Abstract
MTMR2 is a member of the myotubularin family of inositol lipid phosphatases, a large protein-tyrosine phosphatase subgroup that is conserved from yeast to humans. Furthermore, the peripheral neuromuscular disease Charcot-Marie Tooth disease type 4B has been attributed to mutations in the mtmr2 gene. Because the molecular mechanisms regulating MTMR2 have been poorly defined, we investigated whether reversible phosphorylation might regulate MTMR2 function. We used mass spectrometry-based methods to identify a high stoichiometry phosphorylation site on serine 58 of MTMR2. Phosphorylation at Ser58, or a phosphomimetic S58E mutation, markedly decreased MTMR2 localization to endocytic vesicular structures. In contrast, a phosphorylation-deficient MTMR2 mutant (S58A) displayed constitutive localization to early endocytic structures. This localization pattern was accompanied by displacement of a PI(3)P-specific sensor protein and an increase in signal transduction pathways. Thus, MTMR2 phosphorylation is likely to be a critical mechanism by which MTMR2 access to its lipid substrate(s) is temporally and spatially regulated, thereby contributing to the control of downstream endosome maturation events.
Keywords: Endosomes, Mass Spectrometry (MS), Phosphatase, Phosphatidylinositol, Protein Phosphorylation
Introduction
The protein-tyrosine phosphatase superfamily can be divided into distinct subfamilies, which include the receptor tyrosine phosphatases, the intracellular tyrosine phosphatases, and the dual specificity phosphatases, which dephosphorylate phosphoserine/threonine residues, as well as phosphotyrosine residues (1–3). All protein-tyrosine phosphatase family members possess the invariant catalytic sequence C(X)5R and use a thiol phosphate intermediate as a catalytic mechanism (4).
In recent years, other protein-tyrosine phosphatase families have been identified that dephosphorylate phosphatidylinositol phosphates (PIPs)3 as their physiologic substrates. These inositol lipid phosphatases include the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) and the myotubularin-related (MTMR) subgroups (5, 6). PTEN has been widely characterized as a tumor suppressor protein (7, 8), and mutations in three MTMR genes have been identified in distinct human neuromuscular diseases, signifying their importance in fundamental biological processes (9–12).
Phosphatidylinositol is an abundant membrane lipid that is phosphorylated by PI kinases on positions 3, 4, and 5, of the inositol head group in response to various extracellular signals, yielding seven unique PIPs (13, 14). These unique PIPs function to recruit target proteins containing the appropriate PIP binding domains (15) to discrete membrane locations where they can properly respond to extracellular stimuli. These highly specific lipid-protein interactions are subsequently regulated through the phosphorylation/dephosphorylation of the PIPs.
Active MTMRs dephosphorylate the lipid second messengers PI(3)P and PI(3,5)P2 (16–18). These phosphoinositides play key roles in membrane targeting, vesicular trafficking, and regulation of signal transduction pathways (19). In particular, the presence of PI(3)P on early endosomes has recently been shown to facilitate endosome maturation and attenuate growth factor receptor signaling from endocytic structures, indicating that regulators of endosomal PI(3)P levels can directly impact the rate of endocytosis and duration of growth factor signaling (20).
Although the PIP substrates for MTMRs are constituents of membrane bilayers in structures such as endosomes, numerous groups have observed that MTMRs, including MTMR2, do not extensively localize to suborganellar structures containing its lipid substrates (21–23). In this study, we have used mass spectrometry (MS) to identify a high stoichiometry phosphorylation site on MTMR2 that regulates targeting to endocytic structures. An MTMR2 phosphorylation-deficient mutant (S58A) exhibited strong localization to early endocytic structures, where it efficiently depleted PI(3)P. In contrast, wild-type MTMR2 was highly phosphorylated at this site and did not localize to these structures. Furthermore, a phosphomimetic MTMR2 mutant (S58E) exhibited localization identical to that of wild-type MTMR2. Our findings suggest that MTMR2 phosphorylation at Ser58 regulates its targeting to endosomal membranes and access to phosphoinositide substrates. The significance of this effect was underscored by the fact that signaling through pathways known to be sensitive to endosomal PI(3)P levels were dramatically altered in cells expressing the MTMR2 S58A mutant.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The plasmids encoding FLAG-tagged MTMR2 and His-tagged MTMR2 were previously described (24). To generate expression plasmids with the S58A, S58E, and S58A,C417S point mutations, PCR-based site-directed mutagenesis was performed. The forward primer for the generation of S58A used was 5′-CTGCCGACAACTTTGCTCCTGATTTGAGGGTC-3′, whereas the reverse primer was 5′-GACCCTCAAATCAGGAGCAAAGTTGTCGGCAG-3′. The forward primer for the generation of S58E used was 5′-CTTCTGCCGACAACTTTGAGCCTGATTTGAGGGTCC-3′, whereas the reverse primer was 5′-GGACCCTCAAATCAGGCTCAAAGTTGTCGGCAGAAG-3′. The forward primer for the generation of C417S used was 5′-CTGTGGTAGTGCATAGCAGTCATGGTTGGG-3′, whereas the reverse primer was 5′-CCCAACCATCACTGCTATGCACTACCACAG-3′. All PCR-generated constructs were verified by DNA sequencing. The FLAG-tagged MTMR5 construct was a generous gift from Fred Robinson (25). The pEGFP-2xFYVE construct was a generous gift from Harald Stenmark (26).
Cell Culture, Transfections, and Cell Lysis
HeLa and HEK293 cells were grown as a monolayer and maintained at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) fetal bovine serum, 2 mm l-glutamine, and supplemented with antibiotics (100 units/ml of penicillin, 100 μg/ml of streptomycin). Cells were seeded 24 h prior to transfection and were ∼60–70% confluent at the time of transfection. Cells grown on 100-mm culture dishes were transiently transfected with 10 μg of plasmid DNA using polyethylenimine reagent (Sigma) according to the manufacturer's instructions. HeLa cells on culture slides were transfected with 0.5 μg of plasmid DNA at a 3:1 ratio of FuGENE® HD (Roche Applied Science) and DNA following the manufacturer's protocol and processed as indicated under immunofluorescence analysis. At 42 h post-transfection, cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in 0.5 ml (per 100-mm plate) of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 150 mm NaCl, 0.1% SDS) containing protease inhibitors (phenylmethylsulfonyl fluoride, 17.4 mg/ml, and aprotinin, 1 mg/ml). Lysates were cleared by centrifugation at 15,000 × g for 20 min at 4 °C and the protein concentration was determined by a Bradford assay.
Mass Spectrometry
FLAG-tagged MTMR2 was isolated by immunoprecipitation from unlabeled HEK293 cells (∼1 × 107 cells) or ∼2 × 106 cells labeled with [32P]orthophosphate to be used as a radioactive tracer. Following SDS-PAGE, the MTMR2 band was subjected to in-gel trypsin digestion. The total tryptic peptide pool was either directly analyzed by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) MS (Applied Biosystems), or separated by reverse-phase HPLC using a C18 column as previously described (27). HPLC fractions containing radioactive peptides were then analyzed by MALDI-TOF MS. To increase fragmentation and map the precise site of phosphorylation, peptides were sulfonated using the chemical assisted fragmentation (CAF) reagent (Amersham Biosciences) and analyzed by MALDI-MS/MS. Spectra were analyzed using Data Explorer Software (Applied Biosystems).
In Vitro Phosphatase Assays
Phosphatase assays were performed using 50 ng of bacterial recombinant MTMR-His6 proteins purified as described (28). Assays were carried out in reaction buffer (50 mm Tris, 150 mm NaCl, 50 mm BisTris, and 5 mm DTT, pH 7.0) containing 100 μm nonfluorescent di-C8-phosphatidylinositol 3-phosphate and 3,5-bisphosphate derivatives (Echelon). Reactions were carried out at 30 °C for 10 min and terminated using an equal volume of ice-cold 20 mm sodium orthovanadate (Sigma). Phosphate release was determined using a malachite green-based assay system (Echelon) for detecting inorganic phosphate. The absorbance measurements at wavelengths 620/660 nm were measured using a Victor3TM wallac 1420 multilabel counter (PerkinElmer Life Sciences) with free phosphate release determined using linear regression analysis (relative phosphatase activity reported in terms of picomole of inorganic phosphate released).
Co-Immunoprecipitation
HEK293 cells co-transfected with FLAG-MTMR2 and FLAG-MTMR5 were co-immunoprecipitated by pre-binding goat anti-MTMR5 (Santa Cruz) to Protein A-agarose (Invitrogen) (50 ng/μl slurry). Clarified lysate was incubated with 20 μl of antibody-bound Protein A-agarose for 3 h at 4 °C. Immunoprecipitates were washed extensively (50 mm Tris-HCl, pH 7.4, 0.1% Triton X-100, 150 mm NaCl, 0.1% SDS) and suspended in 2× SDS sample buffer.
Immunoprecipitates and lysates were resolved on an 8% SDS-PAGE gel, transferred to polyvinylidene difluoride membrane, and analyzed by immunoblotting. Membranes were blocked with 5% milk solution in 1× Tris-buffered saline (TBS) containing 0.1% Tween 20. Membranes were incubated with FLAG primary antibody (Sigma) for 1 h in 1% skim milk at room temperature. The membranes were washed three times in 1× TBST and incubated with anti-mouse IgG HRP conjugate (Promega) at a 1:5000 dilution in 1% milk for 45 min at room temperature. Proteins were visualized using Super Signal® West Femto reagent (Thermo Scientific). Images were acquired and analyzed using an Alpha Innotech Imager with Chemilmager version 5.5 software and Adobe Photoshop 7.0.
Immunofluorescence Analysis
HeLa cells were seeded in four-chamber tissue culture-treated glass slides (BD Biosciences) in DMEM and cultured as described above. Cells were transiently transfected by using FuGENE® HD according to the manufacturer's instructions. Wortmanin (LC Laboratories) treatments were performed for 1 h at a final concentration of 1 μm. All slides (treated and untreated) were washed twice with PBS (pH 7.4) and fixed with 3.7% paraformaldehyde in PBS for 10 min at room temperature. Where indicated, cells were treated with 0.01% saponin (Sigma) in PBS for 2 min at room temperature prior to fixation. The cells were then washed with TBS and permeabilized with 0.15% Triton X-100/TBS for 2 min. For immunostaining, cells were pre-treated for 1 h with 5% BSA/PBS as a blocking agent, then incubated for 1 h with primary antibodies in 1.0% BSA/PBS. Primary antibodies included anti-FLAG (Sigma; 1:500), anti-Rab5 (Santa Cruz; 1:80), anti-Rab7 (Sigma; 1:100), anti-LAMP1 (Abcam; 1:500), and anti-EGFR (Santa Cruz; 1:50). Cells were then washed with PBS three times for 5 min and incubated with corresponding secondary antibodies for 1 h in 1.0% BSA/PBS. Secondary antibodies employed included Alexa Fluor® 568 goat anti-mouse secondary antibody (Molecular Probes) and affinity purified fluorescein-conjugated goat anti-rabbit IgG (Vector Laboratories).
The cells were then washed and incubated with Hoechst 33342 (Invitrogen) stain (0.5 mg/ml in PBS) for 2 min at room temperature for DNA visualization. Images were captured using a Leica DMIRB inverted fluorescent microscope equipped with FITC/TRITC/DAPI filter cubes and HCX PL S-APO ×40/0.75 objective with a 1300i FAST Cooled Q-Imaging CCD camera (Qimaging Retiga). Additionally, a Zeiss Axiovert 200M inverted fluorescent microscope with FITC/TRITC/DAPI filter cubes and LD Plan-APO-Neofluar ×63/0.75 or EC Plan-Neofluar ×40/1.3 oil objectives equipped with a 1030 × 1300 monochrome CCD camera (Axiovision MRM) was also used. Captured pictures from both microscopes were processed with Northern Eclipse software 6.0 (Empix) and Adobe Photoshop 7.0.
Cell Signaling Analysis
Cells transfected with constructs encoding FLAG-tagged MTMR2 variants (wild type, S58A, and S58E), were incubated in serum-free medium at 37 °C for 30 min. Epidermal growth factor (EGF) (Sigma) was added at a final concentration of 5 ng/ml and incubated at 37 °C for the indicated time periods. The cells were washed, lysed, and immunoblotted as described above using the following antibodies: rabbit PathScan® Multiplex Western Mixture I (phosphorylated signaling proteins) (Cell Signaling Technology) was used at a 1:200 dilution in 5% BSA/TBST overnight at 4 °C, loading control primary antibodies included mitogen-activated protein kinase (MAPK) (Sigma; 1:10,000), EGFR (Santa Cruz; 1:3000), phospho-EGFR (Santa Cruz; 1:200), and actin (Sigma; 1:3000). The secondary anti-rabbit HRP was used at a 1:5000 dilution in 1% skim milk (Bio-Rad). Images were captured as described above.
RESULTS
The N Terminus of MTMR2 Possesses a High Stoichiometric Phosphorylation Site
The localization of MTMR2 under typical cell culture conditions has largely been classified as a diffuse cytoplasmic pattern (21, 22), whereas its substrates, PI(3)P and PI(3,5)P2, reside on suborganellar structures such as endosomes (19, 26). Thus, regulatory mechanisms such as post-translational modifications may play a critical role in controlling localization and thus access of MTMR2 to its physiological substrates.
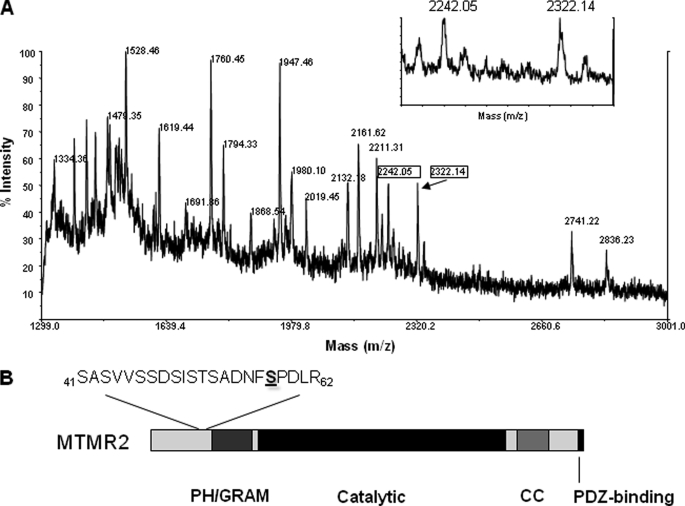
To investigate whether MTMR2 is modified by phosphorylation, FLAG-MTMR2 was immunoprecipitated from transfected HEK293 cells that were radiolabeled with [32P]orthophosphate. Protein samples were resolved by SDS-PAGE and analyzed by autoradiography, revealing that the immunoprecipitated FLAG-MTMR2 was highly phosphorylated. The radiolabeled protein band corresponding to FLAG-MTMR2 was excised from the gel and subjected to in-gel digestion with trypsin for analysis by mass spectrometry. The standard procedure for mapping phosphorylation sites by mass spectrometry involves a purification and/or enrichment step because of the low stoichiometry of phosphorylation sites and their poor ionization during mass spectrometry analysis (29, 30). Surprisingly, when an aliquot of the total tryptic peptide pool was analyzed by MALDI-TOF, we detected a prominent peptide mass at m/z 2322, which corresponds to an N-terminal peptide (amino acids 41–62) containing a single phosphate moiety (Fig. 1A). We also detected the same peptide lacking a phosphate group at m/z 2242. Repeated scans reproducibly yielded these two peptide peaks at roughly the same intensity. Because adding a phosphate moiety to a peptide has been shown to reduce ionization efficiency (29), our results strongly suggested that the stoichiometry of this phosphorylation event on MTMR2 was at least 50%, supporting the notion that a substantial proportion of MTMR2 was phosphorylated under the cellular conditions used.
FIGURE 1.
MTMR2 possesses a high stoichiometry phosphorylation site within its N terminus. FLAG-tagged MTMR2 was isolated from HEK293 cells by immunoprecipitation. MTMR2 was also immunoprecipitated from a small scale culture of cells radiolabeled with [32P]orthophosphate. The labeled and unlabeled protein samples were combined and separated by SDS-PAGE. The MTMR2 band was excised from the gel and subjected to in-gel digestion with trypsin. A, MALDI-TOF linear spectrum of the total pool of MTMR2 tryptic peptides. The m/z values corresponding to the unphosphorylated and phosphorylated forms of peptide Ser41-Arg62 are boxed. This region is also shown as an expanded view. B, schematic diagram of MTMR2 indicating the proximity of the Ser58 phosphorylation site to the PH/GRAM, catalytic, coiled-coil (CC) domains, and the PDZ binding motif.
Identification of Ser58 as the N-terminal Phosphorylation Site
To precisely identify the site of phosphorylation within the N-terminal peptide, we fractionated the total tryptic peptide pool using reverse phase chromatography. The phosphopeptide of interest was located in HPLC fraction 68 and analyzed by MALDI-MS/MS. As shown in Fig. 2A, in addition to the parent ion at m/z 2322, an intense peak at m/z 2224 was observed, which represents the neutral loss of the phosphate group. However, the peptide ion exhibited little additional fragmentation, thereby preventing assignment of the exact site of phosphorylation as this peptide contained eight potential phosphorylation sites. To increase fragmentation and improve MS/MS interpretation, we subjected the peptide to CAF analysis (31). This procedure involves sulfonating the N terminus of a peptide to increase proton mobility thus improving fragmentation at multiple sites along the peptide backbone. MALDI-CAF was successful at producing a superior MS/MS spectrum for analysis (Fig. 2B). Interpretation of the fragment ion series allowed for unambiguous mapping of the phosphorylation site to Ser58, which is ∼11 amino acids away from the regulatory PH/GRAM domain (Fig. 1B).
FIGURE 2.
Mapping the N-terminal phosphorylation site using MALDI-CAF mass spectrometry. MTMR2 tryptic peptides were fractionated by reverse-phase HPLC. A, the HPLC fraction containing the phosphopeptide of interest was further analyzed by MALDI-TOF MS/MS. The neutral loss of the phosphate moiety (−98 m/z) is indicated. B, the same HPLC fraction was subjected to CAF labeling and re-analyzed by MALDI-TOF MS/MS. The fragment ions whose m/z value corresponds to y or b ions with the loss of a phosphate group are indicated. The observed fragment ions are labeled on the spectrum and peptide sequence (phosphorylated Ser58 is underlined and in bold text).
MTMR2 Ser58 Phosphovariants Retain Catalytic Competence and MTMR5 Binding Activity
A common function of reversible phosphorylation of enzymes is to induce a conformational change that affects catalytic activity. To test whether the N-terminal phosphorylation site had an effect on lipid phosphatase activity, the phosphorylation mimetic (S58E) and phosphorylation-deficient (S58A) MTMR2 mutants were generated by site-directed mutagenesis. We purified recombinant wild-type and phosphorylation mutant MTMR2 proteins from Escherichia coli lysates and compared the in vitro phosphoinositide phosphatase activity against water soluble di-C8-PI(3)P and -PI(3,5)P2. As shown in Fig. 3A, MTMR2 phosphovariants retained full catalytic activity toward PI(3)P and PI(3,5)P2 as compared with wild-type MTMR2.
FIGURE 3.
Lipid phosphatase activity and MTMR5 association are unaffected by the phosphorylation status of Ser58. A, phosphatase assays using purified recombinant MTMR2 proteins were carried out at 30 °C for 10 min with 50 ng of enzyme (■, wild-type; [gboc], S58A; dark grey, S58E) and 100 μm of the indicated phosphoinositide substrates as described under “Experimental Procedures.” Phosphate release was measured using a malachite green-based assay for inorganic phosphate. The relative amount of phosphate released by recombinant MTMR2 proteins in these assays is shown in picomoles and represents the mean of triplicate determinations ± S.D. (n = 3). B, co-immunoprecipitation of overexpressed FLAG-tagged MTMR5 and MTMR2 proteins from HEK293 cells. At 42 h post-transfection, cell lysates were prepared and subjected to immunoprecipitation (IP) using MTMR5 antibody conjugated to Protein A-agarose beads. Following SDS-PAGE, immunoprecipitated proteins were detected by immunoblotting (IB) using anti-FLAG antibody. Whole cell lysates were probed for MTMR5 and MTMR2 to verify protein expression.
It has previously been demonstrated that MTMR2 interacts with the catalytically inactive MTMR proteins, MTMR5 and MTMR13, and that the coiled-coiled regions of the MTMR proteins are required for this interaction (22). To determine whether Ser58 phosphorylation regulates MTMR heterocomplex formation with inactive MTMRs, we conducted co-immunoprecipitation experiments using HEK293 cells co-transfected with MTMR5 and MTMR2 phosphomutant expression vectors. As shown in Fig. 3B, the interaction with MTMR5 was comparable between wild-type MTMR2 and the Ser58 phosphovariants. Taken together, these results demonstrate that phosphorylation of Ser58 does not affect the lipid phosphatase activity of MTMR2 or its ability to interact with inactive MTMR proteins.
Phosphorylation of Ser58 Regulates MTMR2 Subcellular Localization
MTMR2 contains an N-terminal PH-GRAM domain that has been implicated as a PIP-binding module and is predicted to be critical for MTMR2 subcellular localization (21). It is likely that modifications affecting PH-GRAM domain function will be critical for regulating MTMR2 function. Because the N-terminal phosphorylation site of MTMR2 is ∼11 amino acids upstream of the PH-GRAM domain, we investigated a potential role for Ser58 phosphorylation in regulating MTMR2 subcellular localization.
MTMR2 localization has previously been reported in various cell types including COS-1 and HEK293 cells, where predominant cytosolic staining with greater intensity in the perinuclear region was observed in transfected cells (21, 22). Notably, localization of MTMR2 to specific subcellular organelle(s) or membrane compartments such as endosomes under normal cell culture conditions has not been observed. In HeLa cells overexpressing MTMR2, we observed a similar localization pattern, with wild-type MTMR2 localizing predominantly to the cytosolic/perinuclear region as previously reported (Fig. 4A). Treatment of cells with saponin prior to fixation and immunostaining has been shown to deplete soluble cytoplasmic proteins, and permit visualization of membrane-localized proteins (32). HeLa cells expressing wild-type MTMR2 exhibited greatly reduced fluorescent staining when pretreated with saponin (Fig. 4B), suggesting that wild-type MTMR2 localization was predominantly cytoplasmic. Furthermore, the phosphomimetic S58E mutant displayed a similar localization pattern as wild-type, with diffuse cytoplasmic staining that largely disappeared following saponin treatment (Fig. 4, E and F). In contrast, the majority of the MTMR2 phosphorylation-deficient S58A mutant was localized to distinct punctate structures, indicative of endosomal localization (Fig. 4C). Accompanying treatment with saponin enhanced this punctate staining pattern, implying direct localization to intact subcellular organelles (Fig. 4D). Thus, our results strongly suggest that phosphorylation of Ser58 regulates MTMR2 subcellular localization by preventing its targeting to endocytic membranes. This finding also indicates that it is the unphosphorylated form of MTMR2 that preferentially localizes to endocytic-like vesicles.
FIGURE 4.
MTMR2 S58A localizes to punctate structures. HeLa cells were transfected with FLAG-MTMR2 phosphorylation mutants: wild-type (A and B), S58A (C and D), S58E (E and F). Following fixation, the cells were immunostained with anti-FLAG monoclonal antibody to localize FLAG-MTMR2 proteins. Saponin treatment (right panels) (0.01%) was used to enhance detection of membrane-localized proteins via depletion of cytosolic contents. Images were collected using ×63 and ×40 HD objectives. The scale bar represents 15 μm (10 μm in expanded views). Boxes denote the regions corresponding to the expanded images (right).
Endocytic Localization of the MTMR2 S58A Phosphorylation-deficient Mutant Greatly Enhances Endocytic PI(3)P Depletion
The observed punctate staining pattern exhibited by the MTMR2 S58A mutant prompted us to examine whether these punctae were also positive for Rab5 and/or Rab7 as markers for early and late endosomes, respectively. These GTPases have been well characterized to associate with early/late endosomes where they facilitate endosome maturation and trafficking processes (33). Immunofluorescence microscopy was performed on HeLa cells overexpressing the different MTMR2 phosphovariants using antibodies for endogenous Rab5. Wild-type MTMR2 and the S58E phosphomimetic mutant showed a diffuse cytosolic staining pattern that lacked co-localization with Rab5 (data not shown). Conversely, a subset of MTMR2 S58A positive vesicles were also positive for Rab5 (Fig. 5). Notably, significant Rab5 and MTMR2 S58A immunostaining were found on neighboring vesicles or opposite sides of the same vesicle (Fig. 5, C and F). In contrast, we did not detect significant co-localization of MTMR2 S58A with Rab7 (data not shown).
FIGURE 5.
Co-localization of MTMR2 S58A with the early endosomal protein Rab5. HeLa cells were transiently transfected with FLAG-MTMR2 S58A for 42 h and analyzed by immunofluorescence microscopy without saponin treatment (A–C), or with saponin treatment (D–F). Cells were probed for endogenous Rab5 (green) (A and D) and FLAG-MTMR2 S58A (red) (B and E). Merged images are shown to indicate partial co-localization (yellow) (C and F). Arrowheads indicate regions of interest and are presented in expanded views. Images were collected using ×63 (top panel) and ×40 oil (lower panel) objectives. The scale bar represents 15 μm (10 μm in expanded views).
The partial staining of MTMR2 S58A and Rab5 may be attributed to the phosphatase activity of MTMR2, as Rab5 associates with PI(3)P-binding proteins on early endosomes. Thus, to better define the MTMR2 S58A endocytic compartment we produced a catalytically inactive MTMR2 variant (S58A,C417S) and performed co-immunofluorescence with various endocytic markers (Fig. 6). We observed strong co-localization with markers for early endosomes including the PI(3)P marker generated from EEA1 (GFP-2xFYVE) and Rab5 (Fig. 6). On the other hand, co-localization was not detected with the late endosome marker Rab7 or the late endosome/lysosomal marker LAMP1 (Fig. 6).
FIGURE 6.
Co-localization of catalytically inactive MTMR2 S58A. C417S with early endosome markers. HeLa cells were transiently transfected with FLAG-MTMR2 S58A alone or co-transfected with EGFP-2xFYVE (upper panel) for 42 h and analyzed by immunofluorescence microscopy with saponin treatment. Cells were probed for FLAG-MTMR2 S58A (red) and endogenous Rab5, Rab7, or LAMP1 (green). Merged images are shown on the right panels with yellow indicative of co-localization. Boxes indicate regions of interest and are presented in expanded views. Images were collected using ×63 objectives. The scale bar represents 15 μm (10 μm in expanded views).
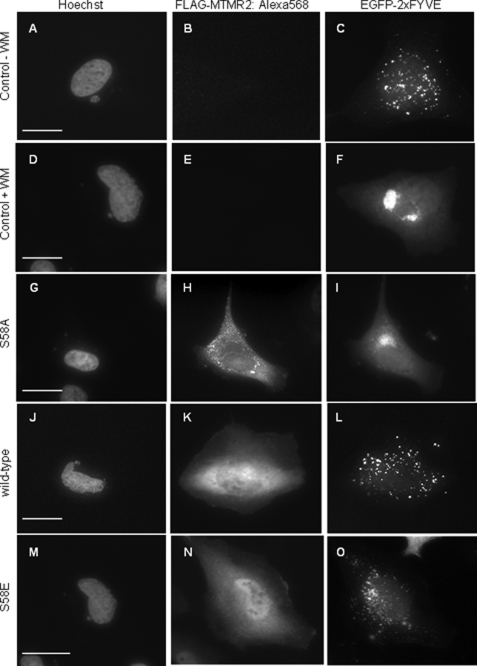
In addition to Rab GTPases, PI(3)P levels are indicative of actively maturing endosomes. The MTMR2 S58A mutant contained lipid phosphatase activity comparable with that of the wild-type enzyme. However, MTMR2 S58A exhibited a marked localization to endosomal structures within transfected cells, whereas wild-type MTMR2 did not. To determine whether MTMR2 S58A affected PI(3)P levels on the surface of these structures, we overexpressed GFP-2xFYVE to localize PI(3)P within these cells. Overexpression of the MTMR2 S58A mutant resulted in efficient depletion of PI(3)P from endosomal structures as indicated by the impaired localization of the GFP-2xFYVE probe (Fig. 7I). This effect was also observed following treatment of the cells with the PI 3-kinase inhibitor wortmanin, which markedly inhibits PI(3)P production (Fig. 7F) (26). Meanwhile, wild-type MTMR2 or MTMR2 S58E had little effect on GFP-2xFYVE endosomal localization (Fig. 7, L and O). Also, the catalytically inactive dephosphorylated MTMR2 (S58A,C417S) did not disperse the GFP-2xFYVE PI3P marker (Fig. 6, upper panel) in contrast to the catalytically active dephosphorylated MTMR2 (S58A) indicating the observed depletion of PI3P requires the phosphatase activity of MTMR2. Collectively, these findings demonstrate that phosphorylation-deficient MTMR2 (S58A) predominantly localizes to early endosomal structures, where it effectively dephosphorylates PI(3)P. Our data further implicates a regulatory mechanism by which reversible phosphorylation of MTMR2 at Ser58 regulates its ability to localize to early endosomes and regulate PI(3)P.
FIGURE 7.
MTMR2 S58A expression depletes PI(3)P from endosomes. HeLa cells were co-transfected with EGFP-2xFYVE and FLAG-MTMR2 phosphorylation mutants. After fixation, the cells were immunostained with anti-FLAG and Alexa Fluor® 568 anti-mouse secondary to detect MTMR2 proteins. Localization of the PI(3)P sensor EGFP-2xFYVE (right panels) on endosomes was detected as a punctate pattern. The punctate pattern is seen in control cells (A–C), and cells expressing wild-type MTMR2 (J–L) and S58E (M–O). The altered localization of the EGFP-2xFYVE marker was observed in both wortmanin-treated cells (D–F) and in cells expressing MTMR2 S58A (G–I). Images were collected using ×63 and × 40 HD objectives. The scale bar represents 15 μm.
MTMR2 Phosphorylation Site Mutant Augments EGF Signaling
The finding that MTMR2 S58A was enriched on early endocytic compartments and depleted localized PI(3)P suggested that PI(3)P-dependent functions in endocytic trafficking and signaling may be attenuated by MTMR2 when unphosphorylated at Ser58. Recent work has shown that depletion of PI(3)P on Rab5-positive early endosomes results in enhanced and prolonged EGF receptor signaling, including activation of the ERK1/2 and Akt signaling pathways (20). We therefore examined whether activation of EGFR signaling pathways might be altered in cells expressing the MTMR2 S58A phosphorylation-deficient mutant as compared with wild-type MTMR2. HeLa cells were transiently transfected with wild-type MTMR2 or S58A mutant, and the phosphorylation of downstream EGFR targets was monitored over several times periods following EGF treatment. A reproducible increase in the phosphorylation of ERK1/2 was observed in cells overexpressing MTMR2 S58A as compared with cells overexpressing wild-type MTMR2 (Fig. 8, A and B). This effect was enhanced at earlier time points (3–5 min). Densitometry analysis revealed an ∼2-fold increase in ERK1/2 phosphorylation with S58A mutant overexpression versus wild-type MTMR2 at the early time periods of 3 and 5 min, respectively (Fig. 8B). Although not as pronounced as the increase in ERK1/2 phosphorylation, phosphorylation of Akt (Ser473), S6 ribosomal protein (Ser235/Ser236), and 90 ribosomal S6 kinase (Ser380) were also increased by MTMR2 expression as compared with wild-type MTMR2 (Fig. 8A). Interestingly, under these conditions, we also observed strong co-localization of MTMR2 S58A with the EGFR (supplemental Fig. S1) and a modest but reproducible increase in EGFR phosphorylation at Tyr1173 (supplemental Fig. S2).
FIGURE 8.
Phosphorylation-deficient MTMR2 increases activation of EGF signaling. A, HeLa cells transfected with wild-type FLAG-MTMR2 and S58A were serum starved for 30 min and treated with 5 ng/ml of EGF for the indicated times. Lysates were immunoblotted (IB) for proteins phosphorylated in response to EGF treatment using the Pathscan® Multiplex antibody to detect phosphorylated proteins. Loading controls included total ERK1/2 and actin. FLAG-MTMR2 was immunostained with anti-FLAG to confirm equal MTMR2 expression levels in each of the samples. B, ERK1/2 phosphorylation levels were quantified by densitometry using Image J, and normalized to total ERK1/2 levels. The values indicate the mean ± S.D. (n = 3).
We further examined EGF-dependent signals in HEK293 and HeLa cells using a single time point (Fig. 9). The level of ERK1/2 phosphorylation/activation was also ∼2–3-fold higher in cells expressing MTMR2 S58A in comparison to control cells (Fig. 9, A and B). In contrast, wild-type MTMR2 and S58E had similar activated ERK1/2 levels when compared with control cells. Therefore, the endosomal localization and subsequent PI(3)P depletion by the MTMR2 S58A mutant is likely responsible for the enhanced EGF signaling detected. These results highlight an important biological consequence of regulating MTMR2 localization by reversible phosphorylation at Ser58.
FIGURE 9.
ERK1/2 activation enhanced by phosphorylation deficient MTMR2. A, HeLa and HEK293 cells were transfected with FLAG-MTMR2 expression vectors for 42 h. The cells were serum starved for 30 min and treated with 5 ng/ml of EGF for 5 min at 37 °C. ERK1/2 phosphorylation was determined by immunoblotting. Total ERK1/2 and actin levels served as loading controls. FLAG-MTMR2 immunoblotting was used to confirm equal transfection between samples. B, ERK1/2 phosphorylation was quantified by densitometry using the Image J program and normalized to total ERK1/2 levels (■, HeLa;  , HEK293). The values are shown as fold-change from control ± S.D. (n = 3).
, HEK293). The values are shown as fold-change from control ± S.D. (n = 3).
DISCUSSION
The results of this study demonstrate that reversible phosphorylation represents a critical mechanism for regulating the subcellular localization of MTMR2. The finding that an MTMR2 phosphorylation-deficient mutant preferentially localized to endocytic structures where it depleted PI(3)P suggests a mechanism by which phosphorylation of Ser58 impairs the localization of MTMR2 to compartments containing one of its physiological substrates, PI(3)P. Because the stoichiometry of MTMR2 Ser58 phosphorylation under normal cell culture conditions was quite high, we hypothesize that extracellular signals are required to induce Ser58 dephosphorylation (Fig. 10). By analogy, attenuation of signals that activate the kinase(s) responsible for phosphorylating MTMR2 at Ser58 would also result in an unphosphorylated MTMR2 and allow its localization to endocytic structures, including those containing PI(3)P. Following depletion of PI(3)P by MTMR2, phosphorylation of Ser58 may be required to dissociate MTMR2 from the vesicle to attenuate signal(s) generated by MTMR2-dependent PI(3)P dephosphorylation. In light of the highly dynamic role of PI(3)P in endocytic processes, the balance between MTMR2 Ser58 phosphorylation and dephosphorylation is likely to contribute to the maturation and signaling capacity of a distinct set of endosomes.
FIGURE 10.
Phosphorylation regulates endosomal targeting of MTMR2. Inhibition of MTMR2 Ser58 phosphorylation results in subcellular targeting to PI(3)P-positive endosomes, which leads to PI(3)P depletion and increased growth factor receptor signaling via endosomal pathways. Phosphorylation of Ser58 by a proline-directed kinase sequesters MTMR2 in the cytoplasm, thus preserving the levels of PI(3)P and promoting endosomal maturation/trafficking.
One of the most striking aspects of the Ser58 phosphorylation site was the apparent high stoichiometry determined by mass spectrometry. Phosphopeptides are rarely detected in unfractionated trypsin digests primarily due to their low stoichiometry, making them underrepresented in a complex mixture and thus subject to suppression by other unphosphorylated peptides present (30). However, the phospho-Ser58 peptide was readily detected in the complex peptide pool, along with the unphosphorylated peptide. These cognate peptides were reproducibly detected at approximately equal intensities. Taking into consideration the widely held view that phosphate addition reduces peptide ionization, we can safely conclude that the stoichiometry of phosphorylation is at least 50% and is likely to be considerably higher under the conditions used in the study. This conclusion is further supported by subcellular localization studies and PI(3)P depletion experiments, where localization of wild-type MTMR2 and endocytic PI(3)P depletion were indistinguishable from the MTMR2 S58E phosphomimetic mutant. In contrast, the unphosphorylatable MTMR2 S58A mutant displayed striking endosomal localization and caused a marked depletion of endosomal PI(3)P. These findings further substantiate our working model that MTMR2 exists in a constitutively phosphorylated state, where it is sequestered away from its physiological substrate until the appropriate signal leads to its dephosphorylation and subsequent targeting to endocytic vesicles, where PI(3)P is localized. It must be taken into consideration that we utilized MTMR2 overexpression throughout these studies. However, during the course of elucidating the functional relevance of the Ser58 phosphorylation site, three independent phosphoproteomic screening studies reported phosphorylation at Ser58 of endogenous MTMR2, thus validating the results obtained in our overexpression studies (34–36). Ongoing efforts of developing a Ser58 phosphospecific antibody will help determine the relative stoichiometry of phosphorylation at Ser58 on endogenous MTMR2 under various conditions.
Inhibitory phosphorylation events that affect membrane binding have been shown to be critical for other inositol lipid phosphatases, including PTEN. Several lines of evidence suggest that phosphorylation of PTEN by casein kinase II and other kinases can inhibit its cleavage by caspase 3 and increase its stability (37, 38). Phosphorylated PTEN also exhibits decreased lipid phosphatase activity and is unable to interact with binding factors that target it to the inner surface of the plasma membrane (39–41). Collectively, these findings have led to a model by which phosphorylation of PTEN induces a closed conformation that is stable, but inactive. PTEN dephosphorylation induces an open conformation that allows PTEN to localize to membrane compartments and act on its lipid substrate, PI(3,4,5)P3. The membrane-associated (unphosphorylated) form of PTEN is more susceptible to proteolytic cleavage, which functions to allow down-regulation of PTEN signaling.
The exact mechanism of MTMR2 phosphorylation remains to be elucidated. However, the sequences surrounding the phosphorylation site can often offer clues to the kinases involved. Ser58 resides in a so-called SP motif, suggesting that MTMR2 is regulated by proline-directed kinases such as members of the MAPK family. Although numerous proline-directed kinases are encoded in the human genome, which represent potential MTMR2 Ser58 kinases, a wide array of chemical inhibitors and proteomic tools are available that will aid in our ongoing efforts to identify the specific kinase(s) that phosphorylate MTMR2 at Ser58.
At this time it is uncertain what extracellular stimuli may be responsible for regulating MTMR2 reversible phosphorylation. However, Berger et al. (21) reported that hypo-osmotic stress in COS-1 cells caused translocation of MTMR2 to membrane vacuoles rich in PI(3,5)P2. Furthermore, a CMT4B disease-associated point mutation in the PH-GRAM domain greatly reduced binding to these vacuoles. We now present compelling data that phosphorylation of a site in close proximity to the PH-GRAM domain regulates endosomal membrane targeting. Because hypo-osmotic stress is known to regulate proline-directed kinases, including MAPK members (42, 43), it is plausible that the localization of MTMR2 to vacuolar structures in response to osmotic stress may involve dephosphorylation of MTMR2 at Ser58. We are currently exploring this possibility as well as systematically identifying regulatory signals that can regulate Ser58 phosphorylation.
Several studies have demonstrated that active MTMRs are positively regulated by inactive MTMR binding partners (22, 25, 44, 45). MTMR2 has been shown to interact with two highly homologous members within the inactive MTMR subtypes, MTMR5 and MTMR13. The mode of interaction between active and inactive MTMRs is thought to be mediated by the coiled-coil domains found in the C termini of the active and inactive binding partners (22, 25). These associations have been suggested to be important for MTMR2 localization. Because we detected no significant difference in the ability of MTMR2 phosphorylation site mutants to interact with MTMR5, and overexpression of MTMR5 did not alter the phosphorylation levels of MTMR2 (data not shown), we conclude that MTMR2 Ser58 phosphorylation is unlikely to regulate its interaction with MTMR5 or MTMR13. Because MTMR13 mutations are also associated with type 4B CMT disease, it will be important to conduct similar experiments focused on MTMR13 to determine whether MTMR2 Ser58 phosphorylation might affect its interaction with MTMR13.
Another interesting observation was the co-localization of MTMR2 S58A and Rab5. We observed a subset of Rab5 positive endosomes that were also positive for catalytically active MTMR2 S58A, suggesting that unphosphorylated MTMR2 is targeted to endocytic like vesicles. However, a considerable amount of MTMR2 S58A positive vesicles were Rab5 negative but often directly adjacent to Rab5 positive endosomes (Fig. 5). This perhaps signifies that targeting of MTMR2 S58A to endocytic structures could affect Rab5 function. Moreover, this putative function would require phosphatase activity as the catalytically inactive variant (MTMR2 S58A,C417S) displayed strong co-localization with Rab5 (Fig. 6). Notably, Rab5 endosomal activities have been linked to PI(3)P levels (33, 46). For example, the critical endosomal effectors Rabenosyn-5 and early endosomal antigen 1 require association with Rab5 and PI(3)P simultaneously to localize to early endosomes (47–49). Furthermore, Rab5 associates with endosomes in its GTP bound form, which is maintained by endosomal guanine exchange factors such as RABX-5. Poteryaev et al. (46) have recently discovered that the endosomal protein Mon1/SAND-1 removes RABX-5 (and subsequently Rab5) from endosomes in a PI(3)P-dependent manner. Therefore, based on our co-localization results and the ability of MTMR2 S58A to effectively deplete endosomal PI(3P), it is conceivable that reversible phosphorylation of MTMR2 could regulate Rab5-dependent trafficking events that are sensitive to PI(3)P. Moreover, the observed co-localization between MTMR2 S58A and EGFR and the MTMR2 S58A-mediated increase in EGF signaling is consistent with a model whereby phosphorylation deficient MTMR2 disrupts the ability of Rab5 to modulate EGFR endocytosis resulting in prolonged receptor signaling.
In conclusion, we have identified a novel mechanism through which the endosomal targeting of MTMR2 is regulated by reversible phosphorylation at Ser58. This discovery provides critical insight into how MTMR2 activity toward its lipid substrates can be spatially and temporally controlled. Furthermore, differential phosphorylation of other MTMR family members may represent a specific means for targeting these enzymes to distinct pools of PI(3)P and PI(3,5)P2. Likewise, understanding the mechanistic details of MTMR phosphorylation may help to clarify how similar gene products (MTM1 and MTMR2) can cause distinct human disorders. Thus, it is of great interest to further investigate whether other MTMR members are phosphorylated and how these modifications regulate their activities.
Supplementary Material
Acknowledgments
We thank Anna Kozarova, Justin Renaud, and Catherine Cheng for technical assistance.
This work was supported, in whole or in part, by a National Institutes of Health National Center for Research Resources grant to the Nebraska Center for Cellular Signaling (to G. S. T.) and Operating Grant 89870 from Canadian Institutes of Health Research (CIHR) (to P. O. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PIP
- phosphatidylinositol phosphate
- MTMR
- myotubularin-related
- CAF
- chemically assisted fragmentation
- PH-GRAM
- pleckstrin homology-GRAM domain
- PI(3)P
- phosphoinositide 3-phosphate
- PI(3,4)-P2
- phosphoinositide 3,5-bisphosphate
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Barford D., Das A. K., Egloff M. P. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 133–164 [DOI] [PubMed] [Google Scholar]
- 2. Fauman E. B., Saper M. A. (1996) Trends Biochem. Sci. 21, 413–417 [DOI] [PubMed] [Google Scholar]
- 3. Guan K. L., Broyles S. S., Dixon J. E. (1991) Nature 350, 359–362 [DOI] [PubMed] [Google Scholar]
- 4. Guan K. L., Dixon J. E. (1991) J. Biol. Chem. 266, 17026–17030 [PubMed] [Google Scholar]
- 5. Taylor G. S., Maehama T., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maehama T., Dixon J. E. (1998) J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 7. Alexiou G. A., Voulgaris S. (2010) Neurol. Neurochir. Pol. 44, 80–86 [DOI] [PubMed] [Google Scholar]
- 8. Whang Y. E., Wu X., Suzuki H., Reiter R. E., Tran C., Vessella R. L., Said J. W., Isaacs W. B., Sawyers C. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5246–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Senderek J., Bergmann C., Weber S., Ketelsen U. P., Schorle H., Rudnik-Schöneborn S., Büttner R., Buchheim E., Zerres K. (2003) Hum. Mol. Genet. 12, 349–356 [DOI] [PubMed] [Google Scholar]
- 10. Azzedine H., Bolino A., Taïeb T., Birouk N., Di Duca M., Bouhouche A., Benamou S., Mrabet A., Hammadouche T., Chkili T., Gouider R., Ravazzolo R., Brice A., Laporte J., LeGuern E. (2003) Am. J. Hum. Genet. 72, 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolino A., Muglia M., Conforti F. L., LeGuern E., Salih M. A., Georgiou D. M., Christodoulou K., Hausmanowa-Petrusewicz I., Mandich P., Schenone A., Gambardella A., Bono F., Quattrone A., Devoto M., Monaco A. P. (2000) Nat. Genet. 25, 17–19 [DOI] [PubMed] [Google Scholar]
- 12. Laporte J., Hu L. J., Kretz C., Mandel J. L., Kioschis P., Coy J. F., Klauck S. M., Poustka A., Dahl N. (1996) Nat. Genet. 13, 175–182 [DOI] [PubMed] [Google Scholar]
- 13. Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. (2001) Annu. Rev. Biochem. 70, 535–602 [DOI] [PubMed] [Google Scholar]
- 14. Balla T. (2005) J. Cell Sci. 118, 2093–2104 [DOI] [PubMed] [Google Scholar]
- 15. Cullen P. J., Cozier G. E., Banting G., Mellor H. (2001) Curr. Biol. 11, R882–893 [DOI] [PubMed] [Google Scholar]
- 16. Nicot A. S., Laporte J. (2008) Traffic 9, 1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suter U. (2007) Cell Mol. Life Sci. 64, 3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Begley M. J., Taylor G. S., Kim S. A., Veine D. M., Dixon J. E., Stuckey J. A. (2003) Mol. Cell 12, 1391–1402 [DOI] [PubMed] [Google Scholar]
- 19. Downes C. P., Gray A., Lucocq J. M. (2005) Trends Cell Biol. 15, 259–268 [DOI] [PubMed] [Google Scholar]
- 20. Zoncu R., Perera R. M., Balkin D. M., Pirruccello M., Toomre D., De Camilli P. (2009) Cell 136, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berger P., Schaffitzel C., Berger I., Ban N., Suter U. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12177–12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim S. A., Vacratsis P. O., Firestein R., Cleary M. L., Dixon J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4492–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laporte J., Blondeau F., Gansmuller A., Lutz Y., Vonesch J. L., Mandel J. L. (2002) J. Cell Sci. 115, 3105–3117 [DOI] [PubMed] [Google Scholar]
- 24. Kim S. A., Taylor G. S., Torgersen K. M., Dixon J. E. (2002) J. Biol. Chem. 277, 4526–4531 [DOI] [PubMed] [Google Scholar]
- 25. Robinson F. L., Dixon J. E. (2005) J. Biol. Chem. 280, 31699–31707 [DOI] [PubMed] [Google Scholar]
- 26. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vacratsis P. O., Phinney B. S., Gage D. A., Gallo K. A. (2002) Biochemistry 41, 5613–5624 [DOI] [PubMed] [Google Scholar]
- 28. Taylor G. S., Dixon J. E. (2004) Methods Mol. Biol. 284, 217–227 [DOI] [PubMed] [Google Scholar]
- 29. Klemm C., Otto S., Wolf C., Haseloff R. F., Beyermann M., Krause E. (2006) J. Mass Spectrom. 41, 1623–1632 [DOI] [PubMed] [Google Scholar]
- 30. Boersema P. J., Mohammed S., Heck A. J. (2009) J. Mass Spectrom. 44, 861–878 [DOI] [PubMed] [Google Scholar]
- 31. Keough T., Lacey M. P., Youngquist R. S. (2000) Rapid Commun. Mass Spectrom. 14, 2348–2356 [DOI] [PubMed] [Google Scholar]
- 32. Lin A., Krockmalnic G., Penman S. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8565–8569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vartak N., Bastiaens P. (2010) EMBO J. 29, 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Sci. Signal. 2, ra46. [DOI] [PubMed] [Google Scholar]
- 35. Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 36. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Birle D., Bottini N., Williams S., Huynh H., deBelle I., Adamson E., Mustelin T. (2002) J. Immunol. 169, 286–291 [DOI] [PubMed] [Google Scholar]
- 38. Xu D., Yao Y., Jiang X., Lu L., Dai W. (2010) J. Biol. Chem. 285, 39935–39942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vazquez F., Grossman S. R., Takahashi Y., Rokas M. V., Nakamura N., Sellers W. R. (2001) J. Biol. Chem. 276, 48627–48630 [DOI] [PubMed] [Google Scholar]
- 40. Wu X., Hepner K., Castelino-Prabhu S., Do D., Kaye M. B., Yuan X. J., Wood J., Ross C., Sawyers C. L., Whang Y. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Y., Dowbenko D., Spencer S., Laura R., Lee J., Gu Q., Lasky L. A. (2000) J. Biol. Chem. 275, 21477–21485 [DOI] [PubMed] [Google Scholar]
- 42. Davenport K. R., Sohaskey M., Kamada Y., Levin D. E., Gustin M. C. (1995) J. Biol. Chem. 270, 30157–30161 [DOI] [PubMed] [Google Scholar]
- 43. Tilly B. C., van den Berghe N., Tertoolen L. G., Edixhoven M. J., de Jonge H. R. (1993) J. Biol. Chem. 268, 19919–19922 [PubMed] [Google Scholar]
- 44. Mochizuki Y., Majerus P. W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9768–9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nandurkar H. H., Layton M., Laporte J., Selan C., Corcoran L., Caldwell K. K., Mochizuki Y., Majerus P. W., Mitchell C. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8660–8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Poteryaev D., Datta S., Ackema K., Zerial M., Spang A. (2010) Cell 141, 497–508 [DOI] [PubMed] [Google Scholar]
- 47. Zerial M., McBride H. (2001) Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 48. Schnatwinkel C., Christoforidis S., Lindsay M. R., Uttenweiler-Joseph S., Wilm M., Parton R. G., Zerial M. (2004) PLoS Biol. 2, E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Merithew E., Stone C., Eathiraj S., Lambright D. G. (2003) J. Biol. Chem. 278, 8494–8500 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.