Abstract
Although sessile, plants are able to grow toward or away from an environmental stimulus. Important examples are stem or leaf orientation of higher plants in response to the direction of the incident light. The responsible photoreceptors belong to the phototropin photoreceptor family. Although the mode of phototropin action is quite well understood, much less is known of how the light signal is transformed into a bending response. Several lines of evidence indicate that a lateral auxin gradient is responsible for asymmetric cell elongation along the light gradient within the stem. However, some of the molecular key players leading to this asymmetric auxin distribution are, as yet, unidentified. Previously, it was shown that phototropin gets autophosphorylated upon illumination and binds to a scaffold protein termed NPH3 (for nonphototropic hypocotyl 3). Using a yeast three-hybrid approach with phototropin and NPH3 as a bait complex, we isolated a protein, termed EHB1 (for enhanced bending 1), with a so far unknown function, which binds to this binary complex. This novel interacting factor negatively affects hypocotyl bending under blue light conditions in Arabidopsis (Arabidopsis thaliana) and thus seems to be an important component regulating phototropism. Interestingly, it could be shown that the gravitropic response was also affected. Thus, it cannot be ruled out that this protein might also have a more general role in auxin-mediated bending toward an environmental stimulus.
Phototropic responses of higher plants are mainly mediated by blue light perceived by photoreceptors of the phototropin family (Briggs and Christie, 2002; Christie, 2007; Holland et al., 2009). Two members of this family have been characterized in Arabidopsis (Arabidopsis thaliana), phot1 and phot2. Both are responsible for phototropism in response to high light (Sakai et al., 2001), while the phototropic response to low blue light fluence rates is exclusively mediated by phot1. It is generally accepted that upon lateral blue light exposure, the shaded side of the stem responds with enhanced longitudinal cell elongation (Iino, 2001). This effect is most likely caused by reorganization of the lateral auxin gradient within the stem to generate an increased capacity of cell elongation on the shaded side (Liscum and Stowe-Evans, 2000). Bending is dependent on the fluence rate and mediated either by phot1 alone or, at higher fluence rates, by both phototropin photoreceptors (Christie et al., 1998). However, many of the components that establish an asymmetric auxin gradient are still unknown. Although several proteins have been identified to interact with phototropin (Sullivan et al., 2009), up to now only two proteins directly interacting with phototropin, namely NONPHOTOTROPIC HYPOCOTYL3 (NPH3; Motchoulski and Liscum, 1999) and ROOT PHOTOTROPISM2 (RPT2), have been intensively characterized. They were reported to be key players involved in further downstream signaling (Sakai et al., 2000). Both proteins were not considered as direct components of the phototropin-mediated blue light signal transduction pathway but rather act as scaffolding elements, allowing other proteins to bind to this complex. Loss-of-function mutants of NPH3 are unable to respond to lateral blue light, although blue light-induced stomatal opening and chloroplast relocation are not affected. RPT2 seems to be required for both phototropin-mediated hypocotyl bending as well as stomatal opening (Inada et al., 2004). However, it is still unclear which component is responsible for the temporary attachment of the phot1/NPH3 complex to the plasma membrane (Sakamoto and Briggs, 2002; Kaiserli et al., 2009). One potential candidate is PHYTOCHROME KINASE SUBSTRATE1 (PKS1), a plasma membrane-associated protein that not only binds to NPH3 but also links blue light-induced phototropism to phytochrome action (Fankhauser et al., 1999; Lariguet et al., 2006; Boccalandro et al., 2008; Molas and Kiss, 2008; Schepens et al., 2008). Further elements of the downstream signaling network were MULTIDRUG RESISTANCE1 (MDR1; Noh et al., 2003), NPH4 and MASSUGU2 (Stowe-Evans et al., 1998; Harper et al., 2000; Tatematsu et al., 2004), and, as recently published, a subunit of PROTEIN PHOSPHATASE2A (Tseng and Briggs, 2010).
We performed a yeast three-hybrid screen with phot1 and NPH3 as a combined bait complex to identify further interacting proteins. With this approach we identified a protein with a C2/CalB calcium-binding domain and an as yet unknown function. Furthermore, we demonstrated that this protein interacts directly with NPH3 in vivo and in vitro. A loss-of-function mutant in Arabidopsis revealed a phenotype exhibiting stronger bending to lateral blue light compared with the wild type. Thus, we have identified a negative-acting element in phototropin signal transduction directly interacting with NPH3, which seemed to be also involved in gravitropic responses of both root and hypocotyl. We named the mutant enhanced bending1 (ehb1) and the corresponding gene EHB1.
RESULTS
Identification of a Novel Protein Binding to NPH3
To identify yet unknown interaction partners of NPH3, we established a yeast three-hybrid approach by coexpressing the GAL4 BD-NPH3 fusion protein together with phot1 as a bait in yeast cells. These cells were cotransformed with a cDNA library obtained from mRNA of Arabidopsis seedlings grown for 6 d in the dark. Transformed cells were grown on quadruple dropout plates either exposed to blue light or kept in darkness for 5 d. Colonies from both plates were collected randomly and analyzed for their cDNA insertion. Only colonies expressing proteins exclusively found on blue light-illuminated plates were further investigated. At first, no known protein involved in blue light signaling could be addressed. However, the most abundant gene we could identify after analyzing approximately 80 yeast colonies encoded a small protein of 25 kD with a yet unknown function. This gene is located on Arabidopsis chromosome 1 between bp 26,702,705 and 26,703,790 (The Arabidopsis Information Resource accession no. At1g70800). Deduced amino acid sequences are presented in Figure 1B. Sequence alignment and database analysis (Finn et al., 2008) of the deduced amino acid sequence revealed that the protein (Fig. 1A) comprises an N-terminal C2/CalB domain (Rizo and Südhof, 1998; Ubach et al., 1998) between amino acids 17 and 88. This is a common Ca2+-binding motif of a vast amount of proteins present in almost all organisms (Nalefski and Falke, 1996; Ponting and Parker, 1996; Nalefski et al., 1997). The N-terminal region consists of a short stretch of several acidic and hydrophobic amino acids, while the C terminus between amino acids 89 and 174 revealed a motif with strong homology to an Arabidopsis protein termed ZINC AND CALCIUM BINDING (ZAC; Jensen et al., 2000). ZAC is a member of the ADP-ribosylation factor GTPase-activating proteins (ARF-GAP; Donaldson and Honda, 2005). Intriguingly, sequence alignment between these two proteins revealed that EHB1 is essentially lacking N terminally the ARF-GAP characteristic zinc-finger motif. This motif is necessary for GTPase activity (Cukierman et al., 1995). Further analysis of the flanking regions revealed a second gene, At1g70810, in close vicinity to At1g70800 and arranged in a tandem array. Alignment of both proteins revealed a similarity of 83% at the amino acid level. The second gene, At1g70810, also encompasses in its sequence a full C2/CalB but lacks the very N terminus of nine amino acids (Fig. 1B). Further data bank analysis revealed a third homolog gene located on chromosome 1 (accession no. At1g23140), indicating that EHB1 is a member of a small gene family.
Figure 1.
Deduced amino acid sequence of At1g70800. Pfam analysis revealed a C2/CalB calcium-binding domain between amino acids 17 and 88. A, Schematic domain structure of the At1g70800 protein. B, Sequence alignment of the deduced amino acid sequences of At1g70800 and At1g70810. Both proteins reveal strong sequence similarity and are arranged as tandem arrays in close proximity to each other (for genome arrangement, see Supplemental Fig. S1).
EHB1 Interacts with NPH3
Further experiments using a yeast two-hybrid approach could show that NPH3 can interact with EHB1 autonomously even without the presence of phot1. Thus, for further interaction analysis, a standard yeast two-hybrid approach with either NPH3 or EHB1 and its neighboring gene At1g70810 as bait was used. In a quantitative analysis, we tested protein-protein interaction by determining β-galactosidase activity (Fig. 2). In this assay, we also analyzed the capability of the protein encoded by the neighboring gene At1g70810 to interact with NPH3. Although At1g70810 interacts with NPH3, this response was much weaker than observed for NPH3 and EHB1. Galactosidase activity for the EHB1/NPH3 couple was 3.4 times higher than for phot1/NPH3 and five times higher than for At1g70810/NPH3. Galactosidase activity of yeast cells expressing either At1g70810 or NPH3 alone was about half as high as observed for phot1/NPH3. Continuative studies were performed to analyze which part of the NPH3 protein binds to EHB1. phot1 was reported to bind to the C terminus of NPH3 containing a coiled-coil domain (Motchoulski and Liscum, 1999). Yeast two-hybrid studies with truncated versions of NPH3 and full-length EHB1 revealed that EHB1 binds preferentially to the N terminus of NPH3, comprising the BTB/POZ domain (Fig. 3). The region between the BTB and coiled-coil domains as well as the coiled-coil domain itself failed to interact with EHB1. Yeast interaction between NPH3 and EHB1 was confirmed by in vitro coimmunoprecipitation assay in the presence or absence of phot1. All proteins were in vitro transcribed/translated and labeled with [35S]Met. NPH3 as well as phot1 were provided with an N-terminal myc tag. Putative interaction in vitro was analyzed by incubation of NPH3 with phot1, EHB1, or both proteins together, followed by immunoprecipitation with anti-myc antibody and protein A-Sepharose. In line with the results obtained in yeast, interaction between EHB1 and NPH3 could be observed, regardless of whether phot1 was present or not (Fig. 4). However, interaction of EHB1 with NPH3 was slightly weaker in the absence of phot1. To test if phot1 alone was able to bind to EHB1, we performed a coimmunoprecipitation assay using phot1-myc in the presence of EHB1. Thus, we could show that phot1 alone was not able to precipitate EHB1 (Fig. 4).
Figure 2.
EHB1 and NPH3 interaction in yeast. Yeast cells expressing the indicated proteins were analyzed for β-galactosidase (β-gal) activity. For comparison and as a positive control, the interaction of NPH3 and phot1 was included. β-Galactosidase activity was more than three times higher in yeast expressing NPH3 with EHB1 instead of phot1. Mean values and sd of eight different experiments are shown.
Figure 3.
Interaction analysis of EHB1 with different NPH3 deletion constructs. NPH3 was truncated as indicated and provided as bait in a yeast two-hybrid system. EHB1 or phot1 served as prey. Transformed yeast cells were grown in liquid double dropout medium, streaked out on agar plates, and analyzed for growth on triple dropout (TDO) and quadruple dropout (QDO) media. NPH3 and its truncated derivatives alone as well as EHB1 alone enabled considerable growth on TDO medium. More severe selection on QDO revealed growth only, if EHB1 was expressed together with the N-terminal part of NPH3 coding for the BTB/POZ domain. –, No growth; +, single colonies visible; ++, moderate growth, single colonies distinguishable; +++, confluent growth.
Figure 4.
Coimmunoprecipitation analysis of myc tag:NPH3 with phot1 and EHB1. Coimmunoprecipitation analysis was performed using 35S-labeled myc:NPH3, myc:phot1, phot1, and EHB1 using anti-myc antibodies and protein A-Sepharose for precipitation. In lanes 1 to 3, pull down of NPH3 is shown, either in the presence of phot1 (lane 2) and EHB1 (lane 3) alone or in the presence of phot1 together with EHB1 (lane 1). As expected, NPH3 binds to phot1 (lane 2). Additionally, NPH3 is able to bind EHB1, regardless if phot1 is present (lane 1) or not (lane 3), while phot1 alone is not able to bind to EHB1 in vitro (lane 4). A myc tag alone cannot pull EHB1 (lane 5).
Blue Light Enhances Hypocotyl Bending in ehb1-2 Mutant Plants
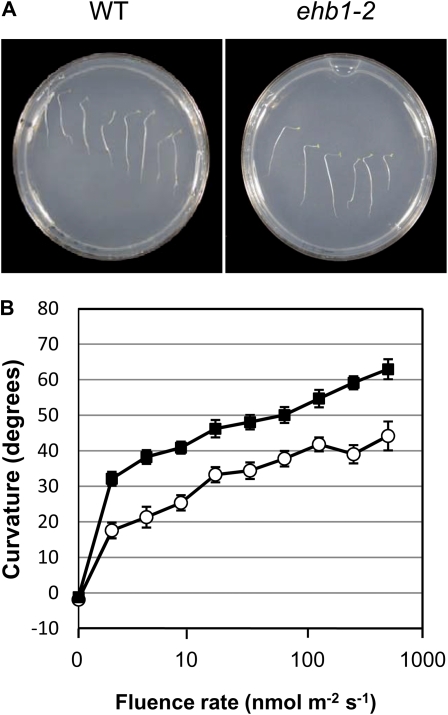
A loss-of-function mutant line (ehb1-2) for At1g70800 was obtained from Syngenta (Syngenta Arabidopsis Insertion Library) and provided by the Arabidopsis Biological Resource Center via The Arabidopsis Information Resource (SAIL 385_C07). The predicted insertion of the T-DNA in exon 3 was verified by PCR analysis with gene- and T-DNA-specific primer combinations and subsequent sequence analysis (Supplemental Fig. S1). The absence of the corresponding transcript was monitored by reverse transcription-PCR using total RNA from dark-grown wild-type and mutant seedlings (Supplemental Fig. S2). Initial experiments using 1 μmol m−2 s−1 of lateral blue light applied for 24 h resulted in an unexpected phenotype. Instead of a reduced or absent response to blue light, mutant seedlings showed increased sensitivity to blue light (Fig. 5). This prompted us to characterize the phenotype of this mutant in more detail.
Figure 5.
Fluence rate response of hypocotyl curvature. A, Representative image of 3-d-old Arabidopsis wild-type (WT) seedlings compared with ehb1-2 mutant seedlings illuminated with 2 μmol m−2 s−1 lateral blue light from the right for 20 h. B, Threshold analysis of wild-type (white circles) and ehb1-2 mutant (black squares) seedlings. Fluence rate threshold was analyzed as described in “Materials and Methods.” For each light intensity tested, bending angles of a minimum of 50 seedlings were determined and se was calculated. Between 0.01 and 0.1 μmol m−2 s−1, a 10 times higher fluence rate has to be applied to obtain a similar bending angle (40°) between ehb1-2 and wild-type seedlings. [See online article for color version of this figure.]
First, we analyzed phototropic bending at different fluence rates in ehb1-1 and a second insertion line, ehb1-2 (Fig. 5B; Supplemental Fig. S3). By doing so, we could clearly show that in both insertion lines, the bending threshold of the mutant was shifted toward lower light intensities, although in line 1-2 the effect was more pronounced. Threshold fluence response, therefore, was further analyzed in the ehb1-2 mutant line (Figs. 5 and 6). While in wild-type seedlings, a fluence rate of 0.1 μmol m−2 s−1 resulted in a bending angle of 40°, in mutant plants, the same bending angle could be achieved with fluence rates 1 order of magnitude less (0.01 μmol m−2 s−1). Although for the sake of clarity in Figure 5B, se values instead of sd were given, statistical analysis confirmed a significant difference in the average bending angles between wild-type and mutant plants at similar fluence rates (P values between 1.44E-04 and 1.41E-09). Additionally, a significant difference between both curves was also validated by ANOVA. Second, the kinetics of the bending response was analyzed by exposing seedlings to different doses of blue light at a constant fluence rate of 0.5 μmol m−2 s−1. Again, the ehb1-2 mutant revealed a much more pronounced bending response than wild-type seedlings. As shown in Figure 6, between 10 and 30 mmol, differences of more than 20° could be obtained, indicating a much quicker response to blue light for the mutant compared with the wild type. After an exposure time of 26 h (50 mmol m−2), wild-type seedlings finally achieved an average bending angle of approximately 90°, while mutant seedlings reached this bending angle approximately 9 h earlier.
Figure 6.
Fluence response relation of wild-type and ehb1-2 seedlings. Six-day-old dark-grown wild-type and ehb1-2 mutant seedlings were exposed for various time periods to unilateral blue light with a fluence rate of 0.5 μmol m−2 s−1. Three independent experiments with 15 seedlings each were analyzed. ehb1-2 mutant seedlings (black squares) bend much stronger and at a lower fluence dose compared with wild-type seedlings (white circles). Again, mutant seedlings respond much more sensitively and quicker to a given blue light stimulus than wild-type seedlings. Error bars indicate sd.
ehb1 Mutant Seedlings Respond Quicker to a Gravitropic Stimulus
In addition to the above-mentioned bending experiments toward a lateral light source, we analyzed the gravitropic response in both lines. For this reason, wild-type and ehb1-2 mutant seedlings were grown on vertical agar plates kept in darkness for 3 d. Up to this point, there was no detectable difference between wild-type and mutant seedlings. The mutant line developed normally and showed no atypical behavior in shoot and root growth and development. The agar plates were then rotated 90° and the reorientation of the stem was monitored over a time period of 48 h (Fig. 7A). Already after 6 h, a significant difference between both bending angles became evident. While wild-type hypocotyls had an average bending angle of 12° (sd = 16.5), ehb1-2 mutant seedlings revealed a bending angle more than twice that of the wild type. This difference was established over the following 42 h. After 48 h, ehb1-2 mutant hypocotyls had an average bending angle of 35° (sd = 21.9), while the corresponding wild-type seedlings had an average bending angle of 26° (sd = 13.2). Additionally, root bending exhibited by wild-type and mutant seedlings was also compared (Fig. 7B). Again, mutant seedlings established a significantly enhanced bending angle of 83° (sd = 20.8) compared with wild-type roots, with an average of 56° (sd = 18.5) after 30 h. In order to rule out any effect induced by 1% Suc added to the medium, the experiment was repeated without adding any sugar. Seedlings were germinated in the dark but after reorientation cultivated under white light conditions from a diffuse light source (Supplemental Fig. S4). Under these conditions, bending occurred much slower and less dramatically, although comparable differences in the bending angle could be detected.
Figure 7.
Gravitropic response of wild-type and ehb1-2 seedlings. A, Time course measurement of the gravity-induced hypocotyl curvature of ehb1 (black squares) and wild-type (white circles) seedlings. Mean angle ± se was calculated from three individual experiments with 25 seedlings each. B, Time course measurement of the root gravitropic response. Wild-type (white circles) and ehb1-2 mutant (black squares) seedlings were analyzed after an initial growth for 3 d in darkness on vertical agar plates. Analysis was performed for the indicated times after turning plates by 90° (for details, see “Materials and Methods”). Mean angle ± se was calculated from five independent experiments with 25 seedlings each.
DISCUSSION
At present, it is generally accepted that the phototropic response to blue light initiates a lateral auxin gradient within the stem. Although several components for stimulus perception and signal transduction have been identified (Kimura and Kagawa, 2006; Christie, 2007; Harada and Shimazaki, 2007), it is only partially clear on the molecular level how lateral blue light irradiation leads to a bending response toward the illuminated side of the stem and how gravity is influencing this process. Several lines of evidence indicate that PINFORMED1 (PIN1) and PIN3 redistribution plays a key role in this tropic response (Friml et al., 2002a, 2002b; Blakeslee et al., 2004). However, a clear picture of how phototropin-mediated light perception is connected to PIN1/3 redistribution is still elusive. Additionally, little is known about signaling elements beyond NPH3 and RPT2 connecting phototropin light perception physically to auxin redistribution. Meanwhile, several proteins directly binding to NPH3 have been identified (Lariguet et al., 2006; Sullivan et al., 2009). PKS1, however, seems to be the only such candidate that not only binds to NPH3 but also connects phototropism and gravitropism to phytochrome action (Lariguet et al., 2006). In order to identify other elements of phototropin signaling, we used a yeast three-hybrid approach offering a NPH3/phot1 complex as bait. We subsequently focused only on yeast colonies containing prey plasmids exclusively found on plates exposed to blue light. One of these candidates, termed EHB1, coded for a small protein with an unknown function. Although we retrieved EHB1 by using an NPH3/phot1 complex as bait in a yeast three-hybrid approach, it became evident that NPH3 can bind to EHB1 without phot1, although to a lesser extent. Further yeast two-hybrid assays using truncated versions of NPH3 as bait revealed that only the N terminus of NPH3 containing the BTB/POZ motif was able to bind EHB1. Two conclusions can be deduced from these results: first, phosphorylation of NPH3 (Pedmale and Liscum, 2007) might not be essential for EHB1 binding to NPH3; and second, EHB1 interaction with the BTB domain might compete with the binding of a Cullin-3-like protein to NPH3 mentioned by Pedmale and Liscum (2007). This would connect EHB1 interaction to the ubiquitin system. Whether interference with CUL3 might serve as an explanation for the observed phenotype of the ehb1 mutant remains to be addressed. Interaction of EHB1 with NPH3 and phot1 was confirmed by a coimmunoprecipitation approach, which revealed that EHB1 could form a ternary complex with phot1 and NPH3. This was also described for the members of the PKS family, namely PKS1 and PKS2 (de Carbonnel et al., 2010), supporting the idea that NPH3 acts as a scaffold, allowing several components to bind to it, depending on the stimulus perceived.
Undoubtedly, the most intriguing outcome of our findings was the unexpected phenotype of corresponding ehb1 loss-of-function mutants. This mutant showed no decrease in but an enhanced bending response toward blue light, suggesting that EHB1 might act as a negative element in phototropin signaling. Interestingly, the previously described mdr1 mutant revealed a similar phenotype (Noh et al., 2003). The corresponding protein, ABC TRANSPORTER B FAMILY19, formerly termed MDR1 or P-GLYCOPROTEIN19 (PGP19), was reported to be involved in auxin distribution by stabilizing the PIN1 protein to the basal membrane, thus enabling auxin redistribution, which is also a prerequisite of cotyledon extension (Petrásek et al., 2006; Blakeslee et al., 2007; Lewis et al., 2009; Titapiwatanakun et al., 2009). However, a direct interaction with phot1 or NPH3 has never been reported. Thus, EHB1 might serve as a first candidate that connects physical interaction to NPH3 with hypocotyl bending.
Additional features of EHB1 might be interesting in this context. (1) Besides binding to NPH3, EHB1 encompasses a putative N-terminal C2 domain, involved in Ca2+ signaling (Rizo and Südhof, 1998) and the association of target proteins with the membranous system (Nalefski et al., 1997). In this context, one has to keep in mind that NPH3 as well as phot1 have no predicted transmembrane anchor but are at least temporarily associated with the plasma membrane (Short et al., 1993; Knieb et al., 2004; Wan et al., 2008). (2) EHB1 contains a Ca2+-binding motif that could serve as a candidate that mediates phot1-driven calcium influx (Baum et al., 1999; Babourina et al., 2002; Folta et al., 2003). (3) EHB1 contains at its C terminus a functionally undefined motif, which has a striking homology to proteins of the ARF-GAP family. These proteins, which were reported to be involved in vesicle redistribution by inactivating ARF proteins and in phospholipid binding (Jensen et al., 2000), encompass besides this C-terminal motif a C2 domain and a GTPase domain, which is surprisingly missing in EHB1 (Donaldson and Jackson, 2000; Hawadle et al., 2002; Donaldson and Honda, 2005). Thus, it is tempting to speculate that EHB1 might compete with a functionally active ARF-GAP protein, like ZAC, for target binding. Some parallels to MDR1/PGP19 are striking. PGP19 is required for the basal accumulation of PIN1 and is involved in the rate and specificity of auxin transport, a process involving ARFs (Geldner et al., 2003; Kleine-Vehn and Friml, 2008). ARF trafficking, however, requires several cofactors like ARF-GAP and ARF-GTP EXCHANGE FACTOR (Donaldson and Jackson, 2000; Hawadle et al., 2002; Donaldson and Honda, 2005). Together with the observations of Sullivan et al. (2009), the above results indicate that EHB1 might be a candidate that on the one hand binds to NPH3 and on the other hand interferes with the vesicular redistribution of PIN proteins. Disturbingly, in literature, the term “ARF” is used for two different protein families, namely the auxin response factor family and the ADP-ribosylation factor involved in vesicular trafficking, which both play a role in this context.
Notably, enhanced bending in ehb1 mutants is also induced by gravitropism (Fig. 7). For this reason, it is possible that the small gene family represented by EHB1 is a common element of both phototropic and gravitropic signal transduction. Another observation analyzing the NAKED PINS IN YUCCA gene family in Arabidopsis points in the same direction (Li et al., 2011). Their findings suggest that gravitropism as well as phototropism use analogous mechanisms involving kinases (e.g. phot1), NPH3-like proteins, and the auxin-binding factor protein family. Members of the EHB gene family might be involved in both pathways, serving as effectors of PIN protein organization required for phototropic and gravitropic bending induced by auxin redistribution. In this context, it will be interesting to see if these proteins can compete with ARF-GAP proteins for ARF binding without having any GTP-exchange capacity.
At present, we cannot rule out the possibility that the gene adjacent to EHB1, At1g70810, has a similar and overlapping function to EHB1. At1g70810 binds to NPH3, as does EHB1 (Fig. 2). Analysis of a potential role of At1g70810 in phototropic curvature is the subject of ongoing studies. Despite a potential redundancy in function between At1g70810 and EHB1, ehb1 mutant seedlings respond to lateral blue light with a phototropic dose-response curve that is 10 times more sensitive than the wild type (Fig. 5B). A comparable effect could be observed if different light doses were applied at a given fluence rate of 0.5 μmol m−2 s−1 (Fig. 6). Thus, the higher sensitivity of the ehb1 mutant corresponds to a faster bending toward blue light. As mentioned above, the alleviative effect of EHB1 on the bending response of Arabidopsis seedlings to light suggests an attenuating function. It remains to be addressed if this role is directly connected to lateral auxin redistribution or to the responsiveness of cells to a given level of auxin.
MATERIALS AND METHODS
Plant Material
Wild-type Arabidopsis (Arabidopsis thaliana) ecotype Columbia and the Salk mutants SAIL 385_C07 (ehb1-2) and SALK_134720.56.00.x (ehb1-1) in the Columbia background were used for all experiments.
Yeast Two- and Three-Hybrid Analyses
For all in vivo interaction analyses in yeast, we used the Matchmaker library construction kit (Clontech) in yeast (Saccharomyces cerevisiae) strain AH109 (Clontech). Bait backbone was either pGBKT7 or pBridge (Clontech), which allows simultaneous but independent expression of two bait proteins. Prey backbone for library construction was the pGADT7 plasmid (Clontech). cDNA was synthesized from total RNA isolated from 6-d-old dark-grown Arabidopsis seedlings. Seedlings were grown on agar-containing standard Murashige and Skoog medium.
NPH3 coding sequence was amplified from cDNA by PCR with oligonucleotide primers TK01 and TK02 (Supplemental Table S1) providing BamHI and EcoRI sites at the 5′ or 3′ end and inserted into the GAL4 DNA-binding domain of pBridge. Correct insertion was verified by sequence analysis. For cDNA library screening, the PHOT1 coding sequence was provided with a NotI site at both ends by PCR using oligonucleotide primer combination TK07/TK08 and inserted behind the MET25 promoter. Yeast cells were transformed and grown according to the manufacturer’s instructions on appropriate dropout medium. After transformation, yeast cells were grown for 5 d under blue light (470 nm; photon fluence rate of 0.18 μmol m−2 s−1) on quadruple dropout plates without Met to induce PHOT1 expression. Control plates were kept in the dark.
All further yeast interaction experiments were carried out by a yeast two-hybrid approach using a standard pGBKT7 (Clontech) vector as bait with inserted NPH3 amplified with TK01 and TK02. Experiments with EHB1, At1g70810, or phot1 as prey were performed using the pGADT7 vector with the cDNA sequence of EHB1 amplified with TK13/TK14, At1g70810 with TK15/TK16, and phot1 with TK03/TK04. All construct were verified by sequence analysis. For testing the interaction of NPH3 deletions, the following oligonucleotide primer combinations were used to amplify the indicated deletions: NPH3ΔN (amino acids 157–747), LP03/LP04; NPH3ΔC (amino acids 1–647), TK01/LP04; NPH3ΔNΔM (amino acids 635–747), LP05/TK02; NPH3ΔCΔM (amino acids 1–163), TK01/LP06; and NPH3ΔCΔN (amino acids 157–647), LP03/LP04. Transformation and ortho-nitrophenyl-β-galactoside assay were performed according to the manufacturer’s instructions (Matchmaker III; Clontech).
In Vitro Interaction Studies
Using a TnT transcription/translation system (Promega), [35S]Met-labeled in vitro-translated and -transcribed proteins of phot1, NPH3, and EHB1 were obtained from the above-described pGBKT7 construct encoding NPH3 and PHOT1 as well as the pGADT7 construct for EHB1 and PHOT1, respectively. This construct provides a myc tag at the N terminus of NPH3 and phot1. Coimmunoprecipitation for phot1 and EHB1 with myc:NPH3 and EHB1 with myc:phot1 were performed by using anti-myc antibodies (Promega) and protein A-Sepharose (Invitrogen). Precipitated proteins were subsequently washed according to the manufacturer’s instructions, boiled in SDS sample buffer, and separated by 12% SDS-PAGE. Dried gels were exposed to an appropriate x-ray film (BioMax MR-1 film; Kodak).
mRNA Isolation and PCR
RNA from 6-d-old dark-grown wild-type and mutant seedlings was extracted using the RNeasy plant mini kit (Qiagen) following the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of total RNA with SuperScript II reverse transcriptase (Invitrogen), RNAse H, and an oligo(dT) primer mix [5′-T(18)A-3′ + 5′-T(18)C-3′ + 5′-T(18)G-3′] for 90 min at 42°C. Subsequent PCR was performed according to standard protocols. Gene-specific oligonucleotide primer sequences are given in Supplemental Table S1.
Blue Light Illumination and Gravitropic Experiments
For threshold analysis, fluence response, and gravitropic bending, Arabidopsis wild-type and mutant seedlings were treated as follows. After 2 d at 4°C, germination was induced by 1 h of illumination with white light (0.5 μmol m−2 s−1). Seedlings were subsequently incubated for 3 d in darkness at 25°C on vertical agar plates containing Murashige and Skoog medium. Wild-type and mutant seedlings were indistinguishable in length and habit. Average length for wild-type seedlings was 14.9 ± 0.6 mm and for mutant seedlings was 14.7 ± 0.4 mm. Fluence-response experiments were performed at 0.5 μmol m−2 s−1 lateral blue light for 20 h or various time intervals for the dose-response analysis. Blue light illumination at 470 nm was performed with Luxeon star light-emitting diodes (LXHL-LB5C; Roithner Lasertechnik). The illumination device was self-constructed with beam-splitting mirrors (threshold box), providing a photon fluence rate from less than 0.01 μmol m−2 s−1 up to 1 μmol m−2 s−1. Light reduction was measured for each mirror and subsequently calculated for each position tested. For analyzing gravitropic bending, plates containing 3-d-old dark-grown seedlings were reoriented by 90°. Hypocotyl bending in response to gravitropism was measured over a time period of 30 h after reorientation of the agar plates. Usually, gravitropic experiments were performed in the presence of 1% (w/v) Suc (Müller et al., 1998). To discriminate if adding of nutritious sugar to the Murashige and Skoog medium seedlings might influence the outcome of our analysis, for some experiments Suc was omitted and seedlings were exposed from all sides to diffuse white light (0.5 μmol m−2 s−1). Under these conditions, plants developed slowly; therefore, root gravitropism was measured over a time period of 148 h. Curvature was analyzed by using the Image tool software (http://ddsdx.uthscsa.edu/dig/download.html). Subsequently, mean values, sd, and se were calculated for each experiment. Significance of differences between wild-type and mutant seedlings was confirmed by using one-way ANOVA, F test (calculated by Excel), and Bonferroni correction.
Sequence data from this article can be found in the EMBL data libraries under accession number BT010960 (gene bank ID 843417).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genomic organization of EHB1 and the adjacent gene At1g70810.
Supplemental Figure S2. Transcript analysis of ehb1 mRNA in wild-type and mutant seedlings.
Supplemental Figure S3. Fluence response curve and root gravitropic bending in a second ehb1 mutant line.
Supplemental Figure S4. Root gravitropism of wild-type and ehb1-2 mutant seedlings grown on plates without sugar.
Supplemental Table S1. Oligonucleotide primer sequences.
Acknowledgments
We are gratefully indebted to Andrea Weisert and Claudia Jung-Blasini for skillful technical assistance, Paul Galland and Alfred Batschauer for many helpful discussions, as well as Katja Forreiter and Lee Herrick for critically reading the manuscript.
References
- Babourina O, Newman I, Shabala S. (2002) Blue light-induced kinetics of H+ and Ca2+ fluxes in etiolated wild-type and phototropin-mutant Arabidopsis seedlings. Proc Natl Acad Sci USA 99: 2433–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum G, Long JC, Jenkins GI, Trewavas AJ. (1999) Stimulation of the blue light phototropic receptor NPH1 causes a transient increase in cytosolic Ca2+. Proc Natl Acad Sci USA 96: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS. (2004) Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol 134: 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccalandro HE, De Simone SN, Bergmann-Honsberger A, Schepens I, Fankhauser C, Casal JJ. (2008) PHYTOCHROME KINASE SUBSTRATE1 regulates root phototropism and gravitropism. Plant Physiol 146: 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM. (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Christie JM. (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282: 1698–1701 [DOI] [PubMed] [Google Scholar]
- Cukierman E, Huber I, Rotman M, Cassel D. (1995) The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 270: 1999–2002 [DOI] [PubMed] [Google Scholar]
- de Carbonnel M, Davis P, Roelfsema MR, Inoue S, Schepens I, Lariguet P, Geisler M, Shimazaki K, Hangarter R, Fankhauser C. (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol 152: 1391–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Honda A. (2005) Localization and function of Arf family GTPases. Biochem Soc Trans 33: 639–642 [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. (2000) Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol 12: 475–482 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. (2008) The Pfam protein families database. Nucleic Acids Res 36: D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Lieg EJ, Durham T, Spalding EP. (2003) Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. Plant Physiol 133: 1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Harada A, Shimazaki K. (2007) Phototropins and blue light-dependent calcium signaling in higher plants. Photochem Photobiol 83: 102–111 [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawadle MA, Folarin N, Martin R, Jackson TR. (2002) Cytohesins and centaurins control subcellular trafficking of macromolecular signaling complexes: regulation by phosphoinositides and ADP-ribosylation factors. Biol Res 35: 247–265 [DOI] [PubMed] [Google Scholar]
- Holland JJ, Roberts D, Liscum E. (2009) Understanding phototropism: from Darwin to today. J Exp Bot 60: 1969–1978 [DOI] [PubMed] [Google Scholar]
- Iino M. (2001) Phototropism in higher plants. Häder DP, Lebert M, , Photomovement. Elsevier Science B.V., Amsterdam, pp 659–811 [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Lykke-Andersen K, Frandsen GI, Nielsen HB, Haseloff J, Jespersen HM, Mundy J, Skriver K. (2000) Promiscuous and specific phospholipid binding by domains in ZAC, a membrane-associated Arabidopsis protein with an ARF GAP zinc finger and a C2 domain. Plant Mol Biol 44: 799–814 [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. (2009) Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell 21: 3226–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kagawa T. (2006) Phototropin and light-signaling in phototropism. Curr Opin Plant Biol 9: 503–508 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Friml J. (2008) Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Knieb E, Salomon M, Rüdiger W. (2004) Tissue-specific and subcellular localization of phototropin determined by immuno-blotting. Planta 218: 843–851 [DOI] [PubMed] [Google Scholar]
- Lariguet P, Schepens I, Hodgson D, Pedmale UV, Trevisan M, Kami C, de Carbonnel M, Alonso JM, Ecker JR, Liscum E, et al. (2006) PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc Natl Acad Sci USA 103: 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Wu G, Ljung K, Spalding EP. (2009) Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 multidrug resistance-like transporter. Plant J 60: 91–101 [DOI] [PubMed] [Google Scholar]
- Li Y, Dai X, Cheng Y, Zhao Y. (2011) NPY genes play an essential role in root gravitropic responses in Arabidopsis. Mol Plant 4: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Stowe-Evans EL. (2000) Phototropism: a “simple” physiological response modulated by multiple interacting photosensory-response pathways. Photochem Photobiol 72: 273–282 [DOI] [PubMed] [Google Scholar]
- Molas ML, Kiss JZ. (2008) PKS1 plays a role in red-light-based positive phototropism in roots. Plant Cell Environ 31: 842–849 [DOI] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E. (1999) Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ. (1996) The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci 5: 2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Slazas MM, Falke JJ. (1997) Ca2+-signaling cycle of a membrane-docking C2 domain. Biochemistry 36: 12011–12018 [DOI] [PubMed] [Google Scholar]
- Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. (2003) Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423: 999–1002 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. (2007) Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J Biol Chem 282: 19992–20001 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Parker PJ. (1996) Extending the C2 domain family: C2s in PKCs delta, epsilon, eta, theta, phospholipases, GAPs, and perforin. Protein Sci 5: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Südhof TC. (1998) C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem 273: 15879–15882 [DOI] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K. (2000) RPT2: a signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR. (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens I, Boccalandro HE, Kami C, Casal JJ, Fankhauser C. (2008) PHYTOCHROME KINASE SUBSTRATE4 modulates phytochrome-mediated control of hypocotyl growth orientation. Plant Physiol 147: 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short TW, Reymond P, Briggs WR. (1993) A pea plasma membrane protein exhibiting blue light-induced phosphorylation retains photosensitivity following triton solubilization. Plant Physiol 101: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans EL, Harper RM, Motchoulski AV, Liscum E. (1998) NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol 118: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Thomson CE, Kaiserli E, Christie JM. (2009) Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett 583: 2187–2193 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Briggs WR. (2010) The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell 22: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, et al. (2009) ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57: 27–44 [DOI] [PubMed] [Google Scholar]
- Ubach J, Zhang X, Shao X, Südhof TC, Rizo J. (1998) Ca2+ binding to synaptotagmin: how many Ca2+ ions bind to the tip of a C2-domain? EMBO J 17: 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YL, Eisinger W, Ehrhardt D, Kubitscheck U, Baluska F, Briggs W. (2008) The subcellular localization and blue-light-induced movement of phototropin 1-GFP in etiolated seedlings of Arabidopsis thaliana. Mol Plant 1: 103–117 [DOI] [PubMed] [Google Scholar]