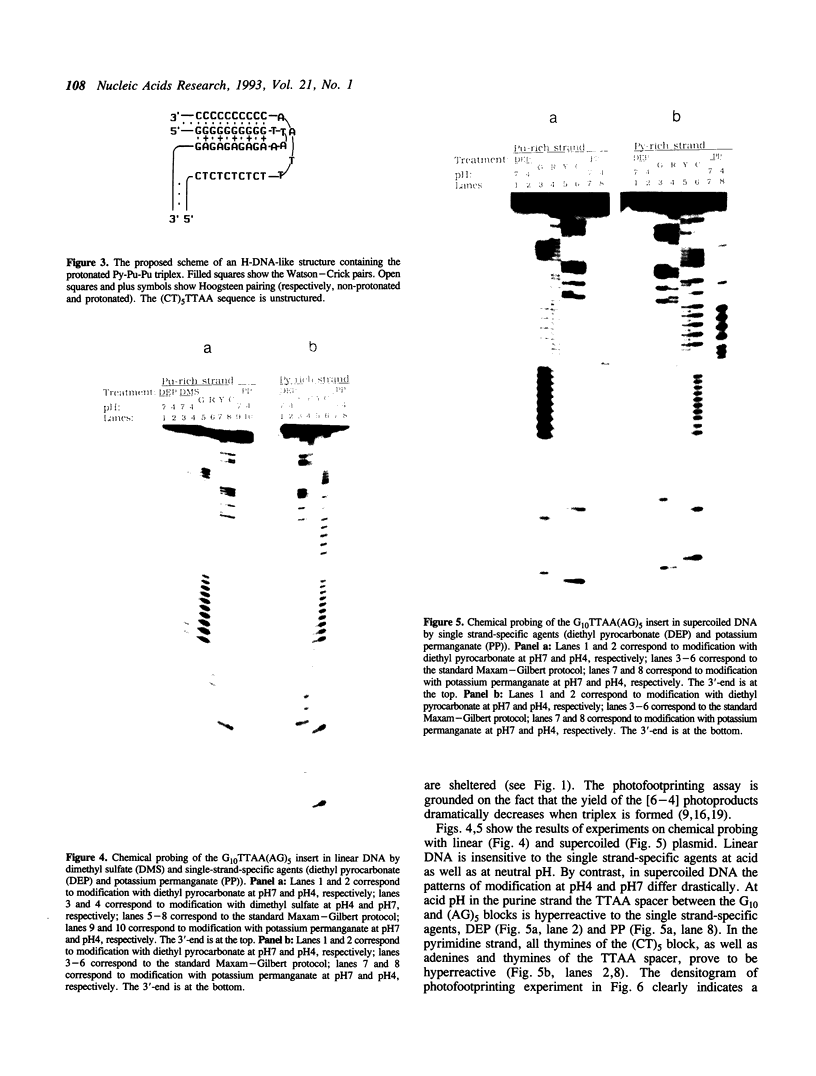
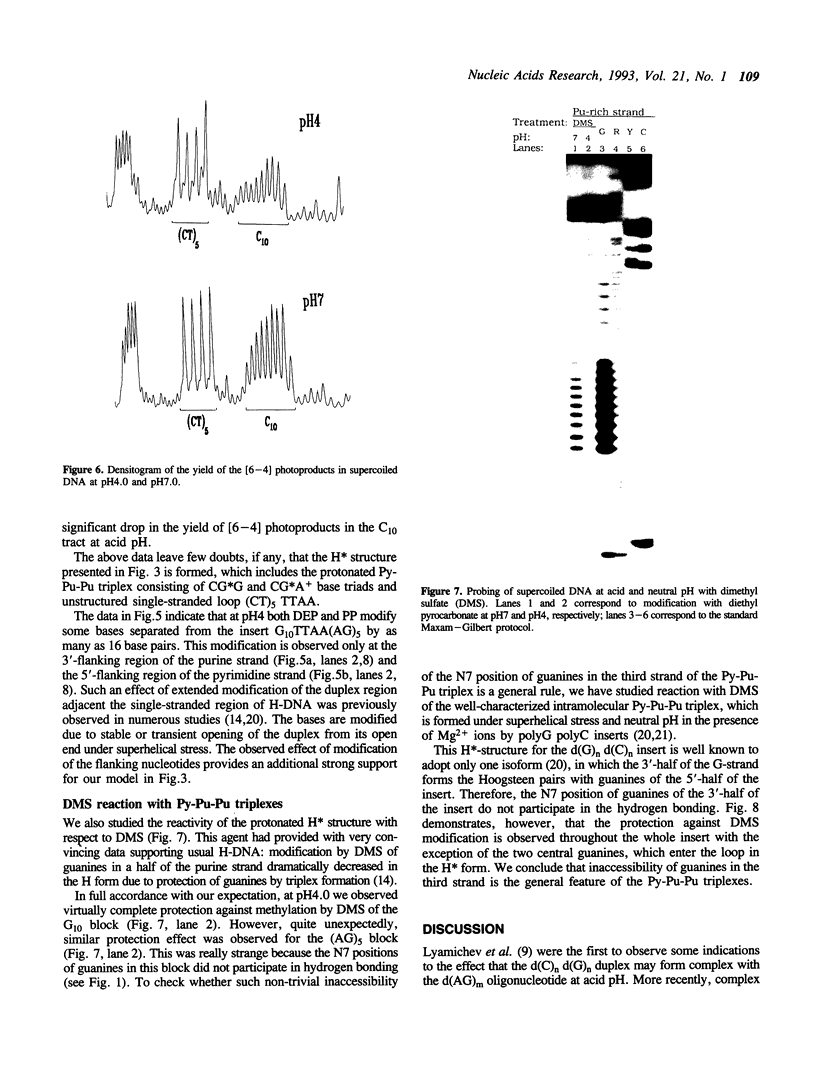
We have studied a protonated pyrimidine-purine-purine (Py-Pu-Pu) triplex, which is formed between the d(C)nd(G)n duplex and the d(AG)m oligonucleotide as the third strand and carries the CG*A+ protonated base-triads. We have observed such an intermolecular complex between a plasmid carrying the d(C)18 d(G)18 insert and the d(AG)5 oligonucleotide without bivalent cations in 200 mM of Na+ at pH4.0. Bivalent cations additionally stabilize the complex. We propose the structures for nearly isomorphous base-triads TA*A, CG*G and CG*A+. To identify the H-DNA-like structure, which includes the triplex between d(C)n d(G)n duplex and the AG-strand, we have cloned in a superhelical plasmid the insert: G10TTAA(AG)5. The data on photofootprinting and chemical modification with diethyl pyrocarbonate, potassium permanganate and dimethyl sulfate demonstrate that the H-like structure with triplex carrying CG*G and CG*A+ base triads is actually formed under acid conditions. In the course of this study we have come across unexpected results on probing of Py-Pu-Pu triplexes by dimethyl sulfate (DMS): the protection effect is observed not only for guanines entering the duplex but also for guanines in the third strand lying in the major groove. We have demonstrated this effect not only for the case the novel protonated Py-Pu-Pu triplex but also for the traditional non-protonated Py-Pu-Pu intramolecular triplex (H*-DNA) formed by the d(C)37 d(G)37 insert in supercoiled plasmid in the presence of Mg2+ ions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. The influence of single base triplet changes on the stability of a pur.pur.pyr triple helix determined by affinity cleaving. Nucleic Acids Res. 1992 Jun 11;20(11):2773–2776. doi: 10.1093/nar/20.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii B. P., Krasilnikova M. M., Veselkov A. G., Frank-Kamenetskii M. D. Kinetic trapping of H-DNA by oligonucleotide binding. Nucleic Acids Res. 1992 Apr 25;20(8):1903–1908. doi: 10.1093/nar/20.8.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii B. P., Veselkov A. G., Filippov S. A., Dobrynin V. N., Mirkin S. M., Frank-Kamenetskii M. D. Formation of intramolecular triplex in homopurine-homopyrimidine mirror repeats with point substitutions. Nucleic Acids Res. 1990 Nov 25;18(22):6621–6624. doi: 10.1093/nar/18.22.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernués J., Beltrán R., Casasnovas J. M., Azorín F. DNA-sequence and metal-ion specificity of the formation of *H-DNA. Nucleic Acids Res. 1990 Jul 25;18(14):4067–4073. doi: 10.1093/nar/18.14.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernués J., Beltrán R., Casasnovas J. M., Azorín F. Structural polymorphism of homopurine--homopyrimidine sequences: the secondary DNA structure adopted by a d(GA.CT)22 sequence in the presence of zinc ions. EMBO J. 1989 Jul;8(7):2087–2094. doi: 10.1002/j.1460-2075.1989.tb03617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain M., Tinoco I., Jr Poly(rA) binds poly(rG).poly(rC) to form a triple helix. Nucleic Acids Res. 1992 Jan 25;20(2):315–318. doi: 10.1093/nar/20.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D., Malkov V. A., Voloshin O. N., Soyfer V. N. Stabilization of PyPuPu triplexes with bivalent cations. Nucleic Acids Symp Ser. 1991;(24):159–162. [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D. Protonated DNA structures. Methods Enzymol. 1992;211:180–191. doi: 10.1016/0076-6879(92)11011-7. [DOI] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Hélène C. Sequence-specific recognition of the major groove of DNA by oligodeoxynucleotides via triple helix formation. Footprinting studies. Nucleic Acids Res. 1988 Dec 23;16(24):11431–11440. doi: 10.1093/nar/16.24.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Ito T., Smith C. L., Cantor C. R. Sequence-specific DNA purification by triplex affinity capture. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):495–498. doi: 10.1073/pnas.89.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y. Cationic metal-specific structures adopted by the poly(dG) region and the direct repeats in the chicken adult beta A globin gene promoter. Nucleic Acids Res. 1989 Jun 26;17(12):4493–4502. doi: 10.1093/nar/17.12.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Frank-Kamenetskii M. D., Soyfer V. N. Protection against UV-induced pyrimidine dimerization in DNA by triplex formation. Nature. 1990 Apr 5;344(6266):568–570. doi: 10.1038/344568a0. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1985 Oct;3(2):327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D., Cantor C. R. A stable complex between homopyrimidine oligomers and the homologous regions of duplex DNAs. Nucleic Acids Res. 1988 Mar 25;16(5):2165–2178. doi: 10.1093/nar/16.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structure of (dG)n.(dC)n under superhelical stress and acid pH. J Biomol Struct Dyn. 1987 Oct;5(2):275–282. doi: 10.1080/07391102.1987.10506393. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Voloshin O. N., Frank-Kamenetskii M. D., Soyfer V. N. Photofootprinting of DNA triplexes. Nucleic Acids Res. 1991 Apr 11;19(7):1633–1638. doi: 10.1093/nar/19.7.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaya R. F., Gilbert D. E., Malek S., Sinsheimer J. S., Feigon J. Structure and stability of X.G.C mismatches in the third strand of intramolecular triplexes. Science. 1991 Oct 11;254(5029):270–274. doi: 10.1126/science.254.5029.270. [DOI] [PubMed] [Google Scholar]
- Malkov V. A., Soyfer V. N., Frank-Kamenetskii M. D. Effect of intermolecular triplex formation on the yield of cyclobutane photodimers in DNA. Nucleic Acids Res. 1992 Sep 25;20(18):4889–4895. doi: 10.1093/nar/20.18.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Roberts R. W., Crothers D. M. Specificity and stringency in DNA triplex formation. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9397–9401. doi: 10.1073/pnas.88.21.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A., Dervan P. B. Single-site enzymatic cleavage of yeast genomic DNA mediated by triple helix formation. Nature. 1991 Mar 14;350(6314):172–174. doi: 10.1038/350172a0. [DOI] [PubMed] [Google Scholar]
- Strobel S. A., Dervan P. B. Site-specific cleavage of a yeast chromosome by oligonucleotide-directed triple-helix formation. Science. 1990 Jul 6;249(4964):73–75. doi: 10.1126/science.2195655. [DOI] [PubMed] [Google Scholar]
- Voloshin O. N., Mirkin S. M., Lyamichev V. I., Belotserkovskii B. P., Frank-Kamenetskii M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature. 1988 Jun 2;333(6172):475–476. doi: 10.1038/333475a0. [DOI] [PubMed] [Google Scholar]
- Young S. L., Krawczyk S. H., Matteucci M. D., Toole J. J. Triple helix formation inhibits transcription elongation in vitro. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10023–10026. doi: 10.1073/pnas.88.22.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]