Abstract
Light regulates multiple aspects of growth and development in plants. Transcriptomic changes govern the expression of signaling molecules with the perception of light. Also, the 26S proteasome regulates the accumulation of positive and negative regulators for optimal growth of Arabidopsis (Arabidopsis thaliana) in the dark, light, or light/dark cycles. BBX22, whose induction is both light regulated and HY5 dependent, is a positive regulator of deetiolation in Arabidopsis. We found that during skotomorphogenesis, the expression of BBX22 needs to be tightly regulated at both transcriptional and posttranslational levels. During photomorphogenesis, the expression of BBX22 transiently accumulates to execute its roles as a positive regulator. BBX22 protein accumulates to a higher level under short-day conditions and functions to inhibit hypocotyl elongation. The proteasome-dependent degradation of BBX22 protein is tightly controlled even in plants overexpressing BBX22. An analysis of BBX22 degradation kinetics shows that the protein has a short half-life under both dark and light conditions. COP1 mediates the degradation of BBX22 in the dark. Although dispensable in the dark, HY5 contributes to the degradation of BBX22 in the light. The constitutive photomorphogenic development of the cop1 mutant is enhanced in cop1BBX22ox plants, which show a short hypocotyl, high anthocyanin accumulation, and expression of light-responsive genes. Exaggerated light responsiveness is also observed in cop1BBX22ox seedlings grown under short-day conditions. Therefore, the proper accumulation of BBX22 is crucial for plants to maintain optimal growth when grown in the dark as well as to respond to seasonal changes in daylength.
Light is one of the major environmental stimuli affecting plant growth and development. Throughout their lives, plants adopt versatile strategies to interpret the environmental light signals in their growth habitat to proceed with the most favorable growth and developmental programs, including skotomorphogenesis, photomorphogenesis, shade avoidance, circadian growth, flowering time control, and eventually senescence.
Skotomorphogenesis and photomorphogenesis are two distinct developmental processes for plants growing in the dark and in the light, respectively. Proper regulation of these two developmental stages is important for plants to optimize their growth and to ensure their success in response to environmental cues (for review, see Casal et al., 2004). Seedlings undergo a sophisticated transcriptomic adjustment during the transition from skotomorphogenesis to photomorphogenesis (for review, see Casal and Yanovsky, 2005). Both positive and negative transcriptional regulators are reported to participate in light-regulated transcriptional modulation in Arabidopsis (Arabidopsis thaliana). Importantly, many of these transcription factors, as well as photoreceptors and other signaling molecules, are subject to posttranslational regulation by light. For example, the 26S proteasome regulates the accumulation of proteins for light perception and signaling (for review, see Henriques et al., 2009). COP1 is a well-known E3 ligase functioning in selective degradation of proteins regulating many aspects of plant development, including photomorphogenesis (Ang et al., 1998; Osterlund et al., 2000b; Holm et al., 2002; Seo et al., 2003, 2004; Jang et al., 2005; Hong et al., 2008), photoperiodic growth (Yu et al., 2008), and flowering time control (Jang et al., 2008; Liu et al., 2008). Although the importance of COP1 in photomorphogenic growth is well documented, the repertoire of light-signaling proteins targeted by COP1 is likely incomplete.
Ample evidence exists for the coordination between light and multiple plant hormones in plant growth and development (Achard et al., 2007; Alabadí et al., 2008; Chen et al., 2008; Feng et al., 2008; Nemhauser, 2008; Vert et al., 2008; Song et al., 2009). Current studies suggest that HY5 is one of the key factors integrating light- and hormone-signaling pathways (Sibout et al., 2006; Chen et al., 2008; Laxmi et al., 2008). However, the molecular mechanisms underlying such cross talk are not fully understood.
BBX22, also known as LZF1 (for light-regulated zinc finger protein 1), STH3 (for salt tolerance homolog 3), and DBB3 (for double B-box zinc finger 3; Chang et al., 2008; Datta et al., 2008; Kumagai et al., 2008; Khanna et al., 2009), is a newly identified signaling component acting in concert with HY5 and BBX21/STH2 to achieve a coordinated deetiolation process in Arabidopsis (Chang et al., 2008; Datta et al., 2008). bbx22 is epistatic to cop1 in the dark, which supports the idea that at least some of the photomorphogenic phenotypes in cop1 are mediated by the accumulation of BBX22 (Datta et al., 2008). Previous results showed that BBX22 could be ubiquitinated in vitro by COP1, and a complex composed of BBX22, HY5, and COP1 might be formed (Datta et al., 2008). However, the expression kinetics of endogenous BBX22 protein and the biological significance of BBX22 degradation have not been carefully characterized.
In this study, we demonstrate that BBX22 is a short-lived protein and contributes to the inhibition of hypocotyl elongation under short-day (SD) conditions. As well, COP1 safeguards the destruction of BBX22 in the dark. The selective degradation of BBX22 ensures a precise skotomorphogenesis process and optimizes seedling growth under SD conditions in Arabidopsis. We also provide the molecular evidence to indicate that BBX22 regulates the expression of genes involved in light and phytohormone pathways, which may contribute to optimal seedling development in Arabidopsis.
RESULTS
Posttranslational Regulation Is Responsible for the Transient Accumulation of BBX22 Protein
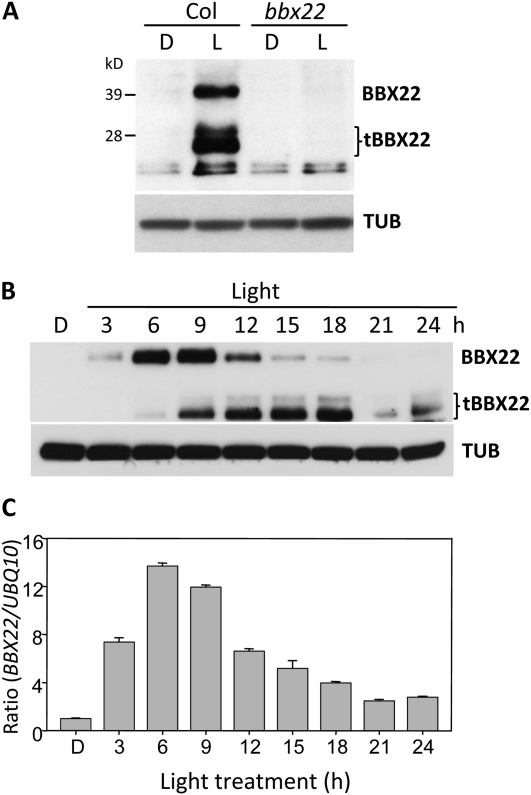
BBX22 functions to convey light signals from HY5 by activating three major photomorphogenic growth features in Arabidopsis: inhibition of hypocotyl elongation, anthocyanin biogenesis, and chloroplast development (Chang et al., 2008; Datta et al., 2008). To further elucidate the action mechanism of BBX22, we sought to characterize the expression pattern of BBX22 protein during the transition from dark to light environments. A BBX22-specific antiserum was generated to monitor the expression of BBX22 protein under both skotomorphogenic and photomorphogenic growth. In wild-type seedlings, full-length (40-kD) BBX22 and a 28-kD truncated BBX22 (tBBX22; see below) could be detected in light-grown seedlings but were barely detectable in dark-grown seedlings (Fig. 1A). Neither form of BBX22 protein could be detected in the bbx22 mutant, which indicates that the antiserum is BBX22 specific (Fig. 1A). The tBBX22 is unlikely to be a product of alternative spliced mRNA because only the full-length BBX22 transcript was observed during the deetiolation process (Supplemental Fig. S1A). When BBX22 was expressed as an N- or C-terminal epitope-tagged form in transgenic Arabidopsis, the tBBX22 showed a mobility shift only when expressed as an N-terminal but not a C-terminal tagged form (data not shown). This finding suggests that, rather than being a product of alternative translational initiation, tBBX22 represents the N-terminal tBBX22.
Figure 1.
Both BBX22 protein and transcript are transiently accumulated. A, A BBX22-specific antibody was generated and used to determine the steady-state protein level by immunoblot analysis. Proteins were isolated from 4-d-old etiolated (dark; D) and 12-h light-treated (L) ecotype Columbia (Col) or bbx22 seedlings. Endogenous α-tubulin was a loading control (TUB). B and C, BBX22 protein (B) and mRNA (C) are transiently accumulated during photomorphogenesis. Immunoblotting was used to detect the accumulation of BBX22 in etiolated seedlings and seedlings illuminated with light for 3 to 24 h. Real-time quantitative RT-PCR was used to monitor the expression of endogenous BBX22 in C. The expression of UBQ10 in each sample was used as an internal control. The BBX22 expression in etiolated seedlings was set to 1. The expression of BBX22 is presented as the amount of increase at each time point relative to that in etiolated seedlings and represented as “ratio.” The means and sd were calculated from three replicates and plotted.
The presence of the N-terminal tBBX22 implies that BBX22 abundance might be regulated by selective degradation. This suggestion prompted us to perform a more detailed examination of BBX22 expression kinetics. Total protein isolated from 4-d-old etiolated seedlings illuminated with 3 to 24 h of light was subjected to immunoblot analysis. As shown in Figure 1B, full-length BBX22 protein peaked at 6 to 9 h after light illumination, then was rapidly degraded. The lag in appearance and the eventual degradation of tBBX22 during the time course examined indicated that the truncated form is an intermediate form temporally accumulated during BBX22 degradation.
Real-time quantitative reverse transcription (RT)-PCR analysis of BBX22 transcripts in samples from these time-course experiments revealed nearly synchronized expression kinetics between BBX22 transcripts and the full-length protein (Fig. 1, B and C). This finding suggests that BBX22 is under highly coordinated regulation at both the transcript and protein levels.
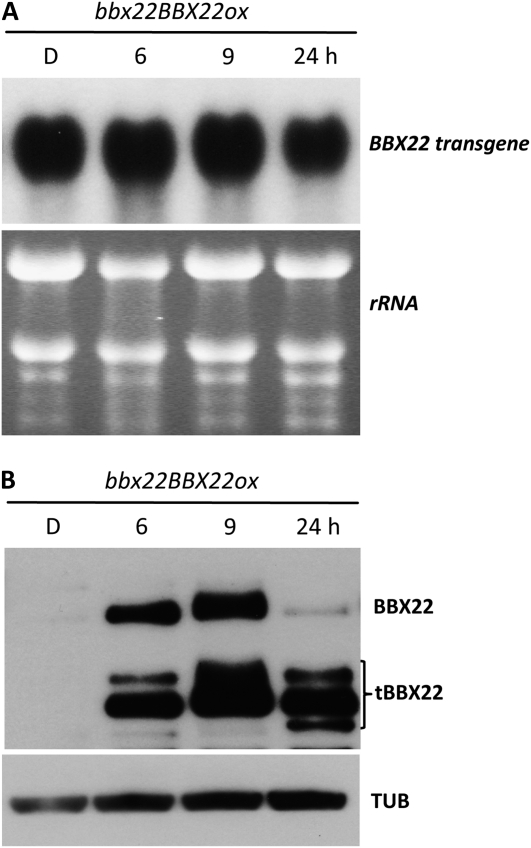
To further assess the impact of posttranslational regulation on BBX22 protein accumulation, we analyzed the BBX22 protein expression pattern in bbx22BBX22ox (lzf1LZF1ox in Chang et al., 2008), which has phenotypes indistinguishable from those of wild-type Arabidopsis (Chang et al., 2008). Because the expression of BBX22 is under the control of the 35S promoter in this transgenic plant, the BBX22 transcripts should accumulate constitutively regardless of light or dark treatment. As expected, transcripts derived from the BBX22 transgene were expressed at high levels at all time points examined (Fig. 2A). Interestingly, the accumulation kinetics of BBX22 in bbx22BBX22ox plants essentially mimic that in wild-type seedlings (Figs. 1B and 2B). No BBX22 or a very low BBX22 level could be detected in dark-grown tissues, and tBBX22 was abundant in seedlings illuminated with 24 h of light (designated as L24 hereafter; Fig. 2B). The increased level of tBBX22 likely reflects the higher level of BBX22 produced from the transgene. These results imply that even when BBX22 is overproduced, its accumulation is under rigid control in both dark-grown and light-illuminated Arabidopsis seedlings. As well, an efficient degradation system exists in Arabidopsis seedlings for the removal of BBX22 protein in a time- and light-dependent manner, which allows for the accumulation of BBX22 only in a restricted window after light illumination.
Figure 2.
Transient accumulation of BBX22 is regulated at the protein level. The stability of BBX22 was determined in 4-d-old bbx22 expressing 35S:BBX22 (bbx22BBX22ox) under the conditions indicated. A, Northern blot showing the overexpression of BBX22 in etiolated (dark; D) bbx22BBX22ox seedlings or etiolated seedlings illuminated with light for 6, 9, or 24 h. The ethidium bromide-stained image was used to show equal loading of the total RNA samples. B, Immunoblot showing the transient accumulation of BBX22 in plant samples used in A. Endogenous α-tubulin was a loading control (TUB).
BBX22 Is a Short-Lived Protein Degraded by the 26S Proteasome
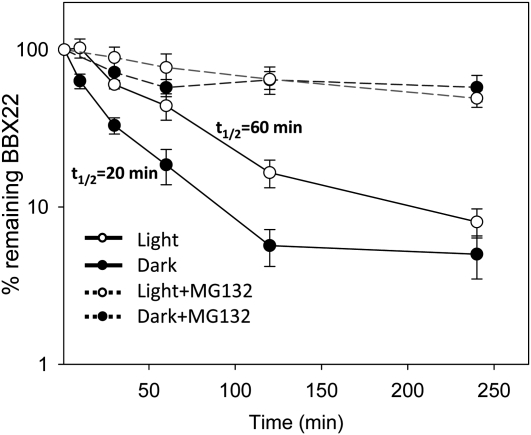
We next examined the capacity for Arabidopsis to degrade BBX22 protein. We used 4-d-old etiolated seedlings treated with 8 h of light for optimal accumulation of BBX22 protein. The seedlings were then incubated with the translation inhibitor cycloheximide and kept under light or transferred to dark treatment for the times indicated. The degradation kinetics of BBX22 protein were determined as a proportion of full-length BBX22 remaining relative to BBX22 expression at 8 h of light treatment (Fig. 3). The half-life of BBX22 was calculated to be 20 min in the dark and 60 min in the light. These results indicate that BBX22 is a short-lived protein and that dark treatment accelerates its degradation.
Figure 3.
BBX22 is a short-lived protein degraded by the 26S proteasome. The degradation kinetics of BBX22 protein were determined as a percentage of full-length BBX22 remaining relative to BBX22 at 8 h of light treatment in the presence of cycloheximide. Half-life (t1/2) was calculated by regression analysis. The half-life of BBX22 is 60 min in the light (white circles, solid lines) and 20 min in the dark (black circles, solid lines). The degradation of BBX22 was blocked by treatment with MG132 under both light (white circles, dashed lines) and dark (black circles, dashed lines) conditions. n = 3.
The involvement of the 26S proteasome in the fast degradation of BBX22 was also examined in parallel by adding the 26S proteasome inhibitor MG132. As shown in Figure 3, as compared with less than 10% of BBX22 detected in the absence of MG132, in the presence of MG132, BBX22 was largely stabilized, as represented by 54.9% or 66.7% residual BBX22 after 4 h of MG1132 treatment under light or dark conditions, respectively. The effective stabilization of BBX22 protein by MG132 indicates that the 26S proteasome is responsible for the degradation of BBX22 under both light and dark conditions.
BBX22 Contributes to the Inhibition of Hypocotyl Elongation under SD Conditions
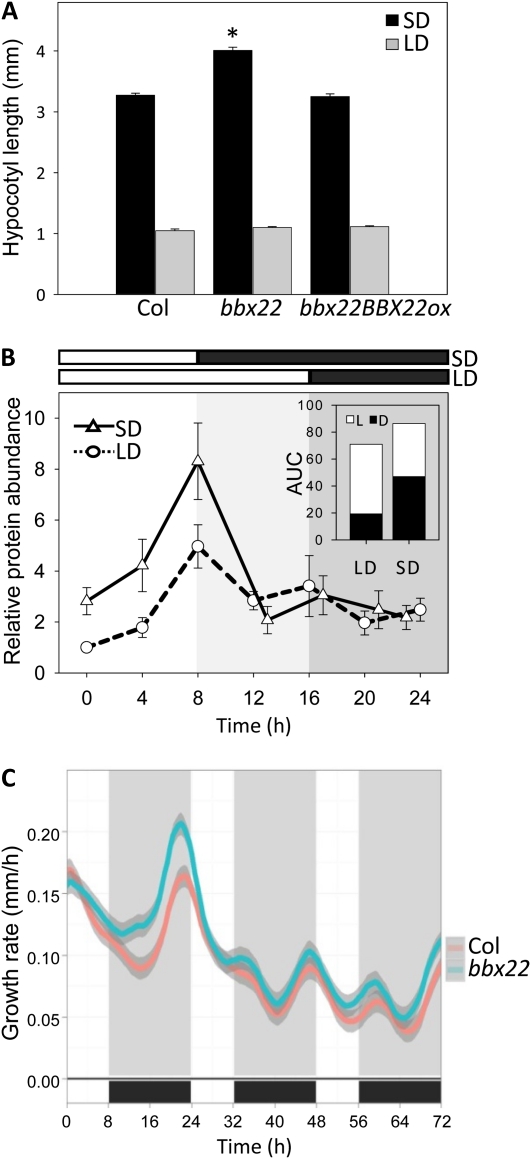
Six-day-old bbx22 exhibits light hyposensitivity (i.e. long hypocotyl) under SD conditions but not long-day (LD) conditions (Datta et al., 2008). We have confirmed this phenotype for 4-d-old seedlings (Fig. 4A). The strict expression regulation of BBX22 prompted us to examine a possible correlation between the BBX22 protein levels and the differential light responsiveness in seedlings grown under SD or LD conditions. As shown in Figure 4B, BBX22 protein showed an expression peak at 8 h after dawn under both SD and LD conditions. After dusk, BBX22 protein level was quickly degraded in both SD- and LD-grown seedlings. Total BBX22 protein in both light and dark periods under SD and LD conditions was estimated, separately, by calculating the areas under the SD or LD protein curves. Results showed that BBX22 accumulates to a higher level under SD than LD conditions (Fig. 4B, top right).
Figure 4.
BBX22 protein functions in the inhibition of hypocotyl elongation under SD conditions. A, The bbx22 mutant is hyposensitive to light only under SD conditions. Hypocotyl length was measured for 4-d-old seedlings grown under SD or LD conditions. * P < 0.01, Student’s t test; n = 38 to 76. B, BBX22 protein accumulates to a higher level in SD-grown Col plants (SD; white triangles, solid lines) compared with that in plants under LD conditions (white circles, dashed lines). BBX22 protein levels at different times of the day were determined by immunoblot analysis. Results are presented as values relative to that at dawn for 4-d-old seedlings grown under LD conditions. White bars indicate light periods and black bars indicate dark periods. n = 4. AUC, Areas under the SD or LD protein curves. C, bbx22 shows an increased growth rate during the dark period of SD conditions, with the most noticeable difference at day 4 after germination. Time 0 indicates dawn of day 4 after germination. Shaded and black areas indicate night (darkness), and white areas indicate day (lights on). Shaded areas around each growth trace (blue and red) show se. n = 29.
Previous studies indicated that the hypocotyl elongation of Arabidopsis seedlings follows a rhythmic pattern (Dowson-Day and Millar, 1999; Nozue et al., 2007). We thus characterized, under SD conditions, whether mutation of BBX22 influences a specific growth phase of hypocotyl elongation by real-time imaging of hypocotyl growth under SD conditions, as described previously (Nozue et al., 2007). As shown in Figure 4C, bbx22 exhibited an increased growth rate during the fast-elongating phase in the dark period. The most noticeable difference was observed on day 4, consistent with the maximal growth capability seen in Arabidopsis seedlings (Gendreau et al., 1997; Nozue et al., 2007).
BBX22 is a positive regulator of the inhibition of hypocotyl elongation during photomorphogenesis (Chang et al., 2008; Datta et al., 2008). The higher level of BBX22 protein under SD than LD conditions may explain its more prominent contribution in the inhibition of hypocotyl elongation under SD conditions. The increased growth rate observed in bbx22 suggests that BBX22 could positively transmit the light signal for attenuated hypocotyl elongation during the night.
COP1 Is Required for Selective Degradation of BBX22 in the Dark
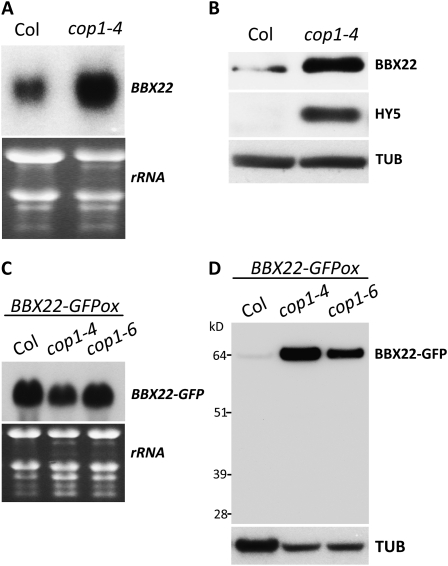
COP1 possesses the ability to ubiquitinate BBX22 in vitro and was proposed to be responsible for the regulation of BBX22 protein (Datta et al., 2008). However, the role of COP1 in BBX22 degradation in vivo has not been examined. The nuclear localization of BBX22 (Datta et al., 2008) prompted us to test whether BBX22 protein could indeed accumulate in cop1 mutants by examining BBX22 abundance in the dark when COP1 is also present in the nucleus (von Arnim et al., 1997; Osterlund et al., 2000a). The direct measurement of BBX22 in the cop1 mutant could be hindered by the accumulation of HY5. HY5 is a direct target of COP1 E3 ligase activity (Osterlund et al., 2000a) and also a direct transcriptional activator of BBX22 expression (Chang et al., 2008). Indeed, both our northern and immunoblot analyses showed an increased accumulation of both BBX22 transcripts and protein in cop1 mutants (Fig. 5, A and B). To bypass the regulation of BBX22 by HY5, we introduced the 35S:BBX22-GFP transgene from a wild-type background (BBX22-GFPox) into the cop1 mutant to directly compare BBX22-GFP protein abundance in wild-type plants and the cop1 mutants cop1-4 and cop1-6. Similar to bbx22BBX22ox plants, BBX22-GFPox plants also showed wild-type phenotypes (see Fig. 7 below). Therefore, the accumulation of BBX22-GFP is likely within the wild-type threshold, even though its expression was driven by a 35S promoter. As expected, although BBX22-GFP transcripts accumulated to high levels (Fig. 5C), only residual BBX22-GFP protein was present in BBX22-GFPox plants grown in the dark (Fig. 5D). In contrast, a high level of BBX22-GFP protein was detected in cop1 mutants (Fig. 5D), which suggests that COP1 is required for the selective degradation of BBX22 in the dark.
Figure 5.
COP1 is required for selective degradation of BBX22 in the dark. A and B, The expression of both BBX22 transcript (A) and BBX22 protein (B) is higher in dark-grown cop1-4 plants than in wild-type (Col) plants. C, Northern-blot analysis was used to confirm the comparable BBX22-GFP transgene expression in these lines. The ethidium bromide-stained image was used to show equal loading of the total RNA samples. D, BBX22-GFP protein accumulates to high levels in cop1 mutants. Endogenous α-tubulin was a loading control (TUB).
Figure 7.
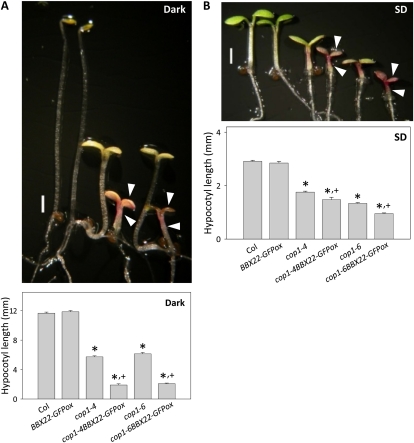
cop1BBX22-GFPox has exaggerated light responsiveness. Exaggerated cop phenotypes (top panels), the extra inhibition of hypocotyl growth and excess anthocyanin accumulating both in cotyledons and hypocotyls (marked by arrowheads), were seen when BBX22-GFP was overexpressed under both dark (A) and SD (B) conditions. Hypocotyl length (bottom panels) was measured in 4-d-old seedlings grown under dark (A) and SD (B) conditions. *,+ Significantly different from Col and the corresponding cop1 allele, respectively (P < 0.01, Student’s t test; n = 20–32). Bars = 1 mm.
HY5 Contributes to Light-Mediated Degradation of BBX22
Although COP1 could mediate a low level of BBX22 ubiquitination in vitro, no direct interaction of COP1 and BBX22 could be detected with yeast two-hybrid or fluorescence resonance energy transfer assay (Datta et al., 2008). When coexpressed with COP1, BBX22 showed a redistribution to nuclear speckles, which implies an indirect interaction between COP1 and BBX22 (Datta et al., 2008). Because HY5 can interact with both COP1 and BBX22 (Datta et al., 2008), COP1, HY5, and BBX22 may form a large complex in Arabidopsis seedlings (Datta et al., 2008). Investigating whether the integrity of this protein complex is essential for the COP1-dependent degradation of BBX22 is of interest.
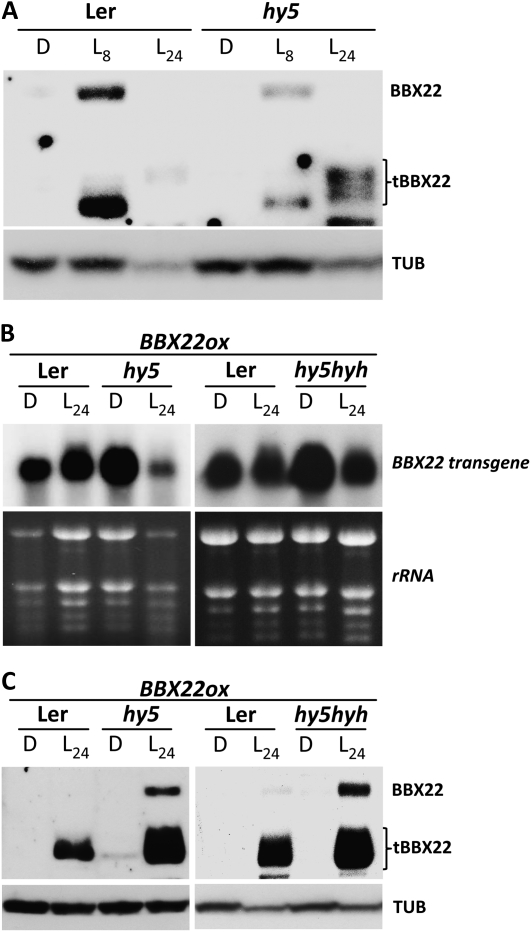
One way to test this hypothesis is to examine the BBX22 degradation patterns in the hy5 mutant, although the results are likely to be compromised because of a HY5-dependent transcriptional activation of BBX22 (Chang et al., 2008). Indeed, our immunoblot analyses revealed decreased BBX22 protein level in hy5 mutants (L8 in Fig. 6A). To circumvent this limitation, we manipulated the overproduction of BBX22 transcripts in the hy5-1 mutant (hy5BBX22ox represents hy5LZF1ox in Chang et al., 2008). For pairwise comparison, the transgene (35S:BBX22) was introduced into the corresponding wild-type Arabidopsis Landsberg erecta (Ler) ecotype by genetic crossing. As shown in Figure 6B, transcripts of the BBX22 transgene accumulated to comparable levels in Ler and hy5 mutants under both dark and L24 conditions. BBX22 protein was effectively degraded in both Ler and hy5 mutants in the dark (Fig. 6C). However, in L24, residual full-length BBX22 protein could be detected in the hy5 mutant but not in Ler (Fig. 6C). This result suggests that the efficient degradation of BBX22 protein is compromised in the light-grown hy5 mutant. An increased level of tBBX22 was also observed in hy5 (L24 in Fig. 6, A and C, left panel), which suggests that HY5 may contribute to the degradation of both full-length BBX22 and tBBX22.
Figure 6.
BBX22 degradation depends in part on HY5 in the light but is independent of HY5 and HYH in the dark. A, BBX22 decreases in hy5 due to a lack of HY5-dependent transcriptional activation of BBX22. B, Northern-blot analysis was used to confirm the comparable expression of BBX22 transgene in Ler, hy5, and hy5hyh plants expressing 35S:BBX22 and grown in the dark (D) for 4 d or in 4-old etiolated seedlings illuminated with light for 24 h (L24). The ethidium bromide-stained image was used to show the amount of total RNA samples loaded in each lane. C, Immunoblotting was used to detect BBX22 protein in the plant samples used in B. The detection of endogenous α-tubulin was performed as a loading control (TUB).
Whether HYH, a HY5 homolog (Holm et al., 2002), substitutes the functions of HY5 in the hy5 mutant was tested by examining BBX22 protein abundance in the hy5hyh double mutant. The hy5 and hy5hyh mutants showed comparable BBX22 degradation patterns (Fig. 6, B and C), which indicates that HYH does not play a decisive role in the degradation of BBX22. Therefore, both HY5 and HYH are dispensable for the degradation of BBX22 in the dark but may contribute to the efficiency of BBX22 degradation in light-illuminated seedlings.
The Selective Degradation of BBX22 Ensures Proper Seedling Development
The results shown above indicate that BBX22 could be effectively degraded even if it is overexpressed, especially in the dark (Figs. 2B, 5D, and 6C). Arabidopsis seedlings have a strong capacity for the removal of BBX22 even at an excess amount. Therefore, BBX22 might have a detrimental role if its level or time of accumulation is not strictly controlled. Our data indicate that the exaggerated accumulation of BBX22 could be achieved only in the cop1 mutant (Fig. 5). To assess whether the overproduction of BBX22 would negatively affect Arabidopsis seedling development, we analyzed the phenotypes of cop1BBX22-GFPox plants under both dark and SD conditions. The parameters measured included the inhibition of hypocotyl elongation and anthocyanin accumulation, both characteristics reported for cop1 mutants (McNellis et al., 1994). Overexpression of BBX22-GFP enhanced the cop phenotype during skotomorphogenesis (Fig. 7A) and light responsiveness under SD conditions (Fig. 7B), including the exaggerated hypocotyl shortening and the excess accumulation of anthocyanin. During skotomorphogenesis, the hypocotyl length of cop1 is about 50% that of the wild type. Overexpression of BBX22-GFP further reduced the hypocotyl length to 13% of that of the wild type (Fig. 7A). Excess anthocyanin accumulation could also be seen in both cotyledons and hypocotyls, as indicated by arrowheads in Figure 7A. Similar results were observed in cop1BBX22-GFPox plants grown under SD conditions (Fig. 7B). In contrast, overexpression of BBX22-GFP in a wild-type background did not result in these phenotypic alterations, which indicates that the presence of COP1 is sufficient to mediate the degradation of excess BBX22-GFP and that the wild-type level of biologically active BBX22-GFP could be properly maintained (Fig. 7, A and B).
The phenotypes of cop1BBX22-GFPox are consistent with the previous observation that bbx22 could partially suppress the short hypocotyl and high anthocyanin level of dark-grown cop1 (sth3cop1 in Datta et al., 2008). These results also emphasize the importance of the COP1-mediated protein surveillance of positive regulators such as BBX22 in optimizing seedling growth under both dark and SD conditions.
BBX22 Influences Genes on Light Signaling and Hormone Responses
Target genes responsible for the exaggerated light phenotype in cop1BBX22-GFPox plants were revealed by comparing transcriptomes among dark-grown wild-type, cop1, and cop1BBX22-GFPox plants. In total, 1,494 genes showed at least 2-fold difference in expression between cop1BBX22-GFPox and wild-type plants at a false discovery rate (FDR) of less than 0.05. These genes were further interrogated for their association with specific biological pathways by use of Gene Ontology analysis. These genes were associated with light-related (P = 2 × 10−4) or hormone-related (P = 3.4 × 10−8) pathways. Among these genes, 207 showed at least 2-fold differential expression in cop1BBX22-GFP compared with cop1 plants at an FDR of less than 0.05 (Supplemental Table S1). These genes were considered to be specifically regulated by BBX22 and were selected for detailed analysis.
Whether BBX22 preferentially participates in specific light or hormone pathways was examined by comparing the 207 BBX22-regulated genes with genes regulated by a specific quality of light or plant hormones during the seedling stage. Table I lists genes retrieved from various experimental conditions treated with light or plant hormones as well as genes associated with the cop1-like phenotype. For each gene list, the percentage representation of the genes in the Arabidopsis genome (ATH1) and in the 207 BBX22-regulated genes was calculated. Fisher’s exact test (http://www.matforsk.no/ola/fisher.htm; Agresti, 1992) was used to evaluate whether BBX22-regulated genes are significantly enriched with any given treatment or genetic material.
Table I. BBX22 regulates light- and hormone-responsive genes.
A total of 207 BBX22-regulated genes were compared with genes regulated by light or plant hormones. Fisher’s exact test was used to evaluate significant enrichment under each given treatment. PHYBY276H, Tyr-to-His mutant allele of phyB; pifq, pif1345 quadruple mutant; 35S:MIF1, MINI ZINC FINGER overexpressor; Wc, continuous white; Rc, continuous red; FRc, continuous far-red; Bc, continuous blue; IAA, indole-3-acetic acid; MJ, methyl jasmonate; ABA, abscisic acid; ACC, 1-amino-cyclopropane-1-carboxylic acid; CK, cytokinin.
| Treatment | ATH1 | BBX22 Regulated | P | Reference |
| % | ||||
| cop1-like | 14.7 | 70.0 | 7.2 × 10−71 | |
| PHYBY276H | 13.1 | 63.8 | 2.7 × 10−63 | Hu et al. (2009) |
| pifq | 4.5 | 30.0 | 1.1 × 10−32 | Leivar et al. (2009) |
| 35S:MIF1 | 0.5 | 3.9 | 1.4 × 10−5 | Hu and Ma (2006) |
| Light | 27.2 | 83.1 | 1.6 × 10−62 | |
| Wc | 15.7 | 61.8 | 2.3 × 10−50 | Jiao et al. (2005); Chang et al. (2008) |
| Rc | 17.2 | 70.0 | 4.6 × 10−62 | Jiao et al. (2005); Leivar et al. (2009) |
| FRc | 8.2 | 42.0 | 5.6 × 10−39 | Jiao et al. (2005) |
| Bc | 11.8 | 42.0 | 1.0 × 10−27 | Jiao et al. (2005) |
| High light | 6.4 | 36.7 | 1.3 × 10−36 | Kleine et al. (2007) |
| Hormone | 20.5 | 41.5 | 9.0 × 10−12 | |
| IAA | 3.5 | 14.0 | 3.3 × 10−10 | Nemhauser et al. (2006) |
| BR | 4.0 | 13.0 | 1.2 × 10−7 | Lisso et al. (2005); Nemhauser et al. (2006); Song et al. (2009) |
| GA | 0.5 | 1.4 | 0.11 | Nemhauser et al. (2006) |
| MJ | 6.7 | 18.4 | 1.7 × 10−08 | Nemhauser et al. (2006) |
| ABA | 12.9 | 24.6 | 5.6 × 10−6 | Nemhauser et al. (2006) |
| ACC | 2.4 | 14.0 | 4.6 × 10−14 | Nemhauser et al. (2006) |
| CK | 2.2 | 4.8 | 0.027 | Nemhauser et al. (2006) |
As shown in Table I, 70% of BBX22-regulated genes were differentially expressed in plants with cop-like phenotypes, which is consistent with the exaggerated phenotype observed for cop1BBX22-GFPox plants (Fig. 7). In total, 83% of BBX22-regulated genes were light-responsive genes as compared with only 27% in ATH1 (P = 1.6 × 10−62). Consistent with being a downstream gene of HY5 (Chang et al., 2008), BBX22 could regulate genes responding to various light qualities (P = 1 × 10−27 to 4.6 × 10−62; Table I). Also, 42% of BBX22-regulated genes are differentially regulated by treatment with multiple plant hormones (P = 9 × 10−12; Table I). The overrepresentation of BBX22-regulated genes in response to light and plant hormones suggested a possible role of BBX22 in regulating light signaling and hormone responses.
DISCUSSION
Selective Degradation of BBX22 Is Important for Proper Transcriptomic Responses in Developing Arabidopsis Seedlings
The optimal responsiveness of plants to the light/dark environment could be achieved by combinations of rapid transcriptional adjustments and posttranslational degradation of both positive or negative factors in light-sensing and -signaling pathways (Casal and Yanovsky, 2005; Henriques et al., 2009). Our results demonstrated that both the transcription of BBX22 and the accumulation of BBX22 protein are strictly controlled (Figs. 1 and 2).
The COP1- and 26S proteasome-mediated selective degradation of BBX22 is crucial to avoid unfavorable seedling development under dark or SD conditions. When overproduced, BBX22 preferentially alters the expression of genes in response to light and multiple plant hormones (Table I). Consistent with the previous observation that BBX22 regulates chloroplast development (Chang et al., 2008), genes regulated by BBX22 include ELIP1, ELIP2, CRY3/CRYD, and SIG5/SIGE, which encode chloroplast proteins (Yao et al., 2003; Tsunoyama et al., 2004; Heddad et al., 2006; Pokorny et al., 2008). ELIP1 and ELIP2, transiently induced by different qualities of light, encode thylakoid proteins functioning in chlorophyll biosynthesis (Casazza et al., 2005; Rossini et al., 2006). Plants overproducing ELIP2 have reduced chlorophyll content (Tzvetkova-Chevolleau et al., 2007). Our results suggest that the selective degradation of BBX22 might fine-tune the expression of ELIP2 for optimal chlorophyll accumulation.
BBX22 also up-regulates genes involved in flavonoid and anthocyanin biosynthesis pathways, including CHS, CHI, F3H, and 4CL3 (Li et al., 1993; Shirley et al., 1995; Ehlting et al., 1999; Raes et al., 2003; Solfanelli et al., 2006; Poustka et al., 2007; Owens et al., 2008; Buer and Djordjevic, 2009). This observation explains the excess accumulation of anthocyanin in etiolated cop1BBX22-GFPox plants (Fig. 7). In addition to being up-regulated by BBX22, the genes CHS and F3H are direct targets of HY5 (Shirley et al., 1995; Lee et al., 2007). The interaction of BBX22 with HY5 (Datta et al., 2008) raises the possibility that BBX22 and HY5 function in a complex for the activation of these two genes.
HY5 and PIF3/4 function to link the light- and hormone-signaling pathways (de Lucas et al., 2008; Lau and Deng, 2010). The enrichment of hormone-responsive genes upon the overexpression of BBX22 implies that BBX22 functions downstream of HY5 in both light and hormone signaling pathways. The overproduction of BBX22 down-regulates hormone-responsive genes and genes involved in cell wall modification (Fig. 8). For example, BBX22 represses EXP3, which, when overexpressed, promotes plant growth (Kwon et al., 2008), and EXP8, which is known be repressed by blue light through cry1 (Kleine et al., 2007). EXP8 is also induced by brassinosteroid (BR; Yin et al., 2002) and auxin through ARF7 (Esmon et al., 2006). Whether BBX22 functions to suppress either auxin- or BR-mediated expression of EXP8 remains to be examined.
Figure 8.
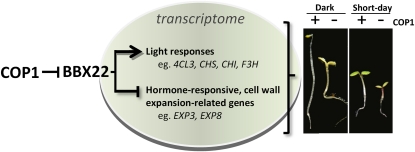
A model illustrating the biological impact of BBX22 degradation on seedling development. Protein abundance of the positive regulator BBX22 is tightly controlled by COP1 to regulate the development of Arabidopsis grown in the dark and under SD conditions. Overaccumulated BBX22 in cop1 gives rise to an exaggerated light responsiveness by altering the expression of genes responsive to light and hormone signals.
Collectively, the efficient degradation of BBX22 in wild-type seedlings ensures adequate skotomorphogenesis of elongated hypocotyl, with minimal pigments accumulating in the dark. Also, degradation of BBX22 could fine-tune hypocotyl length and anthocyanin accumulation under SD conditions. However, the unwarranted accumulation of BBX22 in cop1 or cop1BBX22-GFPox plants under both conditions leads to the exaggerated inhibition of hypocotyl elongation and hyperaccumulation of anthocyanin by activating light-responsive genes and repressing hormone-responsive and cell wall expansion genes (Fig. 8). The short half-life of BBX22 protein (Fig. 3) allows Arabidopsis seedlings to adjust downstream gene expression in a timely manner in response to environmental light changes.
Degradation Mechanism of BBX22 Protein
Our data provide evidence to support that, in the dark, COP1 is required for the selective degradation of BBX22 (Fig. 5). Because BBX22 does not physically interact with COP1 (Datta et al., 2008), identifying any proteins assisting in the targeted degradation of BBX22 by COP1 is of great interest. Possible candidates are early flowering 3 (ELF3) and suppressor of phyA-105, which are COP1-interacting partners and function to facilitate COP1-mediated degradation of GIGANTEA and HY5/CONSTANS, respectively, in the dark (Saijo et al., 2003; Laubinger et al., 2006; Yu et al., 2008). Also, reduced expression of CULLIN4 enhanced the cop phenotype in cop1-4 (Chen et al., 2010), cop10, and det1 (Chen et al., 2006). This finding suggests that, in addition to COP1, CULLIN4 is needed to suppress photomophogenic growth in the dark. Because CULLIN4 is proposed to assist COP1-mediated HY5 degradation (Chen et al., 2006), whether CULLIN4 also accelerates COP1-mediated BBX22 degradation in the dark could be examined.
Although COP1 exists predominantly in the cytoplasm in the light (von Arnim and Deng, 1994), a recent study reported that the nuclear phyB degradation under red light is mediated by COP1 (Jang et al., 2010). BBX22 is also a nuclear protein, and its degradation in the light depends in part on HY5 (Fig. 6). This finding suggests that a small portion of BBX22 protein could be degraded in a protein complex composed of BBX22, HY5, and COP1, as was proposed previously (Datta et al., 2008). However, BBX22 is still largely degraded in hy5 and hy5hyh (Fig. 6). This observation suggests that an unknown factor(s) contributes mainly to BBX22 degradation in the light. Further identification of BBX22-interacting partners by coimmunoprecipitation and proteomic characterization will help in the study of the time-dependent degradation of BBX22 in the light. This mechanism is likely common for the timely elimination of positive regulators in light signal transduction pathways.
The Role of BBX22 in Arabidopsis Rhythmic Growth
bbx22 has a light-hyposensitive phenotype under SD conditions because of an increased hypocotyl elongation rate in the dark phase (Fig. 4). Our results indicate that bbx22 still exhibits rhythmic growth, unlike the arrhythmic growth patterns observed in the light perception and signaling mutants hy2 and hy5 (Nozue et al., 2007). Consistent with BBX22 carrying a branch of HY5-mediated light-signaling outputs, the light-mediated inhibition of hypocotyl elongation during the light phase is intact in bbx22 but absent in hy5. Interestingly, in addition to an increased growth rate at dawn, bbx22 also has an accelerated growth rate in the first half of the dark phase, as was observed in the circadian clock mutants CCA1ox, elf3, and elf4 (Nozue et al., 2007). A previous study indicated that the expression of BBX22 is regulated by the circadian clock (Kumagai et al., 2008). Whether BBX22 conveys the circadian information for the regulation of rhythmic growth is also of interest.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
bbx22, bbx22BBX22ox, and hy5BBX22ox plants were described in our previous study, and LZF1 was the original name (Chang et al., 2008). hy5BBX22ox was backcrossed to Ler to generate BBX22ox plants. hy5BBX22ox was crossed to hy5-ks50hyh (Holm et al., 2002) to generate hy5hyhBBX22ox plants. For generating BBX22-GFPox transgenic plants, the BBX22 coding region was amplified and fused to GFP in frame in a vector of 35S:GFP (Lee et al., 2001). The fragment of p35S:BBX22-GFP was subcloned into pCAMBIA1390 for generating transgenic Arabidopsis (Arabidopsis thaliana) overexpressing BBX22-GFP. Transgenic Arabidopsis expressing 35S:BBX22-GFP was used for crossing with two weak cop1 alleles, cop1-4 and cop1-6 (McNellis et al., 1994).
Plants were grown on one-half-strength Murashige and Skoog medium with 1% Suc and 0.3% gelrite at 4°C for 4 d to synchronize the germination. Seedlings were grown at 22°C under a 16-h/8-h or an 8-h/16-h light (100 μmol m−2 s−1)/dark photoperiod for 4 d for phenotype observation under LD versus SD conditions. For data shown in Figures 1, 2, and 6, 4-d-old etiolated seedlings were illuminated with 100 μmol m−2 s−1 light for the times indicated before RNA and protein extraction.
Growth Rate Analysis
Plants were grown and analyzed essentially as described (Nozue et al., 2007) except that the growth medium was one-half-strength Murashige and Skoog medium with minimal organics with 1% Suc; images were captured with the use of a PixeLINK PL-A781 camera driven by LabView (National Instruments). For growth rate calculations, we did not use a rolling average; instead, change in growth was calculated for each 30-min time, local polynomial regression fitting (Loess) smoothing with smoothing parameter = 0.15 was used in R (R Development Core Team, 2009), and results were plotted with the use of the ggplot2 plotting package (Wickham, 2009).
Immunoblot Analysis and Degradation Kinetics
To generate a BBX22-specific antibody, we used the C-terminal 143 amino acids (C-143) as antigen to reduce cross-recognition of antiserum of BBX22 and other BBX proteins (Khanna et al., 2009). C-143 of BBX22 was constructed into pET28a(+) and expressed in Rosetta2 (DE3) cells (Novagen) for generating rat anti-BBX22 polyclonal antibody. Total protein was isolated as described (Al-Sady et al., 2006) with minor modifications. In brief, seedlings underwent extraction with boiled extraction buffer (4 m urea, 5% SDS, 15% glycerol, 100 mm Tris-HCl, pH 8, with freshly added 10 mm 2-mercaptoethanol, 2 mm phenylmethylsulfonyl fluoride, 2 μg mL−1 apopectin, 1 μg mL−1 pepstatin, 3 μg mL−1 leupeptin, and 1× complete protease inhibitor [Roche]). In total, 30 to 50 μg of protein was separated on 4% to 12% NuPAGE Bis-Tris gels (Invitrogen), and blots were probed with the anti-BBX22 antiserum and then horseradish peroxidase-conjugated anti-rat antiserum (Sigma) as a secondary antibody. Immunoblotting with anti-α-tubulin antiserum (Sigma) and alkaline phosphatase-conjugated anti-mouse antiserum (Santa Cruz Biotechnology) was used to measure endogenous α-tubulin as a loading control. Chemiluminescence horseradish peroxidase substrate (Millipore) was used for signal detection.
For determination of degradation kinetics, seedlings were immersed in 100 μm cycloheximide and incubated under light or dark conditions for the times indicated. The quantity of protein was analyzed by immunoblotting and quantified by the Biospectrum 600 Imaging System (UVP). Protein half-lives were calculated by regression analysis. For determining the proteasome-dependent degradation of BBX22, seedlings were preincubated with 50 μm MG132 (Biomol/Enzo) for 1 h before the addition of cycloheximide.
For determining BBX22 protein levels at different times of day, 3-d-old seedlings grown under LD or SD conditions were harvested at the times indicated starting at dawn of day 4 for protein extraction and immunoblot analysis. BBX22 protein levels were shown as relative values to BBX22 at 0 h under LD conditions. Areas under the BBX22 protein levels versus time curves were obtained by use of the Area below Curves macro (trapezoidal rule) in SigmaPlot 9 (Systat Software).
Real-Time Quantitative RT-PCR and Northern-Blot Analysis
Total RNA was isolated and analyzed by real-time quantitative RT-PCR as described (Chang et al., 2008). A total of 3 μg of RNA was separated on a 1% formaldehyde-agarose gel and transferred to a Hybond N+ membrane (GE Healthcare). The full-length coding region of BBX22 was used to generate a gene-specific probe by incorporating DIG-11-UTP (DIG RNA labeling mix; Roche) by PCR. Hybridization and signal detection were performed as suggested by the manufacturer.
Affymetrix ATH1 Genome Array Hybridization and Data Analyses
Total RNA from 4-d-old etiolated wild-type, cop1-4, and cop1-4BBX22-GFPox seedlings was isolated and applied to the Arabidopsis ATH1 Genome Array (ATH1-121501; Affymetrix) for gene expression analysis as described (Chang et al., 2008). MicroArray Suite 5.0 (Affymetrix) and GeneSpring 7.3 (Agilent Technologies) were used for chip quantification, normalization, and further analysis. In brief, the intensity of all probe sets of each chip was scaled up to 500 for equivalent chip-to-chip comparison. The wild type, cop1-4, and cop1BBX22-GFPox underwent pairwise expression comparison for each probe set. Only genes with FDR of less than 5% (significance analysis of microarrays; Tusher et al., 2001) in triplicate biological repeats were selected for further analyses. Genes with 2-fold or greater or 0.5-fold or less change in expression levels between cop1BBX22-GFPox and the wild type were analyzed for association with the use of Gene Ontology and GeneSpring GX 10. Among those, genes with greater than 2-fold change in level between cop1BBX22-GFPox and cop1-4 were considered BBX22 regulated.
The data sets have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through the Gene Expression Omnibus series accession number GSE22983.
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library with the following locus identifiers: BBX22 (At1g78600), COP1 (At2g32950), HY5 (At5g11260), and HYH (At3g17609).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. BBX22 expresses as a single-sized transcript.
Supplemental Table S1. A list of 207 BBX22-regulated genes.
Acknowledgments
We thank Hsu-Liang Hsieh and Christian Hardtke for providing seeds of cop1-4, cop1-6, and hy5hyh. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy), supported by Academia Sinica.
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP. (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti A. (1992) A survey of exact inference for contingency tables. Stat Sci 7: 131–153 [Google Scholar]
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Buer CS, Djordjevic MA. (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 60: 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Fankhauser C, Coupland G, Blázquez MA. (2004) Signalling for developmental plasticity. Trends Plant Sci 9: 309–314 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Yanovsky MJ. (2005) Regulation of gene expression by light. Int J Dev Biol 49: 501–511 [DOI] [PubMed] [Google Scholar]
- Casazza AP, Rossini S, Rosso MG, Soave C. (2005) Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Mol Biol 58: 41–51 [DOI] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al. (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, et al. (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. (2006) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L. (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103: 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Norén H, Reiser V, Dunaeva M, Andersson B, Adamska I. (2006) Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol 142: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH. (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Kim HJ, Ryu JS, Choi H, Jeong S, Shin J, Choi G, Nam HG. (2008) CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J 55: 361–371 [DOI] [PubMed] [Google Scholar]
- Hu W, Ma H. (2006) Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J 45: 399–422 [DOI] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC. (2009) A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2: 166–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW. (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. (2007) Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T. (2008) The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Kwon YR, Lee HJ, Kim KH, Hong SW, Lee SJ, Lee H. (2008) Ectopic expression of Expansin3 or Expansinbeta1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol Lett 30: 1281–1288 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R. (2008) Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3: e1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim DH, Kim YW, Hwang I. (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J, Steinhauser D, Altmann T, Kopka J, Müssig C. (2005) Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucleic Acids Res 33: 2685–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW. (1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL. (2008) Dawning of a new era: photomorphogenesis as an integrated molecular network. Curr Opin Plant Biol 11: 4–8 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000a) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. (2000b) The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol 124: 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DK, Crosby KC, Runac J, Howard BA, Winkel BS. (2008) Biochemical and genetic characterization of Arabidopsis flavanone 3beta-hydroxylase. Plant Physiol Biochem 46: 833–843 [DOI] [PubMed] [Google Scholar]
- Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO. (2008) Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA 105: 21023–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka F, Irani NG, Feller A, Lu Y, Pourcel L, Frame K, Grotewold E. (2007) A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol 145: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: http://www.R-project.org (April 1, 2010) [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini S, Casazza AP, Engelmann EC, Havaux M, Jennings RC, Soave C. (2006) Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol 141: 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2: e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhou XY, Li L, Xue LJ, Yang X, Xue HW. (2009) Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol Plant 2: 755–772 [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. (2004) Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc Natl Acad Sci USA 101: 3304–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Franck F, Alawady AE, Dall’Osto L, Carrière F, Bassi R, Grimm B, Nussaume L, Havaux M. (2007) The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J 50: 795–809 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Osterlund MT, Kwok SF, Deng XW. (1997) Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol 114: 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer, New York
- Yao J, Roy-Chowdhury S, Allison LA. (2003) AtSig5 is an essential nucleus-encoded Arabidopsis sigma-like factor. Plant Physiol 132: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]