Abstract
In this study, we show that the Arabidopsis (Arabidopsis thaliana) transcription factor MYB46, previously described to regulate secondary cell wall biosynthesis in the vascular tissue of the stem, is pivotal for mediating disease susceptibility to the fungal pathogen Botrytis cinerea. We identified MYB46 by its ability to bind to a new cis-element located in the 5′ promoter region of the pathogen-induced Ep5C gene, which encodes a type III cell wall-bound peroxidase. We present genetic and molecular evidence indicating that MYB46 modulates the magnitude of Ep5C gene induction following pathogenic insults. Moreover, we demonstrate that different myb46 knockdown mutant plants exhibit increased disease resistance to B. cinerea, a phenotype that is accompanied by selective transcriptional reprogramming of a set of genes encoding cell wall proteins and enzymes, of which extracellular type III peroxidases are conspicuous. In essence, our results substantiate that defense-related signaling pathways and cell wall integrity are interconnected and that MYB46 likely functions as a disease susceptibility modulator to B. cinerea through the integration of cell wall remodeling and downstream activation of secondary lines of defense.
The cell wall is a dynamic cell compartment that confers unique and distinctive features to plant cells. Structurally, it has been long recognized as exhibiting specific functions essential to the entire plant (Cassab and Varner, 1988). The construction and architecture of the cell wall vary according to cellular developmental stages (Varner and Lin, 1989). The pliable character of cell walls is directly related to changes in the proportion and degree of assembly of various structural polysaccharides, including cellulose microfibrils, pectin, and hemicellulose polymers (Carpita and McCann, 2000). In addition, cell walls of defined cell types originate due to the deposition of specific polymers with defined functions, such as lignin, or by the presence of different structural proteins. Cell wall synthesis is a process highly regulated at the transcriptional level. A group of transcription factors in the NAC and MYB families have recently been shown as key players in regulating secondary cell wall biosynthesis, including SND1 (Zhong et al., 2006), MYB46 (Zhong et al., 2007; Ko et al., 2009), NST1 and NST3 (Mitsuda et al., 2007), and MYB58 and MYB63 (Zhou et al., 2009), among others.
The plant cell wall is also considered a preformed barrier to pathogen infection (Vorwerk et al., 2004; Hückelhoven, 2007). The failure of pathogenic microorganisms, notably fungal pathogens, to breach plant cell walls constitutes a major component of immunity for nonhost plant species (i.e. species outside the pathogenic host range) and is responsible for a proportion of aborted infection attempts on susceptible host plants (basal resistance; Hückelhoven, 2007). In addition, cell walls are dynamic reservoirs of antimicrobial proteins and secondary metabolites that inhibit the growth of many pathogens (Darvill and Albersheim, 1984). However, little is known about the degree to which the chemical composition of plant cell wall polysaccharides and other constituents are a factor in the outcome of the plant-pathogen interactions. Some pathogens are able to generate enough mechanical force to penetrate cell walls. However, others secrete enzymes that degrade cell walls. This infection strategy facilitates the penetration of pathogenic structures into the interior of the plant cell, ensuring pathogen survival. Consequently, one of the earliest evolutionary responses to pathogen attack was cell walls reinforced by specific cross-linking of cell wall constituents, which were favored by selection, a process mediated by hydrogen peroxide (H2O2) and by cell wall-bound oxide reductase enzyme activities (Wojtaszek, 1997). Therefore, it appears likely that during coevolution with pathogens, plants developed cell wall integrity surveillance systems to sense pathogen-driven perturbations that subsequently alerted the cell to activate defense-related signal transduction pathways.
Extracellular type III peroxidases are key players in secondary cell wall remodeling and have been proposed to act in the polymerization of phenolic monomers into lignin and suberin or to mediate the cross-linking of polysaccharides and proteins in mice (Passardi et al., 2004). More recently, the expression pattern of the Ep5C gene from tomato (Solanum lycopersicum) plants encoding a cell wall type III peroxidase was used as a marker to characterize early transcription-dependent responses controlled by H2O2 following cell wall detection of a pathogen, a mode of gene activation observed to be conserved in tomato and Arabidopsis (Arabidopsis thaliana; Coego et al., 2005a). Notably, the study demonstrated that H2O2 plays a dual role for Ep5C: it functions as a cosubstrate for the encoded peroxidase enzyme and as a gene induction signal. Ep5C::GUS was successfully used as a marker to search for novel defense components participating in plant defense-related pathways. The result was the identification of overexpressor of cationic peroxidase (ocp) mutants in Arabidopsis, which showed a deregulated Ep5C::GUS expression pattern associated with altered disease susceptibility response to pathogens (Coego et al., 2005b; Agorio and Vera, 2007; Ramírez et al., 2009).
In this study, we performed a functional analysis of the Ep5C 5′ promoter region in Arabidopsis to identify cis-regulatory elements important for gene induction and examined the corresponding trans-acting factors. We show that MYB46 modulates the magnitude of Ep5C gene induction following a pathogenic insult. Moreover, we demonstrate that myb46 mutant plants exhibit increased disease resistance to the necrotrophic fungal pathogen Botrytis cinerea, a phenotype that is accompanied by selective transcriptional reprogramming of a set of genes encoding cell wall proteins and enzymes, of which extracellular type III peroxidases are conspicuous.
RESULTS
Identification of a Specific Protein-Binding Site within the Ep5C Promoter Region
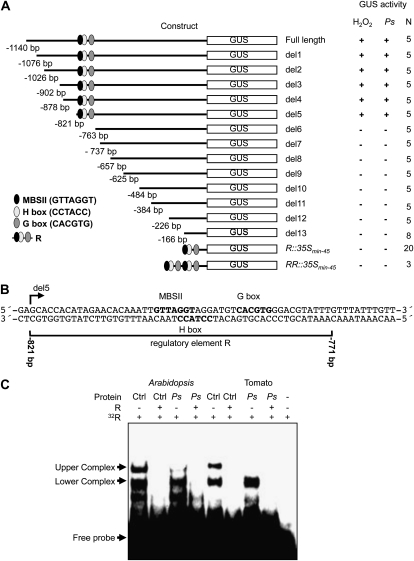
To identify DNA sequence elements important in mediating the induction of Ep5C gene expression, the original 1,140-bp full-length Ep5C promoter region was progressively deleted from its 5′ end, and the resulting deletions, once fused to the uidA gene, were introduced into Arabidopsis by Agrobacterium tumefaciens-mediated stable transformation. Detection of GUS activity was recorded in transgenic plants following leaf infiltration with a 1 mm H2O2 solution. Through a series of 5′ deletions shown in Figure 1A, we observed that 821 bp of promoter sequence (construct del5) maintained complete responsiveness to H2O2, while 763 bp of promoter sequence (construct del6) lost responsiveness, demonstrated by the absence of GUS induction recovery in any of the transgenic lines. Similarly, gene induction was never recovered in any of the successive downstream deletions shown in Figure 1A. A similar responsive effect was observed in the deletion mutant constructs following plant inoculation with the pathogen Pseudomonas syringae DC3000 (PsDC3000). These results indicated that sequence elements adjacent to or between positions –821 and –763 are pivotal for Ep5C gene induction. Inspection of conserved motifs within this promoter interval (Fig. 1B) revealed the presence of a conserved GTTAGGT cis-acting element between positions –806 and –798, which is characteristic of some MYB type II-regulated genes (MBSII; Yang and Klessig, 1996; Gubler et al., 1999). Furthermore, adjacent to this motif, an H box-like sequence was detected (CCTACC; at –797 to –792; Loake et al., 1992). In addition to these two elements and downstream of the H box-like sequence, a G box was identified (CACGTG; at –787 to –782; Schulze-Lefert et al., 1989). The 51-bp sequence (from –821 to –771) was designated regulatory element R (Fig. 1B). A similar regulatory element characterized by the overlapping of a MBSII and an H box element, which may be in proximity to a G box, has been shown to be involved in the transcriptional regulation of some phenylpropanoid pathway genes (Loake et al., 1992; Sablowski et al., 1994; Abe et al., 2003; Hartmann et al., 2005). Interestingly, when a single 510-bp R element or a tandem duplication was fused to a minimal cauliflower mosaic virus (CaMV) 35S promoter (construct R::35Smin-45 and construct RR::35Smin-45 in Figure 1A) to control GUS expression and used to generate transgenic plants, GUS induction could not be recovered in any of the transgenic lines generated upon H2O2 treatment or upon inoculation with PsDC3000. These results indicated that the R element, although important for induction of Ep5C, is not sufficient per se to drive all major aspects of transcriptional regulation mediated by H2O2. It remains possible that other promoter elements downstream of position –763, acting at a distance to the R element, are required for full transcriptional activation of Ep5C.
Figure 1.
Functional analysis of the Ep5C gene promoter in transgenic Arabidopsis plants and R motif identification. A, A series of unidirectional deletions of the Ep5C promoter were transcriptionally fused to the GUS reporter gene and used to generate transgenic Arabidopsis plants. Deletions were named sequentially from del1 to del13. The positions of known cis-acting elements are shown as indicated at bottom left. Constitutive and inducible reporter gene expression was analyzed. + and − symbols denote detection and absence of GUS activity in leaves following infiltration with 1 mm H2O2 or inoculation with PsDC3000 (Ps). The construct containing the 51-bp R sequence, single or duplicated, fused to the 45-bp minimal 35S promoter is shown at the base of the figure. The number of independent lines (N) analyzed for each construct is indicated on the right. B, The nucleotide sequence comprising the region between del5 and del6 of the Ep5C promoter region. The sequence corresponding to the 51-bp (–821 to –771) R element is bracketed below. Numbers denote nucleotide positions relative to ATG. C, EMSA using the R element as a probe and whole-cell extract from mock-inoculated control (Ctrl) leaves and leaves inoculated with PsDC3000 from Arabidopsis and tomato plants. Leaf samples were taken at 4 h.p.i. + and − symbols denote the presence and absence of a 100-fold excess of a nonradiolabeled R competitor. Arrows on the left indicate the appearance of retarded upper and lower complexes.
A potential protein-DNA interaction within the identified Ep5C R element promoter region was tested by performing electrophoretic mobility shift assays (EMSA) with protein extracts derived from control (mock-inoculated) and PsDC3000-infected Arabidopsis plants. It was previously demonstrated that PsDC3000 promotes transcriptional activation of Ep5C::GUS in transgenic Arabidopsis through the generation of H2O2 at infection sites (Coego et al., 2005a). We concurrently inoculated Arabidopsis and tomato leaves with the same bacterial strain, prepared protein extracts, and performed EMSA. A double-stranded oligonucleotide comprising the identified R element was used as a probe. Results are shown in Figure 1C. Two retarded protein complexes (upper and lower complexes in Figure 1C) were detected in Arabidopsis control leaves. The interaction of both complexes was sequence specific, indicated by the prevention of complex formation by adding an excess of unlabeled probe. Interestingly, in EMSA performed using protein extracts derived from PsDC3000-inoculated leaves (at 4 h post inoculation [h.p.i.]), the upper but not the lower complex was dismantled. Similarly, reproducible DNA-binding activity and behavior were detected in EMSA experiments using protein samples derived from tomato plants (Fig. 1C). This congruence indicates that the molecular components mediating this specific binding are highly conserved between the two plant species. Moreover, dismantling the upper complex in either Arabidopsis or tomato leaves coincides with transcriptional activation of Ep5C. This result suggests that the upper complex seems to be formed by the binding to the R element of a constitutively present transcriptional repressor.
The specific sequences within the 51-bp R element involved in the formation of the upper and the lower complexes were identified using competition experiments with different DNA probes (Supplemental Fig. S1). The results indicated that the 31-bp sequence present in the R6 competitor, where the MYBII, H box, and G box motifs remained, are necessary and sufficient to support the formation of the upper protein complexes. Furthermore, the sequence between positions 30 and 51 of the R element is dispensable or at least accessory for the lower complex formation. The remaining competitors exhibiting an effect in the lower complex formation share the DNA sequence between positions 20 and 35, a region that includes the MSBII and H box but not the G box.
Isolation of cDNAs Encoding DNA-Binding Proteins That Recognize the R Motif
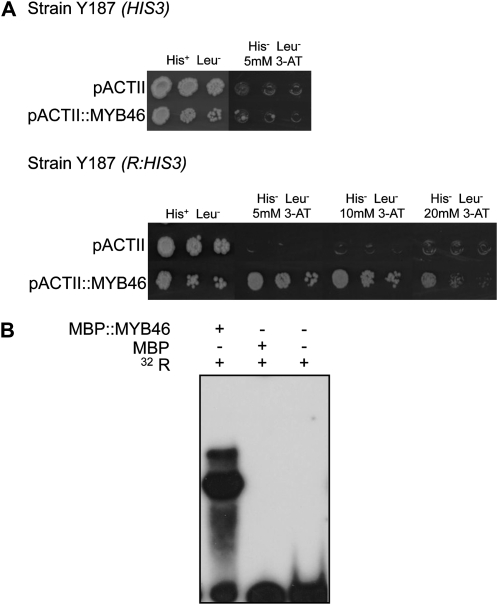
The yeast one-hybrid screening system was used to isolate cDNAs encoding DNA-binding proteins that interact with the R motif. A R::HIS3 gene construct was assembled that carried a single copy of the R element in plasmid pINT1 (Meijer et al., 1998) and used to transform yeast strain Y187 (Fig. 2A). cDNAs encoding DNA-binding proteins of interest were screened by transforming the target reporter Y187(R::HIS3) strain with a GAL4-AD cDNA library from Arabidopsis generated in pACTII (Memelink, 1997) and screened approximately 4.3 × 105 colony-forming units. Six 3-aminotriazole (3-AT)-resistant clones were isolated using this approach. Following sequencing analyses, the cDNA fragments were determined to encode the same protein. BLAST analysis identified the isolated cDNAs as At5g12870, which encodes the MYB46 transcription factor, a MYB member of the R2R3 gene family (Martin and Paz-Ares, 1997; Stracke et al., 2001).
Figure 2.
MYB46 binding to the R element. A, Top, Growth of yeast strain Y187(HIS3) in a complete minimal medium with and without 5 mm 3-AT and carrying MYB46 cDNA in vector pACTII or not carrying the MYB46 cDNA. Bottom, growth of the same yeast carrying the HIS3 gene under the control of the R element [strain Y187(R:HIS3)] in complete medium without 3-AT or with increasing concentrations of 3-AT and carrying the MYB46 cDNA in vector pACTII or not carrying the MYB46 cDNA. B, EMSA indicating the specific binding of affinity-purified recombinant MBP::MYB46 proteins, but not MBP alone, to the R element. + and − symbols denote the absence and presence of the relevant component.
The MYB46 clones were transformed into the Y187 yeast strain carrying HIS3 alone, without the presence of the 51-bp R element. The results showed that the recombinant yeast strain did not grow on the medium lacking His in the presence of 3-AT (Fig. 2A), further supporting the requirement of both the R element and MYB46 to regulate yeast transcription.
Purified MYB46 Binds Specifically to the MBSII Sequence of the R Motif
The in vivo yeast one-hybrid experiment was validated in vitro by testing if MYB46 protein specifically interacts with the R motif. We used Escherichia coli to produce a recombinant MYB46-Maltose Binding Protein (MBP::MYB46) fusion protein (Supplemental Fig. S2). Purified MBP::MYB46 protein and the MBP alone were incubated with the R probe, and the incubation mixtures were analyzed by EMSA (Fig. 2B). Whereas the MBP::MYB46 protein caused a distinct shift in the mobility of the radiolabeled probe, MBP alone produced no such shift (Fig. 2B), indicating that the MYB46 protein specifically binds to the 51-bp R element. A precise identification of the specific binding site of MYB46 within the R element was carried out using a DNA-methylation interference assay, a high-resolution method that identifies critical guanine residues involved in sequence-specific recognition by proteins. Using a top-strand end-labeled R probe, this assay revealed that guanine residues in the upper strand of the R element, at positions 21, 25, and 26, are critical for binding of MYB46 and are precisely located in MBSII, which was identified in the R element (Supplemental Fig. S2). We can infer from this result that MYB46 binds to the R element through the MBSII motif and not to the H box or other sequences in the R1 element. This was further confirmed by site-directed mutagenesis of the R element and resolution of MYB46 binding using EMSA (Supplemental Fig. S3). These assays revealed that direct substitution of the guanine residues by thymine residues in the MBSII motif within the context of the full R element (e.g. the RMBSII probe in Supplemental Fig. S3), but not other mutations (e.g. the RINTER, RGBOX, RCORE, and RFLANK probes), impeded the formation of the DNA-MYB46 complex. Cumulatively, these results suggest that MBSII is the cis-element within the R element for MYB46 binding.
MYB46 Gene Expression Does Not Change following Interaction with Pathogens
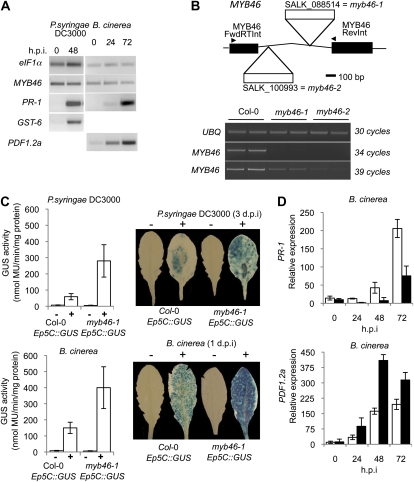
Coego et al. (2005a) reported the activation of Ep5C::GUS gene expression in Arabidopsis by local generation of H2O2 following inoculation with pathogens. MYB46 expression was analyzed by reverse transcription (RT)-PCR under the same conditions following inoculation with the bacterial pathogen PsDC3000 and the fungal pathogen B. cinerea (Fig. 3A). MYB46 was expressed at low levels in leaves of intact plants, and its expression was not affected by infection with either of the two pathogens, whereas disease-related marker genes used to monitor the response of the plant to the bacterial (e.g. PR1 or GST-6) and fungal (PDF1.2a) infection were up-regulated (Fig. 3A). Therefore, MYB46 expression appeared unaltered during the course of plant-pathogen interactions.
Figure 3.
Loss of MYB46 results in faster and increased induction of Ep5C::GUS expression. A, RT-PCR analysis of MYB46, PR-1, GST-6, and PDF1.2 expression in Arabidopsis (Col-0) plant leaves at different times following inoculation with the virulent pathogens PsDC3000 and B. cinerea. B, MYB46 gene structure showing localization of the T-DNA insertion in the SALK_100993 (myb46-1) and SALK_088514 (myb46-2) mutant lines. RT-PCR analysis of MYB46 expression in Col-0 and homozygous myb46-1 and myb46-2 plants is shown; the housekeeping gene UBQ was included as the control for RNA loading. C, Comparative induced expression of GUS activity driven by Ep5C::GUS following PsDC3000 (top) and B. cinerea (bottom) inoculation in Col-0 and myb46-1 backgrounds. PsDC3000 was inoculated by infiltration and B. cinerea by spraying leaves with a spore suspension. Samples were obtained 3 d after inoculation. The left panel depicts a comparative histochemical analysis of GUS activity in fully expanded rosette leaves from a parental wild-type plant (white bars) and a myb46-1 mutant plant (black bars) carrying the Ep5C:GUS transgene. The experiments were repeated three times with similar results. D, Quantitative RT-PCR analysis for PR-1 and PDF1.2a marker gene expression at different h.p.i. with B. cinerea in Col-0 (white bars) and myb46-2 (black bars) plants. Data represent means ± sd (n = 3 biological replicates).
A myb46 Mutant Shows Enhanced Induction of Ep5C::GUS following Pathogen Infection
We reasoned that plants lacking MYB46 should show an altered Ep5C::GUS induction pattern following pathogen inoculation. Consequently, we characterized two T-DNA insertion mutants (Fig. 3B) obtained from the Salk Institute Genomic Analysis Laboratory. Both mutants contained the T-DNA insertion located at different positions in the MYB46 intron. These two mutants were named myb46-1 (SALK_088514) and myb46-2 (SALK_100993; Fig. 2B), with myb46-1 the first to be deposited in the collection. Homozygous plants for each of these two mutations were generated, which did not reveal visible alterations in growth habit or any gross developmental defect. Gene expression analysis by RT-PCR (Fig. 3B) revealed that the two mutants exhibited markedly reduced levels of MYB46 mRNAs; the reduced expression was more pronounced in myb46-2 than in myb46-1 plants.
We subsequently introgressed the myb46-1 mutation into the Ep5C::GUS transgenic line and generated homozygous myb46-1 Ep5C::GUS plants. Similar to the Ep5C::GUS parental plants, myb46-1 Ep5C::GUS plants did not show constitutive expression of the reporter gene (Fig. 3C). However, following leaf inoculation with PsDC3000 or spraying with spores of B. cinerea, myb46-1 Ep5C::GUS plants responded with an enhanced induction of GUS activity markedly higher than that attained by the parental Ep5C::GUS plants (Fig. 3C). The enhanced induction of GUS activity extended along the blade of the inoculated leaves (observed following staining with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid; Fig. 3C). These results indicated that a derepression mechanism is a fundamental aspect of the Ep5C mode of regulation during the course of plant-pathogen interaction and that dismantling of MYB46 facilitates gene induction. Interestingly, the enhanced induction of GUS gene expression following inoculation with B. cinerea was congruent with data reported by Turner et al. (2002), where enhanced induction of PDF1.2a was observed, a marker gene for jasmonic acid that monitors plant responses to infection by necrotrophic fungal pathogens (Fig. 3D). This was not the case for the induction of PR-1, a marker gene for salicylic acid-mediated responses (Ward et al., 1991), which exhibited reduced induction in the mutant relative to the wild-type plants (Fig. 3D). This latter observation is congruent with a demonstrated negative control, mediated by cross-talk mechanisms, performed by the jasmonic acid pathway over the salicylic acid pathway (Koornneef and Pieterse, 2008).
myb46 Mutants Have Enhanced Disease Resistance to B. cinerea
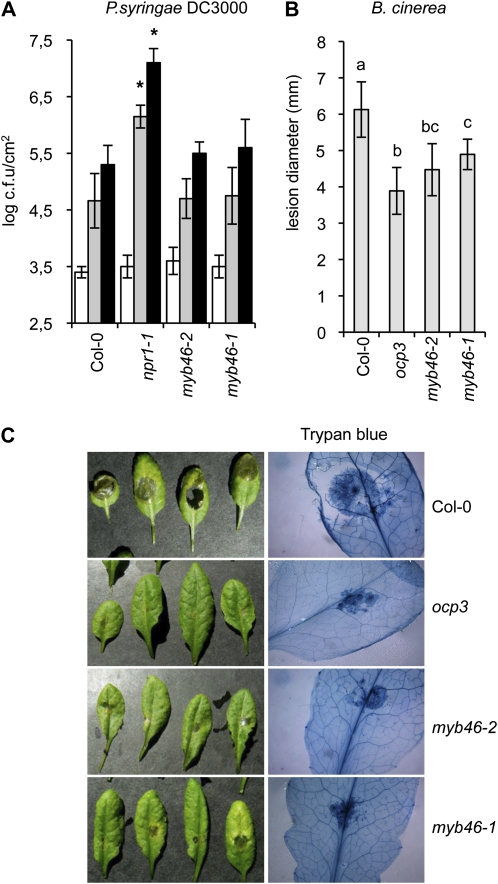
The previous results prompted us to address the presence of a causal link between the increased Ep5C::GUS activation following pathogen inoculation in myb46 plants and disease susceptibility to pathogens. We tested myb46 mutant responses to the virulent bacteria PsDC3000 and the virulent fungal pathogen B. cinerea. The response of myb46-1 and myb46-2 plants to PsDC3000 compared with wild-type plants is shown in Figure 4A. The hypersusceptible mutant npr1-1 (Cao et al., 1997) was used as a control. Bacterial growth rate in extracts from inoculated leaves of ecotype Columbia (Col-0), myb46-1, and myb46-2 plants revealed that the disease susceptibility of myb46 mutants toward PsDC3000 did not vary from the wild type.
Figure 4.
myb46 plants exhibit enhanced resistance to B. cinerea but not to PsDC3000. A, Histogram of PsDC3000 growth rate in Col-0, myb46-1, myb46-2, and npr1-1 plants. The bacterial titer was determined at 0 d (white bars), 3 d (gray bars), and 5 d (black bars) after infection. Data represent means ± se (n = 8 independent individuals). Asterisks indicate significant differences from the control (P < 0.05) using Student’s t test. The experiment was repeated three times with similar results. c.f.u., Colony-forming units. B, Resistance response of Col-0, ocp3, myb46-1, and myb46-2 plants to B. cinerea was evaluated 3 d post inoculation by determining the average lesion diameter on three leaves from 15 plants each. Data points represent average lesion size ± se of measurements. An ANOVA was conducted to assess significant differences in disease symptoms with a 0.05 level of significance. Different letters indicate significant differences between genotypes. C, Lactophenol-trypan blue staining of representative leaves from ocp3, myb46-1, and myb46-2 plants at 4 d post inoculation with a 6-μL droplet of B. cinerea spores (2.5 × 104 conidia mL−1; as in B).
However, results demonstrated that the myb46 mutations elicited changes in B. cinerea disease susceptibility, with a significant shift from susceptibility to resistance. Plants were scored for disease symptoms by following the level of necrosis appearing in the inoculated leaves (Fig. 4B). The enhanced disease resistance mutant ocp3 (Coego et al., 2005b; Ramírez et al., 2009) was used as an internal control. As expected, wild-type plants were highly susceptible to B. cinerea, and inoculated plants showed extensive necrosis accompanied by widespread proliferation of the fungal mycelia revealed by trypan blue staining (Fig. 4C). In contrast, myb46-1 and myb46-2 plants showed a significant reduction in the extent of necrosis (Fig. 4C); relative to myb46-1, myb46-2 plants consistently exhibited a slightly enhanced disease resistance to B. cinerea. In addition, proliferation of fungal mycelia was notably inhibited in the two mutants. As expected, ocp3 plants showed similar and even increased disease resistance (Fig. 4C). These results indicated that disease susceptibility to B. cinerea is a characteristic trait linked to MYB46.
Overexpression of MYB46 Does Not Alter Disease Susceptibility to B. cinerea
We investigated whether overexpression of MYB46 resulted in changes in plant response to pathogen infection. We generated transgenic plants showing increased MYB46 expression under the control of the CaMV 35S promoter (MYB46ox; Supplemental Fig. S4A). Several homozygous overexpression lines (all MYB46 overexpression lines showed similar results, so only data obtained with one or two of these lines are shown) were compared with wild-type plants for their responses to B. cinerea and PsDC3000 (Supplemental Fig. S4, B and C). Levels of disease resistance to both pathogens were quantified and indicated that MYB46 overexpression did not differ from the normal disease susceptibility of Col-0 plants. These results suggested that the normal endogenous level of MYB46 present in wild-type plants is sufficient to accomplish its normal role during disease. Congruently, the genetic introduction of one of the MYB46ox insertions (i.e. MYB46ox4.2), in the transgenic background containing the Ep5C::GUS gene, did not cause an alteration in the normal pattern of GUS induction following PsDC3000 or B. cinerea inoculation (Supplemental Fig. S4D).
myb46-2 and MYB46ox4.2 Plants Show Differences in the Deposition of Lignin in Stems
MYB46 is proposed to function as a transcriptional activator of secondary cell wall biosynthesis and lignin deposition in xylem fibers and vessel elements (Zhong et al., 2007). In fact, dominant repression of MYB46 in transgenic plants, due to fusion with the EAR-dominant repression domain, resulted in an apparent reduction in secondary cell wall thickening of the interfascicular fibers and concomitant atypical developmental alterations (Zhong et al., 2007). However, it should be noted that the phenotypes caused by the dominant repression experiments might not directly reflect those of knockout or knockdown mutants, such as those characterized here for myb46-1 and myb46-2. Zhong et al. (2007) noted that dominant repressors should be evaluated with caution, as the repressor inhibits not only the functions of the transcription factors targeted for repression but also their homolog functions by competing with their binding to the same cis-element or interacting proteins.
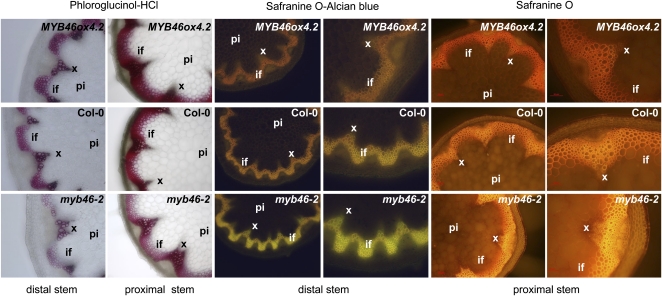
To test that the knockdown myb46 mutants coregulate increased disease resistance to B. cinerea in leaves with a defect in secondary cell wall deposition in stem fibers and vessels, we performed comparative histological examinations of stems in myb46-2, wild-type, and MYB46ox4.2 plants. myb46-2 plants exhibited a weak but detectable reduction in secondary wall thickening of interfascicular fibers, confirmed following phloroglucinol staining of cross-sections and visualization using transmitted light (Fig. 5, left panel). The reduction in phloroglucinol staining was noticeable in the distal stem and became progressively less evident in the proximal stem. Alternatively, the proximal stem of MYB46ox4.2 plants and not the distal stem exhibited enhancement of lignin deposition in the secondary cell walls of interfascicular fibers following staining with phloroglucinol. These differences in lignin deposition can be verified more succinctly by staining cross-sections of the stem with the fluorescent dye Safranin O (Stockert et al., 1984). Due to changes in fluorescence emission, Safranin can differentiate regions of high and low lignin more accurately than phloroglucionol; regions of high lignin fluoresce red/orange, and regions with low lignin fluoresce yellow. Alternatively, the differential levels of secondary cell wall deposition can be more easily visualized by counterstaining Safranin with a nonfluorescent dye such as alcian blue. This counterstaining strategy (Fig. 5, central panels) would enable us to clearly observe the differential deposition of lignin between the distal stem tissues. A differential fluorescence emission was characterized that ranges in myb46-2 plants from bright yellowish in poorly lignified portions of the distal stem, to a pale orange emission in similar portions of wild-type plant stems, to even more orange-like emission in similar distal portions of MYB46ox4.2 plant stems. The differences in fluorescence emission were also observed after staining only the proximal positions of the stems with Safranin O (Fig. 5, right panel). This result demonstrates that despite the absence of any cellular or tissue-specific anomalies, the vascular cylinder, and particularly the interfascicular fibers, of myb46-2 plants emitted yellow fluorescence, which is in marked contrast to the orange-type emission observed in MYB46ox4.2 plants. This result indicates that MYB46ox4.2 plants accumulate more stem lignin than the myb46-2 plants.
Figure 5.
Overexpression and down-regulation of MYB46 leads to changes in lignin deposition in the stem. Stem sections were stained with phloroglucinol-HCl, Safranin O, or Safranin O plus alcian blue for the detection of lignin. if, Interfascicular fibers; pi, pith parenchyma; xy, xylem. Left panels, phloroglucinol-HCl-stained (red color) distal or proximal stem sections, showing lignin deposition in the walls of the interfascicular fibers and xylem cells as observed with the light microscope. Top, middle, and bottom images show stem sections derived from MYB46ox4.2, wild-type (Col-0), and myb46-2 plants, respectively. Central panels, Safranin O-stained and alcian blue-counterstained distal stem sections, indicating an orange-to-yellow change in the staining pattern in the interfascicular fibers and xylem as observed by fluorescence microscopy. Right panels, Safranin O-stained proximal stem sections, indicating a red-to-orange change in the staining pattern in the interfascicular fibers and xylem as observed by fluorescence microscopy.
Leaves of myb46-2 Plants Do Not Show Changes in Major Polymer Constituents of Cell Walls
How can MYB46 behave as a positive regulator of secondary cell wall deposition in the stem and at the same time regulate disease susceptibility to B. cinerea in the leaf? We hypothesized that leaves of myb46 and Col-0 plants possess molecular differences related to cell wall fortification and that the differences are responsible for the observed changes in B. cinerea disease susceptibility. Consequently, we isolated cell walls from fully expanded leaves and proceeded to quantify the total amount of phenolic acids and determine how much of these phenolics were in the form of lignin. Table I indicates the total phenolic acids present in cell walls in the form of alcohol-insoluble residue (AIR) and the amount of lignin (as nonesterified phenolics) present in cell wall preparations. Significant differences were not detected in the two cell wall components of leaf samples derived from Col-0, myb46-2, and MYB46ox4.2 plants. A small increase in lignin content was observed in the cell wall preparations derived from MYB46ox4.2 leaves. This provides support to previous data demonstrating that excessive MYB46 overexpression leads to ectopic deposition of secondary walls in leaf epidermal cells (Zhong et al., 2007; Ko et al., 2009).
Table I. Total phenolic acids and lignin in leaves from Col-0, myb46-2, and MYB46ox4.2 plants.
| Plant | Total | Lignin |
| μg/mg | ||
| Col-0 | 6.8 ± 0.4 | 3.9 ± 1.4 |
| myb46-2 | 7.1 ± 0.9 | 3.8 ± 0.6 |
| MYB46ox4.2 | 6.9 ± 0.9 | 5.0 ± 0.6 |
The absence of differences prompted us to further investigate altered cell wall composition using Fourier transformed infrared (FTIR) spectroscopy, a powerful tool for plant cell wall analysis (Chen et al., 1998; Vogel et al., 2002). The FTIR absorption spectra of AIRs from Col-0, myb46-2, and MYB460x4.2 leaves between 1,800 and 900 cm−1, the IR region characteristic for cell wall polysaccharides and phenolic material, including cellulose, hemicellulose, pectin, and lignin, are shown in Supplemental Figure S5. Significant differences in the spectra of the three different genotypes were not detected. Furthermore, a principal components analysis of FTIR spectra from Col-0, myb46-2, and MYB46ox4.2 leaf samples was performed using five principal components that accounted for 99.60% of the cumulative variance. The results did not discern any significant differences among the three groups. All observations indicated that in the leaves of myb46-2 plants, cell walls do not carry alterations in the amount and proportion of the major polymer constituents that could provide an explanation for the observed enhanced disease resistance to B. cinerea.
Transcriptomic Analysis of myb46-2 Plants
The lack of differences in major cell wall polymers does not eliminate the possibility that other minor constituents of the cell wall, which might be under direct or indirect control by MYB46, are responsible for enhanced disease resistance of myb46 plants to B. cinerea. Consequently, whole-transcriptome analysis using two-color long-oligonucleotide microarrays was performed to identify genes differentially expressed in myb46-2 leaves. A total of 189 genes with 2-fold or greater change in myb46-2 versus Col-0 and P < 0.05 were identified; of these, 32 genes were up-regulated and 157 genes were down-regulated in myb46-2. The list of genes differentially expressed is provided in Supplemental Table S1. Validation of microarray data was achieved using quantitative RT-PCR. Supplemental Figure S6 shows the expression of all randomly chosen genes constitutively up-/down-regulated in myb46-2 leaves, which correlated well with microarray data.
A salient feature of the transcriptional analysis was that few genes were up-regulated in leaves when MYB46 was down-regulated. Of the 32 overexpressed genes, only PCC1, NIMIN1, ACD6, and WRKY20 correlated with disease resistance, particularly in the regulation of salicylic acid-dependent defense responses (Lu et al., 2003; Li et al., 2004; Sauerbrunn and Schlaich, 2004; Weigel et al., 2005). Gene expression encoding enzymes of the phenylpropanoid pathway was not altered in myb46-2 plants, this being consistent with the lack of variation in phenolic acid amounts deposited in the cell walls of myb46-2 leaves. In contrast, the majority of genes identified in the myb46-2 microarray corresponded to down-regulated genes. A functional classification of the genes using Gene Ontology databases (Supplemental Fig. S7) revealed a ranking of functionalities, where a significant number of genes were related to cell wall metabolism and extracellular matrix remodeling. Among them, some genes possessed encoded proteins related to biotic stress that function extracellularly in plant defense; among these were different secreted pathogenesis-related proteins such as PR-1 and a chitinase, seven lipid transfer proteins and proteinase inhibitors, seven DEFL and PDF1.2-like defensins, allergenic proteins, and a RNase. These genes were easily identified and reminiscent of a classical PR gene asset. Despite the down-regulation observed in the microarray, defense-related genes were highly induced upon inoculation of myb46-2 plants with B. cinerea and reached levels of expression similar to those attained in wild-type plants (Supplemental Fig. S8).
Among the genes more directly involved in cell wall metabolism and remodeling were different expansins (EXPA7, EXPA18, EXLB3), arabinogalactan proteins (AGP3, AGP22, AGP24), a Gly-rich protein (GRP3), Pro-rich proteins (PRP1, PRP3), a pectin methylesterase (SKS16) and its inhibitor (UNE11), two polygalacturonases, and also different enzymes involved in xyloglucan biosynthesis and modification (Supplemental Table S1). In relationship to cell wall remodeling was the presence of nine genes encoding cell wall-bound class III peroxidases (PER; Tognolli et al., 2002; Passardi et al., 2004, 2005), which appeared down-regulated in myb46-2 plants (Table II; Supplemental Table S1).
Table II. Comparative expression level of PER genes in Col-0 and myb46-2 plants under resting conditions and as derived from microarray analysis, and induction pattern and fold change at 72 h.p.i. with B. cinerea as determined by quantitative RT-PCR.
N.D., Not determined.
| Microarray Data | Quantitative RT-PCR Data | |||||
| Arabidopsis Genome Initiative No. | Protein Name | Fold Change (myb46-2 versus Col-0) | Fold Induction in Col-0 after B. cinerea Infection (72 h.p.i.) | Fold Induction in myb46-2 after B. cinerea Infection (72 h.p.i.) | Fold Change (myb46-2 versus Col-0) after B. cinerea Infection (72 h.p.i.) | |
| At1G05240 | PER1 | −2.08 | 1.1 ± 0.2 | 1.3 ± 0.3 | 1.2 | |
| At1G14540 | PER4 | −3.45 | 1.3 ± 1.8 | 8.4 ± 1.2 | 6.5 | |
| At1G71695 | PER12 | N.D. | −185.0 ± 1.0 | −112.4 ± 0.9 | −1.7 | |
| At2G37130 | PER21 | N.D. | 60.1 ± 17.9 | 168.1 ± 24.1 | 2.8 | |
| At3G49120 | PER34 | N.D. | 4.6 ± 0.5 | 6.9 ± 0.9 | 1.5 | |
| At4G08770 | PER37 | N.D. | 8.3 ± 1.0 | 16.0 ± 1.7 | 1.9 | |
| At4G11290 | PER39 | −2.07 | 1.7 ± 0.5 | 1.5 ± 0.2 | −1.1 | |
| At4G30170 | PER45 | −2.04 | 0.8 ± 0.3 | 4.9 ± 0.7 | 6.1 | |
| At5G17820 | PER57 | −2.04 | 1.0 ± 0.7 | 1.1 ± 0.9 | 1.1 | |
| At5G39580 | PER62 | −2.06 | 184.6 ± 18.7 | 984.9 ± 131.9 | 5.3 | |
| At5G64100 | PER69 | −2.15 | 31.1 ± 18.7 | 267.7 ± 55.2 | 8.6 | |
| At5G64120 | PER71 | −2.62 | 4.8 ± 0.7 | 16.6 ± 1.0 | 3.5 | |
| At5G67400 | PER73 | −2.03 | 0.8 ± 0.5 | 4.4 ± 11.3 | 5.5 | |
Rapid Induction of PER Genes in myb46 Plants following Inoculation with B. cinerea
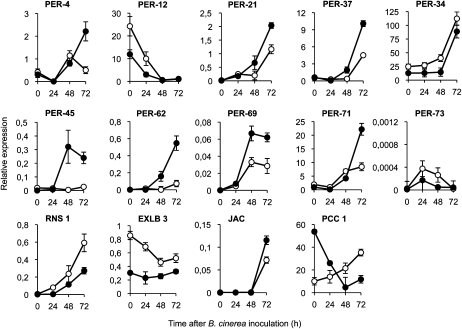
Overexpression of some peroxidases that were detected as down-regulated in myb46-2 plants (i.e. PER21, PER62, and PER71) conferred enhanced disease resistance to B. cinerea in Arabidopsis (Chassot et al., 2007). Consequently, we studied the transcriptional regulation of the nine deregulated PER genes identified in the myb46-2 microarray following inoculation with B. cinerea (Table II; PER1, PER4, PER39, PER45, PER57, PER62, PER69, PER71, and PER73). We extended this survey to other PER members that, although not present in the myb46-2 microarray, were derived from a bibliographic survey, including PER12, PER21, PER34, and PER37. Table II indicates the expression levels measured by quantitative RT-PCR for all sampled PER genes at 72 h.p.i. with B. cinerea in Col-0 and myb46-2 plants. Among the entire set of genes, a 184-fold induction was observed for PER62 in Col-0 following infection with B. cinerea. The induction was increased 984-fold in the myb46-2 background. The up-regulation observed for PER69 (31-fold in Col-0 versus 267-fold in myb46-2) was also a notable induction change. Likewise was the induction observed for PER21 (168-fold in myb46-2) and to a lesser extent the changes in PER4, PER34, PER37, PER45, or PER71.
Reexamining transcript abundance more accurately during a 72-h time course following inoculation with B. cinerea revealed different transcriptional response patterns: early, transient, and late (Fig. 6). Early responsive genes (i.e. PER12) coincided with a repression of gene expression already apparent at 24 h.p.i. Transient responsive genes (PER45, PER69, and PER73) exhibited a temporary induction that peaked between 24 and 48 h.p.i. Finally, late responsive genes (PER4, PER34, PER37, PER21, PER62, and PER71) featured a sustained response that initiated at 24 to 48 h.p.i. and continued throughout 72 h.p.i. The last three genes in this group have been shown to confer individual resistance to B. cinerea when individually overexpressed in transgenic plants (Chassot et al., 2007). Other randomly chosen genes exhibited up- or down regulation in myb46-2 plants (i.e. PCC1, JAC, RNS1, and EXLB3) and behaved differentially (Fig. 6). Therefore, a gradual coordinated release of a set of cell wall peroxidases is a characteristic response set in motion following the recognition of B. cinerea in Arabidopsis, a response heightened in myb46-2 plants.
Figure 6.
Expression of PER genes in leaves at early stages of B. cinerea infection. Relative expression was assayed over a 72-h time course by quantitative RT-PCR on total RNA from leaves of Col-0 (white circles) or myb46-2 (black circles) plants following inoculation with a spore suspension of B. cinerea. Three general transcription responses to B. cinerea were observed: early (as in PER-12), transient (as in PER45, PER69, and PER73), and late (as in PER4, PER21, PER34, PER37, PER21, PER62, and PER71). Expression of the nonperoxidase genes RNS1, EXLB3, JAC, and PCC1 was concurrently assayed as internal controls and used for comparison. Data represent means ± sd (n = 3 biological replicates).
myb46-2 Plants Exhibit Accelerated Peroxidase Deployment following B. cinerea Detection
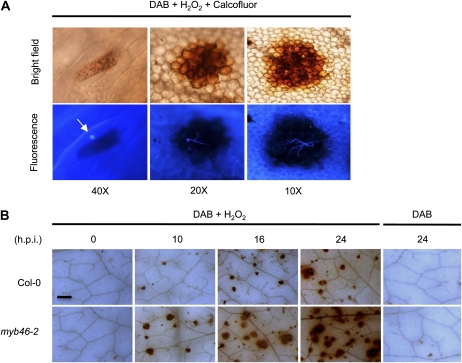
The enhanced induction of PER genes led us to investigate an in situ biochemical correlation between induced peroxidase activity and confinement of this peroxidase activity to the B. cinerea infection site. Our protocol included staining Arabidopsis leaves with 3,3′-diaminobenzidine (DAB), a histochemical reagent that when combined with H2O2 acts as a substrate for peroxidases and specifically detects the center of peroxidase activity using light microscopy (Thordal-Christensen et al., 1997; Bestwick et al., 1998). Alternatively, counterstaining of DAB-stained leaves with calcofluor white and inspection with a fluorescence microscope resulted in the detection of fungal structures associated with the center of peroxidase activity.
Brown staining due to DAB oxidation was negligible in the leaves of control plants, with the exception of occasional staining in the cells at the base of trichomes. Spray inoculation of leaves with a spore suspension of B. cinerea induced strong DAB oxidation throughout the leaf blade. The staining pattern ranged from single cells early postinoculation to collections or clusters of plant cells at later stages postinoculation (Fig. 7A). The size of cell clustering indicating peroxidase activity correlated with the degree of fungal structure development, as ascertained by calcofluor staining in the same tissue preparations. Single cells with DAB staining corresponded to early stages of conidium interaction with epidermal cells (Fig. 7A). Over time, fungal appressorium and germ tube development was observed on the inoculated leaf surface. DAB oxidation in reacting cells increased and continued until the brown coloration due to DAB oxidation was easily detectable in association with the growth of secondary hyphae (Fig. 7A). Larger sections of DAB deposition generally corresponded to advanced stages of infection and became visible to the naked eye (i.e. DAB deposition from different areas of infection fused). In Col-0 and myb46-2 plants, the appearance of peroxidase activity was always associated with fungal structures, suggesting that induction is a consequence of local interaction of the plant cell with the fungus. Interestingly, recording the induction of peroxidase activity following inoculation with B. cinerea spores indicated that myb46-2 plants react earlier and more aggressively than Col-0 plants (Fig. 7B). At 10 h.p.i., myb46-2 plants initiated intensive peroxidase activity. The difference in peroxidase activity in myb46-2 plants was even more evident at 24 h.p.i., where large areas of the inoculated leaf surface were nearly covered by oxidized DAB (Fig. 7B). The sustained peroxidase activity observed at these early stages of infection was a cellular mechanism in response to B. cinerea, a pathogen with the potential to cause cell death. Furthermore, staining with trypan blue did not occur at these early stages of fungal infection, and in the absence of added H2O2, DAB staining was absent (Fig. 7B), indicating that peroxide levels were limited at early stages of infection.
Figure 7.
Local initiation of peroxidase activity in response to inoculation with B. cinerea spores. A, DAB + H2O2 staining at the site of spore inoculation, showing progressive enhancement of peroxidase activity from a single reacting epidermal cell (left) to clusters of cells that increase in size as infection progresses over time. The bottom row shows the same leaf sections after counterstaining with calcofluor to reveal fungal structures associated with the peroxidase-reacting cells. The arrow indicates an example of a single epidermal cell reacting to the presence of a conidium. B, DAB + H2O2 staining of inoculated leaves from Col-0 and myb46-2 plants was performed at the indicated times after inoculation with B. cinerea. Shown are representative sections of leaves selected from each plant genotype. Bar = 50 μm.
These results indicate that conditional peroxidase-mediated cell wall modifications are part of a cell wall toolkit activated by plant cells upon detection of B. cinerea. The early and more abrupt initiation of cell wall peroxidases in myb46-2 plants may be the basis to explain increased disease resistance to B. cinerea.
DISCUSSION
Young (1926) provided one of the pioneer reports on the response of plants to fungal challenges, and modifications to the plant cell wall were already recognized as a potential mechanism of resistance. Decades of subsequent research have demonstrated that in fungi that use penetration of a plant cell wall as an essential part of the initial pathogenesis cycle, controlling the fungus during the initial phases of cellular penetration can be an effective means of plant defense. Not surprisingly, while fungal microbes try to breach this barrier for colonization, plant cells respond to attempted penetration by a battery of wall-associated defense responses and cell wall modifications (Hückelhoven, 2007). In this study, we showed that the Arabidopsis MYB46 transcription factor is required for disease susceptibility to the virulent necrotrophic fungal pathogen B. cinerea and, in turn, that mutations in the MYB46 gene confer strong disease resistance to the same pathogen. MYB46 was identified in this study by its ability to bind to a cis-element, here characterized and named the R element, located in the promoter region of the type III peroxidase Ep5C gene originally identified in tomato plants. When MYB46 is down-regulated, enhanced induced expression of promEp5C::GUS activity occurs following pathogen inoculation. Consequently, a conditional heightened induction in gene expression for a cell wall-related peroxidase gene and enhanced disease resistance to B. cinerea were linked in myb46 plants, suggesting that MYB46 functions as a negative regulator of this characteristic. However, and in contrast, MYB46 was previously identified as a positive regulator of the formation of secondary cell walls in the vascular tissue of the Arabidopsis stem. This suggest that MYB46 may be a bifunctional transcription factor that acts mainly as an activator during vascular development but becomes a conditional repressor when involved in the regulation of genes important for disease resistance to B. cinerea. A similar independent function in response to phytopathogens with a parallel role in development was described for the MYB-related gene AS1 (Nurmberg et al., 2007). A dual role for other transcription factors, acting as activator and repressor depending on the target gene and biological conditions, has been described and seems to be a common feature in plants (i.e. WUSCHEL [Ikeda et al., 2009], Pti4 [González-Lamothe et al., 2008], and WRKY53 [Miao et al., 2004]) and animals (Adkins et al., 2006). Unidentified factors that interact with MYB46 might convert it from an activator to a repressor and vice versa. Future research in this direction will facilitate clarifying the dichotomy observed for MYB46.
Transcriptome analysis supports the cell wall as one of the cellular compartments affected in myb46 plants, as a high percentage of the deregulated genes encode cell wall structural proteins or proteins related to cell wall dynamics. However, changes in cell wall constituents due to the loss of function of MYB46 must be very specific, precise, and/or targeted at B. cinerea and therefore may escape detection by FTIR. As B. cinerea attempts to recognize specific signatures in the host cell wall during the infection process, these changes may represent a mosaic of discrete cell wall modifications that represent a variety of obstacles preventing the fungus from initiating the infection process. A coordinated deregulation of all modifications (the result of invoking mutations in the MYB46 locus) may represent a superior form of disease resistance. Furthermore, some of these individual cell wall modifications have a demonstrated positive impact in resistance against B. cinerea. For example, the simultaneous suppression of polygalacturonase (LePG) and expansin (LeExp1) gene expression, but not suppression of LePG or LeExp1 alone, in tomato dramatically reduces B. cinerea disease susceptibility (Cantu et al., 2008). Similarly, the ectopic expression of a polygalacturonase-inhibiting protein (pPGIP) from pear (Pyrus communis) fruit reduces the disease susceptibility to B. cinerea infection (Powell et al., 2000). Overexpression of pectin methylesterase protein inhibitors PMEI-1 and -2 from Arabidopsis (Lionetti et al., 2007) results in increased B. cinerea disease resistance. The fact that these genes appear similarly deregulated in myb46 plants, as revealed by microarray analysis, contributes to a better understanding of why mutant plants show an enhanced disease resistance to B. cinerea. Therefore, very precise and discrete changes in cell wall constituents might have a direct effect on the integrity of the cell wall fabric and in turn may alter substrate accessibility to pathogen cell wall-degrading enzymes, with an immediate effect in reducing fungal growth. However, other possible mechanisms may contribute to alterations in pathogen susceptibility in myb46 plants. In fact, Ellis and coworkers demonstrated that cell wall changes associated with unbalanced cellulose biosynthesis, although detrimental to normal plant development and exhibiting different pleiotropic effects in plants, could activate novel defense pathways (Ellis and Turner, 2001; Ellis et al., 2002; Hernández-Blanco et al., 2007). However, this seems not to explain the observed enhanced disease resistance to B. cinerea, since cellulose accumulation appears uncompromised in myb46 plants, as deduced from FTIR analyses.
Reinforcement of the cell wall fabric by direct cross-linking of its structural proteins through the formation of dityrosine and isodityrosie linkages (Fry, 1986; Bradley et al., 1992; Domingo et al., 1999) by the action of cell wall-bound type III peroxidases, or even heightening the cross-linking of the phenolic network of secondary cell walls by the same peroxidases, may serve as a basis to explain the enhanced disease resistance to B. cinerea in myb46 plants. PER activity was found associated with cross-linking of phenolic acids at Botrytis allii infection sites in onion (Allium cepa) cell walls (McLusky et al., 1999). In addition, in bean (Phaseolus vulgaris) leaves, B. cinerea was shown to suppress PER activity, supporting a role for PER in plant resistance as scavengers of harmful reactive oxygen species (Tiedemann, 1997). Moreover, overexpression of different PER genes (e.g. PER21, PER62, or PER71) in transgenic Arabidopsis plants confers a notable disease resistance to B. cinerea (Chassot et al., 2007). Interestingly, a set of nine PER genes, including PER21, PER62, and PER71, were found coregulated in myb46-2 plants. These PER genes were down-regulated under resting conditions and were highly induced locally following the recognition of B. cinerea. The initiation of peroxidase activity and the substantial overinduction of peroxidase in myb46-2 plants once B. cinerea structures were in contact with the epidermal cell walls also served to explain the heightened disease resistance phenotype of myb46-2 plants. A more rapid cross-linking of cell wall structural components at sites of fungal entry may pose additional difficulties for B. cinerea to penetrate the cell wall during its infection cycle. These inducible changes, in concerted action with the repertory of other existing changes in cell wall constituents, may have altered the cell wall in such a way that the pathogen no longer possesses the effective tools to disassemble the plant cell wall. Moreover, the alterations in cell wall composition resulting from mutations in MYB46 may further confer on the mutant plant cell increased sensitivity to respond faster to the presence of the pathogen (i.e. defense-related genes are correspondingly activated sooner). In fact, myb46 plants respond to B. cinerea with an earlier and heightened activation of the jasmonic acid-regulated plant defensin gene PDF1.2a that is effective against necrotrophic fungal pathogens. This further substantiates that defense-related signaling pathways and cell wall integrity are interconnected, and MYB46 likely functions as a disease susceptibility modulator to B. cinerea through the integration of cell wall remodeling and downstream activation of secondary lines of defense.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a growth chamber (19°C–23°C, 85% relative humidity, 100 mE m−2 s−1 fluorescent illumination) on a 10-h-light/14-h-dark cycle.
Transgenic Plants and Mutants
A 5′ deletion series of a 1,140-bp-long Ep5C promoter region was generated by PCR using specific primers where XbaI or SmaI restriction sites were incorporated by site-directed mutagenesis. For each construct, the ep2 reverse primer (5′-CCCCGGGTGTTATGTACG-3′) was used with each of the following forward primers: del1 (5′-CCTACCTTTTCTCTAGACAAC-3′), del2 (5′-CCCAAACACTCTAGAAAATTCA-3′), del3 (5′-GAATCTAGATAAGTAGCT3-3′), del4 (5′-CTCTAGATAGAGTTGAACAAG-3′), del5 (5′-ACCATCTAGAACACAAATTG-3′), del6 (5′-GTTCTAGATTATACAAGTTTG-3′), del7 (5′-GTCTAGATATGCATACTCAAAG-3′), del8 (5′-GCCTCTAGATTAATTACAAAC-3′), del9 (5′-GAGTCTAGACAAAACAAGG-3′), del10 (5′-TTCTAGACATACAACATC-3′), del11 (5′-GTCTAGATGCTTGCCAAC-3′), del12 (5′-ATCTAGAGCGTATAGGCC-3′), and del13 (5′-CTTCTAGACTCTTACTCC-3′). Each DNA-amplified fragment was cloned upstream of the GUS gene in pBI101.1 (Clontech), within XbaI and SmaI sites. To generate the R::35Smin-45:GUS construct, a double-stranded R fragment was obtained by R1 (5′-TCGACACCACATAGAACACAAATTGTTAGGTAGGATGTCACGTGGGACGTATTTGC-3′) and R2 (5′-TCGAGCAAATACGTCCCACGTGACATCCTACCTAACAATTTGTGTTCTATGTGGTG-3′) oligonucleotide hybridization. This R DNA fragment, which contains SalI and XhoI extensions, was cloned upstream of the 35Smin-45:GUS gene in pBT10-GUS, within SalI and XhoI sites. The R::35Smin-45:GUS construct was subcloned into the pBI101.3 (Clontech) BamHI and SacI sites. The RR::35Smin-45:GUS construct was generated in a similar way, except that R was cloned twice into pBT10-GUS. All the constructs were introduced into Arabidopsis Col-0 via Agrobacterium tumefaciens transformation. First-generation transformants were assayed for GUS activity by an in situ assay using 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Jefferson et al., 1987).
The myb46-1 (SALK_088514) and myb46-2 (SALK_100993) mutants of Arabidopsis Col-0 were obtained from T-DNA insertion lines from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/). For PCR-based genotyping, the following primers were used to identify homozygous mutants: MYB46RevInt (5′-GTAAAACCCTAAGGAAGAGAAG-3′) and LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) for myb46-1 and MYB46FwdRTInt (5′-CATTCCATCCTCGGCAACAG-3′) and LBb1 for myb46-2. The wild-type allele was detected by using MYB46RevInt and MYB46FwdRTInt primers.
The myb46-1 Ep5C::GUS plants were generated by the genetic cross of myb46-1 and Ep5C::GUS transgenic plants. The presence of Ep5C::GUS was detected by PCR using EP5C300Fwd (5′-GTGACATACTACACGCAGAGC-3′) and EP5C534Rev (5′-GCGTCCTTGTTTTGTCTCGTC-3′) primers.
For the MYB46-overexpressing construct, a full-length cDNA for MYB46 was amplified by PCR using Pfu DNA polymerase (Stratagene) and cloned into the EcoRI and XhoI sites of the binary vector pC1300intB-35SnosBK (GenBank accession no. AY560326) under the control of the constitutive CaMV 35S promoter. This construct was introduced into Arabidopsis Col-0 via Agrobacterium transformation. MYB46 expression of single-insertion homozygous transgenic plants was analyzed by RT-PCR.
Tissue Staining
Staining for the presence of peroxidase activity via the DAB uptake method in the presence of H2O2 was performed as described (McLusky et al., 1999). Staining for the presence of GUS activity was performed as described previously (Jefferson et al., 1987). Staining with lactophenol-trypan blue was performed as described by Mayda et al. (2000). Staining with 0.01% calcofluor white was performed as described previously (Hughes and McCully, 1975) and observed with a UV fluorescence microscope. Thin sections of the stem were stained with phloroglucinol-HCl, which was shown as bright red using standard microscopy. Staining with Safranin O of thin sections was performed as described (Bond et al., 2008) and observed with a Nikon Eclipse fluorescence microscope. Stained sections were excited at 492 nm and imaged using a B-2A filter (520 nm).
EMSA
Leaf whole-cell extracts were prepared as described previously (Carrasco et al., 2003). The DNA fragments used as probes were double-stranded oligonucleotide radioactively labeled by filling in with [α-32P]dCTP using the Klenow DNA polymerase. Binding reactions (20 μL) containing 20,000 cpm of probe, 10 μg of leaf protein or 50 ng of recombinant MBP::MYB46 protein, 10 mm Tris-HCl, pH 8, 40 mm KCl, 0.1 mm EDTA, 2 mm dithiothreitol, 0.2 μg mL−1 bovine serum albumin, 0.05% Nonidet P-40, 10% glycerol, and 40 μg mL−1 poly(dI-dC)·poly(dI-dC), in the presence or absence of competitors, were incubated on ice for 30 min. DNA-protein complexes were separated from free DNA probe by electrophoresis on native 6% (38:2) polyacrylamide gels run in 0.5 Tris-borate/EDTA at 4°C.
Yeast One-Hybrid Screening
An Arabidopsis Col-0 cDNA expression library constructed in the λ-ACTII/pACTII′ system (Memelink, 1997) was used for the yeast one-hybrid screening, which was carried out as described by Ouwerkerk and Meijer (2001). The R element cloned in the pHIS3NX/pINT1 plasmid system (Ouwerkerk and Meijer, 2001) was used as a bait sequence. The double-stranded R fragment (see above), with SalI and XhoI extensions, was cloned in pBluescript II KS (Stratagene) and subsequently cloned into pHIS3NX (GenBank accession no. AF275030) within the BamHI and SmaI sites. The R::HIS3 fusion was subcloned into pINT1 (GenBank accession no. AF289993) within the NotI and XbaI sites. The resulting plasmid pINT1(R::HIS3) was introduced into yeast strain Y187 (Clontech). The cDNA expression library screening was performed using yeast strain Y187(R::HIS3) on medium lacking His and Leu and in the presence of 5 mm 3-AT as described (Ouwerkerk and Meijer, 2001). Colony PCR using the primers ATH (5′-CCCCACCAAACCCAAAAAAAG-3′) and 3′AD (5′-GTTGAAGTGAACTTGCG-3′) was performed to amplify the cDNA inserts of positive clones. Amplification products were then sequenced.
Production of Recombinant MYB46
The coding region of MYB46 was released from one of the pACTII′ clones obtained in the yeast one-hybrid screening by EcoRI and XhoI digestion and subcloned into the EcoRI and SalI sites of pMAL-c2 (New England Biolabs; www.neb.com). The MBP::MYB46 fusion protein was expressed in Escherichia coli Rosettagami(DE3)pLys cells (Novagen; www.merck-chemicals.com) by induction with 0.15 mm isopropylthio-β-galactoside at 28°C for 4 h. The recombinant MBP::MYB46 was purified by affinity chromatography using amylose resin (New England Biolabs) following the manufacturer’s instructions.
DNA Methylation Interference
DNA methylation interference was carried out according to Green et al. (1987). Primer R1 was 5′ end labeled using [γ-32P]ATP and T4 polynucleotide kinase and used to produce a top-strand end-labeled R probe by annealing with primer R2. A total of 27 pmol of the labeled probe was partially methylated with dimethyl sulfate (Green et al., 1987), ethanol precipitated, and purified using a Qiaquick purification kit. Gel retardation assays were then carried out with the methylated probe (60,000 cpm) and 1 μg of MBP::MYB46 recombinant protein, as described above. Following native gel electrophoresis, the unbound probe (F) and protein-DNA complexes (B) were located by autoradiography and excised. The DNA was purified and exposed to 1.25 m piperidine at 90°C for 30 min. The piperidine-treated unbound and protein-bound DNAs were resolved on an 8% sequencing gel.
RNA Isolation, RT-PCR, and Quantitative RT-PCR Analysis
RNA was isolated with Trizol (Invitrogen; www.invitrogen.com). RNA was quantified with a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies; www.nanodrop.com). RNA quality was assessed with a 2100 Bioanalyzer from Agilent Technologies (www.agilent.com). For RT, RevertAid Moloney murine leukemia virus reverse transcriptase (Fermentas; www.fermentas.com) was used according to the manufacturer’s instructions. Semiquantitative RT-PCR and quantitative RT-PCR were carried out with gene-specific primers designed using the Primer Express 2.0 software (Applied Biosystems; www.appliedbiosystems.com): PR-1, 5′-AAGGGTTCACAACCAGGCAC-3′ and 5′-CACTGCATGGGACCTACGC-3′; PDF1.2a, 5′-CTTGTTCTCTTTGCTGCTTTC-3′ and 5′-CATGTTTGGCTCCTTCAAG-3′. A list containing the rest of the genes for which quantitative RT-PCR analyses have been performed is provided in Supplemental Table S1. Quantitative RT-PCR was performed using the SybrGreen PCR Master Mix (Applied Biosystems) in an ABI PRISM 7000 sequence detector. Cycle threshold (Ct) was calculated using the 7000 System SDS Software Core Application Version 1.2.3 (Applied Biosystems), and the data were transformed with the equation 2^(40-Ct). Quantitative RT-PCR and RT-PCR analyses were performed at least three times using sets of cDNA samples from independent experiments.
Microarray Analysis
RNA obtained from leaves was amplified with the MessageAmp aRNA amplification kit from Ambion (www.ambion.com) following the instruction manual. To allow later labeling with Cy fluorophores, aminoallyl UTP (Ambion) was added to the mix of the T7 RNA polymerase-driven aRNA amplification reaction. The amount and quality of aRNA obtained were assessed as before. The aminoallyl-labeled aRNA (10 μg) was incubated in 1 m Na2CO3 with 8 nmol of dye monofunctional NHS ester (Cy3/Cy5) RPN 5661 (Amersham Biosciences; www.gehealthcare.com) at room temperature in the dark for 1 h. Then, 35 μL of 0.1 m sodium acetate, pH 5.2, was added and incubated for a further 5 min in the dark. The Cy-labeled aRNA was purified with the Megaclear kit from Ambion and measured with the Nanodrop ND-100 spectrophotometer. Three biological replicates were independently hybridized for transcriptomic comparison.
Microarray slides were composed of synthetic 70-mer oligonucleotides from the Operon Arabidopsis Genome Oligo Set version 1.0 (Qiagen; www.qiagen.com) spotted on aminosilane-coated slides (Telechem; www.telechem.com) by the University of Arizona. Slides were rehydrated and UV cross-linked according to the supplier’s Web site (http://ag.arizona.edu/microarray/methods.html). The slides were then washed twice for 2 min in 0.1% SDS and in ethanol for 30 s. Arrays were drained with a 2,000-rpm spin for 2 min. Slides were prehybridized in 6× SSC, 0.5% (w/v) SDS, and 1% (w/v) bovine serum albumin at 42°C for 1 h, followed by five rinses with milliQ water. Excess water was drained with a 2,000-rpm spin for 2 min.
For the hybridization, equal amounts of dye of each aRNA labeled with either Cy3 or Cy5, ranging from 200 to 300 pmol, were mixed with 20 μg of poly(A) and 20 μg of yeast tRNA (Sigma-Aldrich; www.sigmaaldrich.com) in a volume of 9 μL. To this volume, 1 μL of RNA fragmentation buffer was added (RNA fragmentation reagents; Ambion), and after 15 min at 70°C, 1 μL of stop solution. Formamide, 20× SSC, 50× Denhardt’s, and 20% SDS were added to a final concentration of 50% formamide, 6× SSC, 5× Denhardt’s, and 0.5% SDS. This mix was boiled for 3 min at 95°C and then added to the prehybridized slide. Hybridization took place overnight at 37°C in a hybridization chamber. Arrays were then washed for 5 min at 37°C in 0.5× SSC and 0.1% SDS, twice for 5 min at room temperature (21°C) with 0.5× SSC and 0.1% SDS, three times with 0.5× SSC at room temperature, and 5 min with 0.1× SSC. The slides were then drained with a 2,000-rpm spin for 2 min. The slides were stored in darkness until they were scanned.
The scanning was done with a GenePix 400B scanner (Molecular Devices; www.moleculardevices.com) at 10-μm resolution. The images were quantified with GenePix Pro 5.1. Images from Cy3 and Cy5 channels were equilibrated and captured with a GenePix 4000B (Axon; www.axon.com), and spots were quantified using GenePix Pro 5.1 software (Axon). The data from each scanned slide were first scaled and normalized using the Lowess method before being log transformed. The mean of the three replicate log-ratio intensities and their sd values were generated.
The expression data were normalized and statistically analyzed using the LIMMA package (Smyth and Speed, 2003). LIMMA is part of Bioconductor, an R language project (Ihaka and Gentleman, 1996). First, the data set was filtered based on the spot quality. A strategy of adaptive background correction was used that avoids exaggerated variability of log ratios for low-intensity spots. For local background correction, the “normexp” method in LIMMA to adjust the local median background was used. The resulting log ratios were print-tip Lowess normalized for each array (Smyth and Speed, 2003). To have similar distribution across arrays and to achieve consistency among arrays, log-ratio values were scaled using as scale estimator the median absolute value (Smyth and Speed, 2003).
Linear model methods were used for determining differentially expressed genes. Each probe was tested for changes in expression over replicates using an empirical Bayes-moderated t statistic (Smyth, 2004). To control the false discovery rate, P values were corrected using the method of Benjamini and Hochberg (1995). The expected false discovery rate was controlled to be less than 5%. Genes were considered to be differentially expressed if the corrected P values were less than 0.05. In addition, only genes with more than 2-fold change were considered for further analysis.
Pathogen Infection
PsDC3000 inoculation, by leaf infiltration of 105 colony-forming units mL−1, and quantification were described previously (Coego et al., 2005b). Data are reported as means and sd of the log (colony-forming units cm−2) of eight replicates. Botrytis cinerea inoculation and symptom measurements were described previously (Coego et al., 2005b). Five-week-old-plants were inoculated by applying 6-μL droplets of spore suspension of B. cinerea (2.5 × 104 conidia mL−1) to three fully expanded leaves per plant. The plants were maintained at 100% relative humidity, and disease symptoms were evaluated 3 d after inoculation by determining the average lesion diameter on three leaves of 15 plants each. For the rest of the experiments, plants were inoculated by spray.
Lignin Determination
Leaves were harvested, boiled in ethanol for 5 min, and washed with ethanol until pigments were removed. The dry leaves were finely homogenized and considered as the AIR. The AIR was treated with 2 m NaOH for 1 h under N2 atmosphere to remove the esterified phenolic material. The lignin contents of the hole AIR and of the deesterified AIR were measured by the acetyl bromide/acetic acid method (Johnson, 1961).
FTIR Analysis
For FTIR analysis, potassium bromide pellets were prepared by grinding the AIR with solid potassium bromide. The FTIR spectra were obtained at a resolution of 4 cm−1 and 64 interferograms coadded for a high signal-to-noise ratio, using a Bomem spectrometer (Hartman and Braun). The WinDas informatic application (John Wiley and Sons) was used for data analysis.
The microarray data have been submitted to the Gene Expression Omnibus databases under accession number GSE25838.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence specificity of competition analysis interaction.
Supplemental Figure S2. Methylation interference assay.
Supplemental Figure S3. Specificity of MYB46 binding to the R element.
Supplemental Figure S4. MYB46ox plants exhibit no effect on resistance to P. syringae DC3000 and B. cinerea.
Supplemental Figure S5. FTIR spectroscopy.
Supplemental Figure S6. Validation of microarray data.
Supplemental Figure S7. Functional classification of genes.
Supplemental Figure S8. Expression of some pathogenesis‐related genes.
Supplemental Table S1. Microarray data.
Acknowledgments
We thank J. Memelink for providing the Arabidopsis cDNA library and Ana Lopez for helpful discussions.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins NL, Hagerman TA, Georgel P. (2006) GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol 84: 559–567 [DOI] [PubMed] [Google Scholar]
- Agorio A, Vera P. (2007) ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bond J, Donaldson L, Hill S, Hitchcock K. (2008) Safranine fluorescent staining of wood cell walls. Biotech Histochem 83: 161–171 [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. (1992) Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70: 21–30 [DOI] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell AL. (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci USA 105: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC. (2000) The cell wall. Buchanan BB, Gruissem W, Jones R , Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 52–109 [Google Scholar]
- Carrasco JL, Ancillo G, Mayda E, Vera P. (2003) A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J 22: 3376–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab GI, Varner JE. (1988) Cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 39: 321–353 [DOI] [PubMed] [Google Scholar]
- Chassot C, Nawrath C, Métraux JP. (2007) Cuticular defects lead to full immunity to a major plant pathogen. Plant J 49: 972–980 [DOI] [PubMed] [Google Scholar]
- Chen L, Carpita NC, Reiter WD, Wilson RH, Jeffries C, McCann MC. (1998) A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J 16: 385–392 [DOI] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Ellul P, Mayda E, Vera P. (2005a) The H2O2-regulated Ep5C gene encodes a peroxidase required for bacterial speck susceptibility in tomato. Plant J 42: 283–293 [DOI] [PubMed] [Google Scholar]
- Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P. (2005b) An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17: 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvill AG, Albersheim P. (1984) Phytoalexins and their elicitors: a defense against microbial infection in plants. Annu Rev Plant Physiol 35: 243–275 [Google Scholar]
- Domingo C, Saurí A, Mansilla E, Conejero V, Vera P. (1999) Identification of a novel peptide motif that mediates cross-linking of proteins to cell walls. Plant J 20: 563–570 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG. (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1986) Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol 37: 165–186 [Google Scholar]
- González-Lamothe R, Boyle P, Dulude A, Roy V, Lezin-Doumbou C, Kaur GS, Bouarab K, Després C, Brisson N. (2008) The transcriptional activator Pti4 is required for the recruitment of a repressosome nucleated by repressor SEBF at the potato PR-10a gene. Plant Cell 20: 3136–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua NH. (1987) Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6: 2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57: 155–171 [DOI] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, et al. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R. (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Hughes J, McCully ME. (1975) The use of an optical brightener in the study of plant structure. Stain Technol 50: 319–329 [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. (1996) R: a language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Moore WE, Zank LC. (1961) The spectrophotometric determination of lignin in small wood samples. Tappi 44: 793–798 [Google Scholar]
- Ko JH, Kim WC, Han KH. (2009) Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 60: 649–665 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CM. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake GJ, Faktor O, Lamb CJ, Dixon RA. (1992) Combination of H-box [CCTACC(N)7CT] and G-box (CACGTG) cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proc Natl Acad Sci USA 89: 9230–9234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT. (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. (1997) MYB transcription factors in plants. Trends Genet 13: 67–73 [DOI] [PubMed] [Google Scholar]
- Mayda E, Mauch-Mani B, Vera P. (2000) Arabidopsis dth9 mutation identifies a gene involved in regulating disease susceptibility without affecting salicylic acid-dependent responses. Plant Cell 12: 2119–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLusky SR, Bennett MH, Beale M, Lewis MJ, Gaskin P, Mansfield J. (1999) Cell wall alterations and localized accumulation of feruloyl-3′-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant J 17: 523–534 [Google Scholar]
- Meijer AH, Ouwerkerk PBF, Hoge JH. (1998) Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast 14: 1407–1415 [DOI] [PubMed] [Google Scholar]
- Memelink J. (1997) Two yeast/Escherichia coli lambda/plasmid vectors designed for yeast one- and two-hybrid screens that allow directional cDNA cloning. Trends Genet 13: 376 [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmberg PL, Knox KA, Yun BW, Morris PC, Shafiei R, Hudson A, Loake GJ. (2007) The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc Natl Acad Sci USA 104: 18795–18800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk PBF, Meijer AH. (2001) Yeast one-hybrid screening for DNA-protein interactions. Curr Protoc Mol Biol August Chapter 12, Unit 12.12 [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24: 255–265 [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540 [DOI] [PubMed] [Google Scholar]
- Powell AL, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM. (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact 13: 942–950 [DOI] [PubMed] [Google Scholar]
- Ramírez V, Coego A, López A, Agorio A, Flors V, Vera P. (2009) Drought tolerance in Arabidopsis is controlled by the OCP3 disease resistance regulator. Plant J 58: 578–591 [DOI] [PubMed] [Google Scholar]
- Sablowski RW, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M. (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J 13: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerbrunn N, Schlaich NL. (2004) PCC1: a merging point for pathogen defence and circadian signalling in Arabidopsis. Planta 218: 552–561 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Becker-André M, Schulz W, Hahlbrock K, Dangl JL. (1989) Functional architecture of the light-responsive chalcone synthase promoter from parsley. Plant Cell 1: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Stockert JC, Cañete M, Colman OD. (1984) Histochemical mechanism for the orthochromatic staining and fluorescence reaction of lignified tissues. Cell Mol Biol 30: 503–508 [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Tiedemann A. (1997) Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol Mol Plant Pathol 50: 151–166 [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P. (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138 [DOI] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. (2002) The jasmonate signal pathway. Plant Cell (Suppl) 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner JE, Lin LS. (1989) Plant cell wall architecture. Cell 56: 231–239 [DOI] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC. (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S, Somerville S, Somerville C. (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA. (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C. (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. (1997) Oxidative burst: an early plant response to pathogen infection. Biochem J 322: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Klessig DF. (1996) Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc Natl Acad Sci USA 93: 14972–14977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PA. (1926) Penetration phenomena and facultative parasitism in Alternaria, Diplodia, and other fungi. Bot Gaz 81: 258–279 [Google Scholar]
- Zhong R, Demura T, Ye ZH. (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]