Abstract
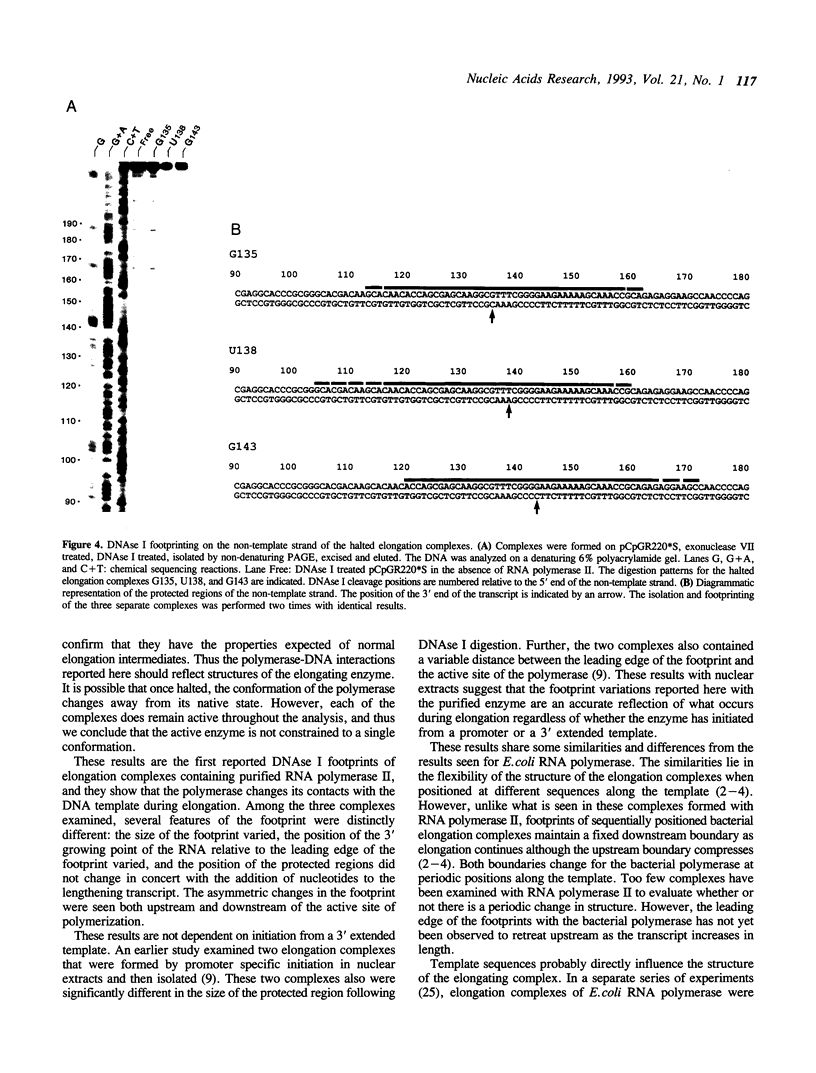
Elongation complexes of RNA polymerase II, RNA-DNA-enzyme ternary complexes, are intermediates in the synthesis of all eukaryotic mRNAs and are potential regulatory targets for factors controlling RNA chain elongation and termination. Analysis of such complexes can provide information concerning the structure of the catalytic core of the RNA polymerase and its interactions with the DNA template and RNA transcript. Knowledge of the structure of such complexes is essential in understanding the catalytic and regulatory properties of RNA polymerase. We have prepared and isolated complexes of purified RNA polymerase II halted at defined positions along a DNA template, and we have used deoxyribonuclease I (DNAse I) to map the interactions of the polymerase with the DNA template. DNAse I footprints of three specific ternary complexes reveal that the enzyme-template interactions of individual elongation complexes are not identical. The size of the protected region is distinct for each complex and varies from 48 to 55 bp between different complexes. Additionally, the positioning of the protected region relative to the active site varies in different complexes. Our results suggest that RNA polymerase II is a dynamic molecule and undergoes continual conformational transitions during elongation. These transitions are likely to be important in the processes of transcript elongation and termination and their regulation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buratowski S., Hahn S., Guarente L., Sharp P. A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989 Feb 24;56(4):549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Cai H., Luse D. S. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation, and elongation complexes. J Biol Chem. 1987 Jan 5;262(1):298–304. [PubMed] [Google Scholar]
- Cai H., Luse D. S. Variations in template protection by the RNA polymerase II transcription complex during the initiation process. Mol Cell Biol. 1987 Oct;7(10):3371–3379. doi: 10.1128/mcb.7.10.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. W., Lynch E. C., Leason K. R., Court D. L., Shapiro B. A., Friedman D. I. Functional importance of sequence in the stem-loop of a transcription terminator. Science. 1991 Nov 22;254(5035):1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- Dedrick R. L., Chamberlin M. J. Studies on transcription of 3'-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry. 1985 Apr 23;24(9):2245–2253. doi: 10.1021/bi00330a019. [DOI] [PubMed] [Google Scholar]
- Hodo H. G., 3rd, Blatti S. P. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1977 May 31;16(11):2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3'----5' direction in the presence of elongation factor SII. Genes Dev. 1992 Jul;6(7):1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Analysis of the signals for transcription termination by purified RNA polymerase II. Biochemistry. 1990 Jan 9;29(1):269–278. doi: 10.1021/bi00453a037. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry. 1989 Sep 19;28(19):7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992 May 20;225(2):239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992 May 20;225(2):221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- Linn S. C., Luse D. S. RNA polymerase II elongation complexes paused after the synthesis of 15- or 35-base transcripts have different structures. Mol Cell Biol. 1991 Mar;11(3):1508–1522. doi: 10.1128/mcb.11.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger W., Schickor P., Heumann H. A cinematographic view of Escherichia coli RNA polymerase translocation. EMBO J. 1989 Sep;8(9):2745–2754. doi: 10.1002/j.1460-2075.1989.tb08416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natori S. Stimulatory proteins of RNA polymerase II from Ehrlich ascites tumor cells. Mol Cell Biochem. 1982 Aug 6;46(3):173–187. doi: 10.1007/BF00239666. [DOI] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992 Feb 25;267(6):3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Reines D., Ghanouni P., Li Q. Q., Mote J., Jr The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992 Aug 5;267(22):15516–15522. [PMC free article] [PubMed] [Google Scholar]
- Reynolds R., Chamberlin M. J. Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J Mol Biol. 1992 Mar 5;224(1):53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- Rice G. A., Kane C. M., Chamberlin M. J. Footprinting analysis of mammalian RNA polymerase II along its transcript: an alternative view of transcription elongation. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4245–4249. doi: 10.1073/pnas.88.10.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Huet J., Fromageot P. Similar binding site for P37 factor on yeast RNA polymerases A and B. Biochem Biophys Res Commun. 1980 Sep 16;96(1):258–264. doi: 10.1016/0006-291x(80)91208-5. [DOI] [PubMed] [Google Scholar]
- Sluder A. E., Greenleaf A. L., Price D. H. Properties of a Drosophila RNA polymerase II elongation factor. J Biol Chem. 1989 May 25;264(15):8963–8969. [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G., Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988 Sep 9;241(4871):1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]