Abstract
The transforming growth factor-β (TGF-β) superfamily of ligands signals along two intracellular pathways, Smad2/3-mediated TGF-β/activin pathway and Smad1/5/8-mediated bone morphogenetic protein pathway. The C terminus of Hsc70-interacting protein (CHIP) serves as an E3 ubiquitin ligase to mediate the degradation of Smad proteins and many other signaling proteins. However, the molecular mechanism for CHIP-mediated down-regulation of TGF-β signaling remains unclear. Here we show that the extreme C-terminal sequence of Smad1 plays an indispensable role in its direct association with the tetratricopeptide repeat (TPR) domain of CHIP. Interestingly, Smad1 undergoes CHIP-mediated polyubiquitination in the absence of molecular chaperones, and phosphorylation of the C-terminal SXS motif of Smad1 enhances the interaction and ubiquitination. We also found that CHIP preferentially binds to Smad1/5 and specifically disrupts the core signaling complex of Smad1/5 and Smad4. We determined the crystal structures of CHIP-TPR in complex with the phosphorylated/pseudophosphorylated Smad1 peptides and with an Hsp70/Hsc70 C-terminal peptide. Structural analyses and subsequent biochemical studies revealed that the distinct CHIP binding affinities of Smad1/5 or Smad2/3 result from the nonconservative hydrophobic residues at R-Smad C termini. Unexpectedly, the C-terminal peptides from Smad1 and Hsp70/Hsc70 bind in the same groove of CHIP-TPR, and heat shock proteins compete with Smad1/5 for CHIP interaction and concomitantly suppress, rather than facilitate, CHIP-mediated Smad ubiquitination. Thus, we conclude that CHIP inhibits the signaling activities of Smad1/5 by recruiting Smad1/5 from the functional R-/Co-Smad complex and further promoting the ubiquitination/degradation of Smad1/5 in a chaperone-independent manner.
Keywords: E3 Ubiquitin Ligase, Protein Degradation, Protein Structure, Protein-Protein Interactions, Signal Transduction
Introduction
Transforming growth factor-β (TGF-β) signaling, subdivided into TGF-β/activin pathway and bone morphogenetic protein (BMP)2 pathway, regulates a variety of essential cellular responses including cell proliferation, differentiation, apoptosis, and specification of developmental fate (1–3). The Smad family proteins are key components for the intracellular signaling cascade and are classified into three functional groups: the receptor-regulated Smads (R-Smads), the co-mediator Smads (Co-Smads), and the inhibitory Smads. Upon ligand binding, the specific heteromeric transmembrane serine/threonine kinase receptor complexes undergo phosphorylation/activation and subsequently phosphorylate the two Ser residues in the C-terminal SXS motif of specific R-Smads, Smad1/5/8 for BMP pathway and Smad2/3 for TGF-β/activin signaling. The activated R-Smads then associate with Co-Smad, Smad4. The heteromeric complexes translocate into the nucleus, where they bind to DNA directly or indirectly to regulate the transcription of specific genes. The inhibitory Smads (Smad6/7) play an opposing role by counteracting the effects of R-Smads (4).
Dysfunction of TGF-β signaling is associated with many types of human cancer (5, 6). Different regulatory mechanisms are involved in controlling the TGF-β pathway, especially via down-regulating the Smad proteins. Recent studies have shown that Smad proteins constantly shuttle between the cytoplasm and nucleus, and the exported R-Smads are dephosphorylated by specific R-Smad phosphatases (7–10). In addition, Smad proteins are subjected to proteasomal degradation by various classes of E3 ubiquitin ligases (11–13). Smad ubiquitination regulatory factor (Smurf) of the homologous to the E6-AP C terminus (HECT) family provides the first link between TGF-β signaling and the ubiquitination system, which induces the degradation of not only Smad proteins but also activated receptors and transcriptional co-repressors (14–16). In a yeast two-hybrid screen, the C terminus of Hsc70-interacting protein (CHIP) was isolated as a Smad1-interacting protein and was shown to induce Smad1 degradation (17).
CHIP is highly conserved across species and plays a crucial role in the protein quality control system (18–20). CHIP comprises three domains, an N-terminal tetratricopeptide repeat (TPR) domain, a C-terminal Ubox domain, and an intervening coiled-coil region. The TPR domain of CHIP interacts with the molecular chaperones Hsc70/Hsp70 and Hsp90, as CHIP was originally identified as a co-chaperone in protein folding (21). In addition, CHIP has the Ubox-dependent ubiquitin ligase activity and triggers degradation of chaperone client proteins by the proteasome while abrogating their folding. For instance, CHIP can facilitate the chaperone-dependent degradation of a number of signature proteins involved in neurodegenerative diseases (22). Accumulated evidence has also linked CHIP to tumor development, as several oncogenic proteins are its substrates (23–26). A recent study observed that expression of CHIP suppressed tumor cell growth and metastasis in breast cancer, suggesting that CHIP may as well have a role in controlling tumor progression (27). Therefore, CHIP acts as a link between the folding-refolding machinery and the degradation pathway and might help the development of innovative therapeutic interventions for neurodegenerative disorders and cancers.
In addition to target Smad1 for degradation, CHIP was also found to mediate ubiquitination of other R- and Co-Smad proteins (17, 28, 29). However, the molecular mechanism for CHIP to regulate Smad activity is unclear. In particular, it is vital to understand how Smad proteins are recognized by CHIP and how the binding of CHIP to Smads suppresses TGF-β signaling. To address these questions, we carried out structural and functional analyses. We found that CHIP ubiquitinates Smad1/5, which depends on the direct interaction between the TPR domain of CHIP and the extreme C-terminal signature sequence ISSVS of Smad1/5. The crystal structures of CHIP-TPR in complexes with the phosphorylated or pseudophosphorylated Smad1 C-terminal peptides and with an Hsc70/Hsp70 peptide revealed that the three peptides bind into the same groove on CHIP-TPR, and subsequent competition experiments showed that the molecular chaperones antagonize CHIP-mediated Smad1 ubiquitination. Remarkably, the C-terminal hydrophobic residues are not conserved among Smads, which are the determinants for the distinct binding affinities of CHIP to different Smad proteins. Moreover, CHIP competes with Co-Smad Smad4 for Smad1 binding and, therefore, disrupts the crucial R-/Co-Smad complex. Together, our data suggest that CHIP plays important roles in down-regulating Smad1/5 activity via sequestering Smad1/5 from the core signaling complex, further mediating its ubiquitination in a chaperone-independent manner.
EXPERIMENTAL PROCEDURES
Constructs, Mutagenesis, and Protein Purification
Plasmids of human Smad proteins were provided by Dr Sheng-Cai Lin (Xiamen University). Fragments, mutants, and chimeric proteins of Smads and CHIP were generated by PCR-based strategies and inserted into pET21b or pGEX4T vectors with a C-terminal His6 tag or N-terminal GST tag. The authenticity of each construct was confirmed by nucleotide sequencing. All proteins, overexpressed in Escherichia coli strain BL21(DE3) at room temperature, were first isolated over nickel-nitrilotriacetic acid columns (Qiagen) or glutathione-Sepharose columns (GE Healthcare) and further purified by anion exchange and gel filtration chromatography (Source-15Q and Superdex-200, GE Healthcare). The affinity tag was proteolytically removed from the fusion proteins when indicated. The identities of all proteins were confirmed by mass spectroscopy. Detailed procedures for protein purification are summarized in supplemental Table S1. Hsp70/Hsc70-C peptide (NH3+-GPTIEEVD-COO−), Hsp90-C peptide (NH3+-SRMEEVD-COO−), and pseudophosphorylated Smad1-DVD peptide (NH3+-SPHNPISDVD-COO−) were synthesized by Beijing Scilight Biotechnology LLC. The phosphorylated Smad1 peptide (NH3+-SPHNPISpSVpS-COO−) was synthesized by Shanghai GL Biochem Ltd.
Gel Filtration Assays
Size exclusion chromatography using a Superdex 200 10/300 column on an ÄKTA FPLC (GE Healthcare) was carried out to assess the interaction between CHIP and Smad proteins at 4 °C. All protein samples were incubated at 4 °C for at least 30 min to allow equilibrium to be reached. The column was equilibrated with the buffer containing 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 2 mm dithiothreitol (DTT) at a flow rate of 0.5 ml/min and calibrated with molecular mass standards. All fractions were collected at 0.5 ml each, and aliquots of relevant fractions were subjected to SDS-PAGE followed by Coomassie Blue staining.
GST-mediated Pulldown Assays
GST-mediated pulldown assay was carried out to assess the interaction between CHIP and Smad proteins at 4 °C. The experiments were initiated with the binding of 0.4 mg of recombinant GST-tagged Smad proteins to 0.2 ml of glutathione-Sepharose 4B resin (GE Healthcare). To remove excess unbound Smad or other contaminants, the resin was washed 5 times with 1.0 ml of buffer containing 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 2 mm DTT. Then, 0.6 mg of non-tagged CHIP was allowed to flow through the resin (GE Healthcare). After extensive washing, the bound proteins were eluted with 5 mm reduced glutathione. Samples were subjected to SDS-PAGE, and proteins were visualized by Coomassie Blue staining.
Plasmids, Cell Culture, Transfection, and Co-immunoprecipitation
pCS2+-FLAG-Smads and pCDNA-HA-CHIP were generated according to standard molecular techniques and subjected to DNA sequencing. HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone), 100 μg/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified, 5% CO2 incubator. Cells were co-transfected with FLAG-tagged Smad and HA-tagged CHIP using VigoFect (Vigorous) according to the manufacturer's instructions and lysed at 48 h after transfection in a buffer containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 25 mm sodium fluoride, 2 mm sodium orthovanadate, 1 mm EDTA, and protease inhibitors. The lysates were incubated on ice for 10 min and then centrifuged at 13,000 rpm for 10 min at 4 °C. The supernatants were immunoprecipitated with mouse monoclonal anti-FLAG antibody (Sigma) and protein A/G plus agarose (Santa-Cruz Biotechnology) and rotated overnight at 4 °C. After spinning and washing 3 times with the lysis buffer, the beads were mixed with 2× SDS sample buffer, boiled, and subjected to 12% SDS-PAGE. The samples were transferred to PVDF membranes (Millipore) and immunoblotted with anti-HA and anti-FLAG antibodies (Sigma).
Ubiquitination Assays
The ubiquitination assay was performed according to the method of Li et al. (29) with modification. The reaction mixture (20 μl) containing 5 μm GST-tagged Smad proteins, 0.1 μm E1, 2.5 μm UbcH5a, 5 μm CHIP, and 2 μg/μl ubiquitin in the ATP regenerating system was incubated for 4 h at 30 °C. The ATP regenerating system contains 50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm DTT, 1 mm ATP (Sigma), 10 mm creatine phosphate (Fluka), 4 mm magnesium acetate (Sigma), and 10 unit/ml creatine kinase (Sigma). Samples were analyzed by SDS-PAGE and then subjected to immunoblot with anti-GST antibody.
Crystallization, Data Collection, and Structure Determination
Crystals of CHIP-TPR in complex with phosphorylated Smad1 peptide were grown by the hanging-drop vapor diffusion method by mixing the protein/peptide complex (∼7 mg/ml) with an equal volume of reservoir solution containing 100 mm Bistris propane, pH 7.0, 40% polyethylene glycol monomethyl ether 2000, 1% polyethylene glycol monomethyl ether 550, and 50 mm magnesium acetate at 21 °C. Crystals of CHIP-TPR in complex with pseudophosphorylated Smad1-DVD peptide appeared after 2–3 days with a reservoir solution containing 100 mm Bistris propane, pH 7.0, 41% polyethylene glycol monomethyl ether 2000, 50 mm magnesium acetate, and 50 mm proline. Crystals of CHIP-TPR in complex with Hsp70/Hsc70 peptide were obtained with a reservoir solution containing 100 mm Bistris propane, pH 7.0, 40% polyethylene glycol monomethyl ether 2000, and 3% (w/v) xylitol. The crystals were cryoprotected in the reservoir solutions supplemented with 5% glycerol and flash-frozen under cold nitrogen stream at 100 K. The diffraction data were collected at 0.97924 Å at beamline 17U at Shanghai Synchrotron Radiation Facility and processed using the HKL2000 (30). The structures were solved by molecular replacement using Phaser with the structure of mouse CHIP bound to Hsp90 peptide (PDB code 2C2L) as search model (31). Standard refinement was performed with the programs Phenix (32) and Coot (33). The data processing and refinement statistics were summarized in Table 1. All structural representations were prepared using PyMOL. The atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession codes 3Q4A for the complex of CHIP-TPR and phosphorylated Smad1 peptide, 3Q47 for that of CHIP-TPR and pseudophosphorylated Smad1-DVD peptide, and 3Q49 for that of CHIP-TPR and Hsp70/Hsc70-C peptide.
TABLE 1.
Data collection and refinement statistics
| CHIP-TPR in complex with |
|||
|---|---|---|---|
| Phosphorylated Smad1 peptide | Smad1-DVD peptide | Hsp70/Hsc70-C peptide | |
| Data collectiona | |||
| Space group | P21212 | P21212 | P21212 |
| Cell dimensions | |||
| a, b, c (Å) | 46.1, 77.4, 36.5 | 46.0, 77.4, 36.1 | 46.0, 78.0, 37.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0-1.54 (1.57-1.54)b | 99.0-1.70 (1.73-1.70) | 50.0-1.54 (1.57-1.54) |
| Rmerge | 5.3 (17.1) | 7.2 (26.5) | 6.2 (19.5) |
| I/σ (I) | 57.7 (11.8) | 41.1 (6.9) | 52.2 (11.2) |
| Completeness (%) | 99.4 (97.9) | 97.0 (82.4) | 99.8 (99.7) |
| Redundancy | 7.7 (7.1) | 7.1 (5.5) | 7.5 (6.6) |
| Refinement | |||
| Resolution (Å) | 33.0-1.54 | 32.7-1.7 | 33.7-1.54 |
| No. reflections | 19,797 | 14,183 | 20,035 |
| Rwork/Rfree | 16.8/20.8 | 17.3/19.2 | 16.8/20.7 |
| No. atoms | |||
| Protein | 1,114 | 1,118 | 1,121 |
| Water | 185 | 167 | 198 |
| B-factors | |||
| Protein | 18.6 | 22.1 | 17.8 |
| Water | 31.5 | 36.8 | 27.2 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.01 | 0.01 | 0.01 |
| Bond angles (°) | 1.03 | 1.14 | 0.87 |
| Ramachandran plot | |||
| Most favored | 115 (93.5%) | 115 (93.5%) | 114 (91.9%) |
| Additionally allowed | 8 (6.5%) | 8 (6.5%) | 10 (8.1%) |
| Generously allowed | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Disallowed | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
a All data sets were collected from single crystals.
b Values in parentheses are for the highest resolution shell.
RESULTS
The C-terminal SXS Motif of Smad1 Is Indispensable for CHIP-Smad1 Interaction
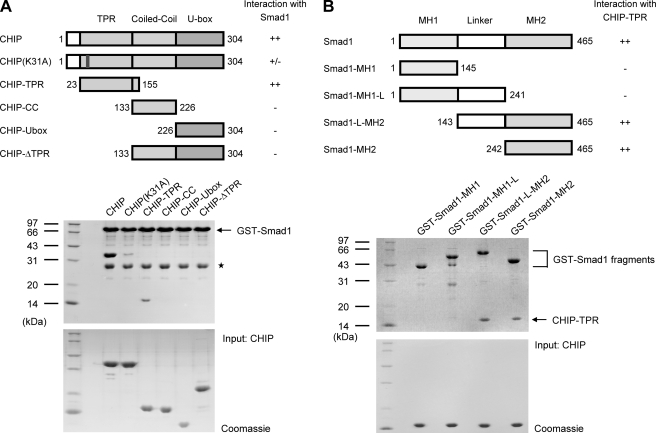
CHIP was linked to the TGF-β signaling as a Smad1-interacting protein (17). To identify domains responsible for the Smad1-CHIP interaction, we performed GST-mediated pulldown assays using various CHIP fragments and GST-tagged full-length Smad1. The results showed that Smad1 binds to full-length CHIP and the TPR domain (CHIP-TPR) but not to other isolated domains, indicating that CHIP-TPR is necessary and sufficient for the interaction of CHIP with Smad1 (Fig. 1A). Smad proteins are highly conserved within the N-terminal Mad homology 1 (MH1) domain and the C-terminal MH2 domain. As shown in Fig. 1B, the MH2 domain of Smad1 (Smad1-MH2) is able to sufficiently interact with CHIP-TPR. These data demonstrate that the CHIP-Smad1 interaction is mainly mediated by CHIP-TPR and Smad1-MH2 domains, which is consistent with a previous study (17).
FIGURE 1.
Determination of domains required for CHIP-Smad1 interaction. A, identification of the Smad1 binding domain on CHIP is shown. Glutathione beads immobilized with wild type GST-Smad1 were used to pull down CHIP mutants. Schematic diagrams on the top depict the CHIP proteins used, and a summary for their interaction with Smad1 is also listed on the right. The middle panel is the electrophoretic pattern of CHIP and Smad1 proteins after GST pulldown assays. The protein amounts of CHIP mutants used are shown at the bottom. The asterisk indicates the GST contaminant. CC, coiled-coil. B, shown is the determination of the Smad1 sequence critical for CHIP binding. The GST-mediated pulldown assays were carried out with various GST-tagged Smad1 fragments and CHIP-TPR. The panels are arranged as in panel A.
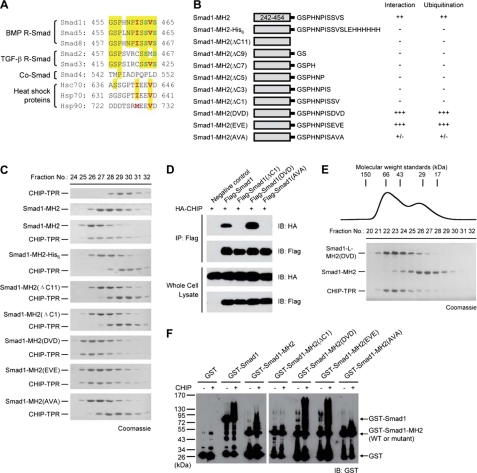
To analyze the interaction of Smad1 with CHIP using gel filtration assays, we initially generated a C-terminal His6-tagged protein, Smad1-MH2-His6 (Fig. 2, A and B). Surprisingly, this fusion protein failed to interact with CHIP, suggesting that the C terminus of Smad1 may play a vital role in the interaction with CHIP (Fig. 2C). We then truncated 11 residues at the C terminus as these residues are flexible in the structures of Smad-MH2 domains (34, 35). As expected, this mutant, Smad1-MH2(ΔC11), failed to bind to CHIP-TPR. We also generated a series of C-terminal truncation mutants in the background of Smad1-MH2, and none of the mutants interacted with CHIP-TPR (Fig. 2B). Notably, deletion of the very last Ser residue of Smad1 abrogates its ability to bind to CHIP-TPR (Fig. 2C). These results clearly demonstrate that the extreme C terminus of Smad1 is indispensable for the formation of the CHIP-Smad1 complex.
FIGURE 2.
Smad1 C-terminal sequence plays a crucial role in CHIP interaction and CHIP-mediated ubiquitination. A, shown is alignment of the C-terminal sequences from human R-Smads, Co-Smad, and heat shock proteins. The phosphorylatable Ser residues in the C-terminal SXS motif of R-Smads are shown in green, and the critical hydrophobic residues differentiate BMP R-Smads from TGF-β R-Smads and Co-Smad, highlighted in red. B, shown is a schematic illustration of Smad1-MH2 proteins used. A summary for their CHIP interaction and CHIP-mediated ubiquitination results is also listed. C, representative results for size exclusion chromatography analyzing the interaction between Smad1-MH2 mutants and CHIP-TPR are shown. The fractions of CHIP-TPR alone and Smad1-MH2 alone are shown as the top two panels followed by the fractions of protein mixtures. D, co-immunoprecipitation (IP) assays for the interaction between CHIP and Smad1 is shown. HEK293T cells were co-transfected with 2 μg of FLAG-Smad1 and 1 μg of HA-CHIP plasmids as indicated. Lysates were subjected to anti-FLAG immunoprecipitation followed by anti-HA immunoblotting (IB). E, competition between wild type Smad1 and pseudophosphorylated Smad1(DVD) for CHIP binding is shown. The pre-equilibrated mixture of the three proteins was subjected to size exclusion chromatography. Mutant Smad1-L-MH2(DVD) was used to distinguish the pseudophosphorylated Smad1 form wild type Smad1-MH2 on SDS-PAGE. Molecular weights (kDa) were indicated. F, shown is in vitro CHIP-mediated ubiquitination of Smad proteins in the presence of recombinant E1, E2 (UbcH5a), ubiquitin, and ATP regenerating system. Smads here and in subsequent ubiquitination assays are all used as GST fusion proteins so that they can be conveniently immunoblotted with anti-GST antibody.
CHIP Prefers to Bind to and Ubiquitinate Phospho-Smad1
Smad1 carries the characteristic C-terminal SXS motif common to all R-Smads, and the phosphorylation of the two serine residues is crucial for the intracellular signal transduction. To assess the significance of the two Ser residues, we generated three mutants by replacing the SVS motif of Smad1 with DVD, EVE, and AVA and examined their affinities to CHIP (Fig. 2, B and C). Mutant Smad1-MH2(AVA), where both polar Ser residues were substituted by hydrophobic alanine, barely retained any affinity to CHIP-TPR, demonstrating the importance of these serine residues. By contrast, the DVD and EVE mutants, where the Ser side chains were replaced with Asp or Glu to mimic the structural and electrostatic properties of phospho-Ser, appeared to interact with CHIP-TPR even more strongly than the wild type Smad1-MH2. To verify the importance of the SXS motif, we generated various mutations on the C terminus of full-length Smad1 and examined their effect on the CHIP-Smad1 interaction. Consistently, the wild type Smad1 protein was co-immunoprecipitated by CHIP, whereas the mutants ΔC1 and AVA failed to bind to CHIP (Fig. 2D). Notably, the pseudophospho-Smad1(DVD) mutant pulled down more CHIP than the wild type protein. To compare their affinities, we then performed a competitive binding assay between the unphosphorylated Smad1 and its pseudophosphorylated DVD mutant. When the pre-equilibrated, stoichiometric mixture of CHIP-TPR, Smad1-MH2, and Smad1-MH2(DVD) was subjected to the size exclusion chromatography and subsequent SDS-PAGE analysis, the wild type Smad1-MH2 protein was separated from the complex of the DVD mutant and CHIP-TPR, indicating that the pseudophosphorylated Smad mutant has higher affinity to CHIP than the wild type Smad1 (Fig. 2E).
We next performed in vitro ubiquitination assays with recombinant Smad1 and CHIP proteins. Both full-length Smad1 and Smad1-MH2 domain were polyubiquitinated in the presence of CHIP (Fig. 2F). Remarkably, the ability of the C-terminal Smad1 mutants undergoing polyubiquitination was correlated precisely with their respective affinity to CHIP (Fig. 2B). No significant polyuibiquitination was observed when the interactive C terminus of Smad1 was depleted. For instance, Smad1-MH2(ΔC1) exhibited little polyubiquitination. Importantly, the pseudophosphorylated DVD and EVE mutants displayed an enhanced CHIP-mediated ubiquitination due to stronger affinities to CHIP, whereas mutant Smad1-MH2(AVA) that barely associated with CHIP was less ubiquitinated (Fig. 2F). Together, these results demonstrate that the C-terminal SXS motif of Smad1 is strictly required for CHIP binding and CHIP-mediated ubiquitination and that the C-terminal phosphorylation of Smad1 can promote the interaction and ubiquitination.
Crystal Structures of CHIP-TPR in Complex with Two Smad1 C-terminal Peptides
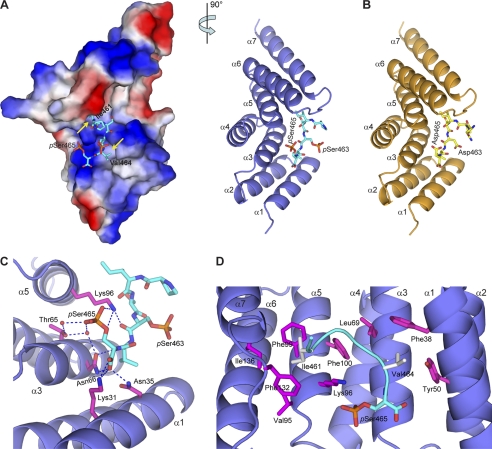
To elucidate the molecular mechanism of CHIP-Smad1 interaction, we synthesized several Smad1 C-terminal peptides and determined the crystal structures of CHIP-TPR in complex with a double-phosphorylated Smad1 peptide and with a pseudophosphorylated Smad1-DVD peptide (Fig. 3, A and B, and Table 1). CHIP-TPR contains three TPR motifs, each folding into a typical helix-turn-helix pattern and a seventh helix packing against the third TPR (supplemental Fig. S1). The Smad1 peptides nestle into a groove on the concave surface of CHIP-TPR, resulting in the burial of an approximate 1000 Å2 of exposed surface area.
FIGURE 3.
Smad1 C-terminal peptides bind to CHIP-TPR. A, shown is the crystal structure of CHIP-TPR in complex with phosphorylated Smad1 peptide. The two views of the structure are related by a 90° rotation around a vertical axis. The Smad1 peptide is shown as cyan sticks. In the left panel, CHIP-TPR is shown in surface representation and colored according to electrostatic potential (positive, blue; negative, red). The yellow arrows indicate the hydrophobic pockets on CHIP-TPR accommodating Ile-461 and Val-464 of Smad1. In the right panel, CHIP-TPR is colored in slate, and the α-helices are labeled α1 to α7 from the N to C termini. B, shown is a schematic representation of CHIP-TPR (orange) in complex with pseudophosphorylated Smad1(DVD) peptide (yellow). C, hydrogen bond networks at the interface of CHIP-TPR and Smad1 phosphopeptide are shown. The residues on CHIP-TPR are highlighted in magentas sticks. Hydrogen bonds among oxygen (red) and nitrogen (blue) atoms and water molecules (red) are indicated by blue dashed lines. D, van der Waals contacts between CHIP-TPR and Smad1 phosphopeptide are shown. The Smad1 peptide is shown as a cyan ribbon, and the hydrophobic residues are highlighted in gray sticks.
In the complex of CHIP-TPR with phosphorylated Smad1 peptide, the last six residues and the covalently bound phosphates on Ser-463 and Ser-465 are well resolved in the electron density (supplemental Fig. S2). The last phosphorylated Ser residue of Smad1, phospho-Ser-465, plays a central role in the complex formation by nucleating a mass of hydrophilic interactions (Fig. 3C). The phosphate group donates two hydrogen bonds to Lys-96 on Helix α5 and three water-mediated hydrogen bonds to Thr-65 and Asn-66 from Helix α3 of CHIP. Moreover, the carboxylate of phospho-Ser-465 directly coordinates with three residues, Lys-31 and Asn-35 from Helix α1 and Asn-66 on Helix α3 of CHIP, which is reminiscent of the “two carboxylate clamp” mechanism observed in other TPR-peptide complexes (36–38). In contrast, the phosphate group of phospho-Ser-463 makes no observable interaction with CHIP, and only its carbonyl group forms one intermolecular hydrogen bond with Lys-96 (Fig. 3C). The last phospho-Ser residue of Smad1, particularly the phosphate group and the free carboxylate at the very C terminus, might be the most significant contributor(s) to the specific recognition of Smad1 by CHIP. These structural analyses are consistent with the biochemical data that deletion of the very last Ser-465 (Smad1-MH2-ΔC1) and the blockage of the free carboxylate (Smad1-MH2-His6) completely abolished formation of the Smad1-CHIP complex (Fig. 2C). Therefore, CHIP-TPR most likely recognizes the pSXpS or (D/E)X(D/E) motif at the extreme C terminus of a protein.
In addition to the hydrophilic contacts, the Smad1 C-terminal tail makes massive van der Waals contacts with CHIP-TPR (Fig. 3D). Ile-461 penetrates into a hydrophobic pocket formed by six residues, Val-95, Lys-96, Phe-99, Phe-100, Phe-132, and Ile-136 from CHIP-TPR, and Val-464 makes hydrophobic contacts with Phe-38, Tyr-50, and Leu-69 from helices α1-α3. Mutation of Ile-461 or Val-464 to a polar residue abolished the formation of the CHIP-Smad1 complex (data not shown). Taken together, these results indicate that the last serine in the SXS motif plays a significant role in the Smad1-CHIP interaction and that the hydrophobic residues exert auxiliary functions.
The pseudophosphorylated DVD peptide of Smad1 is similarly accommodated via massive hydrophilic and hydrophobic contacts (supplemental Fig. S3). It is noteworthy that the side chain carboxyl group of Asp-465 mimics the phosphate group of phospho-Ser-465 and mediates similar hydrogen bonds, which clearly demonstrates that the DVD mutant is a reliable functional substitute of the phosphorylated Smad1.
Peptides from Smad1 and Hsp70/Hsc70 Bind in the Same Groove of CHIP-TPR
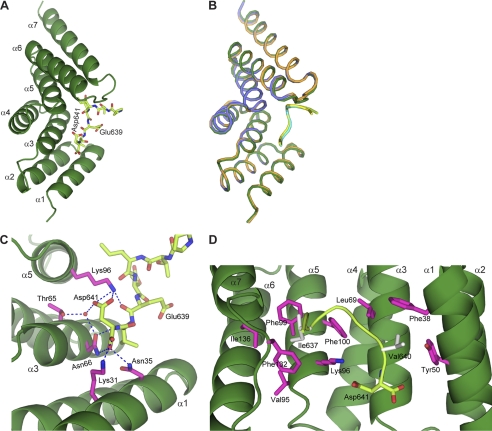
Usually, CHIP mediates the ubiquitination of client proteins in a chaperone-dependent manner (22–26). However, CHIP directly binds to Smad1 and mediates Smad1 polyubiquitination in the absence of molecular chaperones Hsp70/Hsc70 and Hsp90. Interestingly, the CHIP-Smad1 structures are readily superposed to the counterparts in the asymmetric homodimer of full-length CHIP in complex with an Hsp90 C-terminal peptide (37) (supplemental Fig. S4). Moreover, the peptides from Smad1 and Hsp90 bind in the same groove with essentially the same binding mode. To validate the binding mode between CHIP and molecular chaperone, we also determined the complex structure of CHIP-TPR and the C-terminal octopeptide of Hsp70/Hsc70 (Hsp70/Hsc70-C peptide), which adopts almost identical conformation to the CHIP-Smad1 complexes (Fig. 4, A and B, and Table 1). Strikingly, the last three amino acids of Hsc70/Hsp70 and Hsp90 are EVD (Fig. 2A), highly conserved with the phosphorylated Smad1 C terminus in terms of both amino acid sequence and structural characteristics (Fig. 4, C and D). In addition, the mutant CHIP(K31A), which was incapable to interact with Hsp70/Hsc70 and Hsp90 (23), barely associated with Smad1 (Fig. 1A) because Lys-31 from CHIP-TPR interacts with the free carboxylate of either Smad1 or heat shock peptides (Figs. 3C and 4C). These data together raise a possibility that molecular chaperones and Smad1 compete with each other for CHIP interaction.
FIGURE 4.
CHIP-TPR interacts with Hsp70/Hsc70 peptide. A, shown is a schematic representation of CHIP-TPR (forest) in complex with the Hsp70/Hsc70-C peptide (lemon). B, superposition of the three complex structures of CHIP-TPR with Smad1 and Hsp70 peptides is shown. The molecules are colored the same as in Figs. 3, A and B, and 4A, respectively. C and D, hydrophilic (C) and hydrophobic (D) contacts between CHIP-TPR and the Hsp70/Hsc70-C peptide are shown. The two views were oriented same as the ones in Fig. 3, C and D, respectively.
Molecular Chaperones Antagonize CHIP-mediated Smad1 Ubiquitination
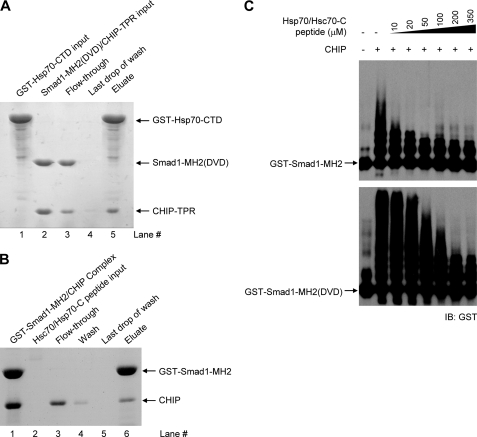
To examine whether molecular chaperones affect CHIP-mediated Smad1 ubiquitination, we first investigated the interaction between Hsp70 and a CHIP-Smad1 complex (Fig. 5A). If the molecular chaperone binds to CHIP without disrupting the CHIP-Smad interaction, GST-tagged Hsp70 is expected to pull down the CHIP-Smad1 binary complex and form a ternary complex. Our results showed that when the preformed complex of CHIP-TPR and Smad1-MH2(DVD) was applied to the immobilized C-terminal domain of Hsp70 (GST-Hsp70-CTD, residues 383–641), all Smad1 proteins flowed through the resin (Fig. 5A, lane 3), whereas the majority of CHIP-TPR proteins were retained on column and formed binary complex with Hsp70 (lane 5). Hsp70-CTD also disrupted the complex of CHIP with wild type Smad1-MH2 (supplemental Fig. S5). These results demonstrate that Smad1 and Hsp70 mutually exclude each other from the association with CHIP. This was further validated by a competition assay with a synthesized Hsp70/Hsc70-C peptide, which recruited CHIP from the immobilized CHIP-Smad1 (DVD) complex (Fig. 5B). In consistent with the competitive binding ability, the Hsp70/Hsc70-C peptide impressively inhibited CHIP-mediated polyubiquitination of Smad1 and its DVD mutant in a dose-dependent manner (Fig. 5C). A Hsp90-C peptide similarly disrupted the CHIP-Smad1 complex and concomitantly suppressed Smad1 ubiquitination (supplemental Fig. S6). Therefore, in contrast with the previous chaperone-dependent model, we found that molecular chaperones compete with Smad1 for CHIP interaction, suppress the CHIP-mediated ubiquitination, and hence, protect Smad1 from futile degradation.
FIGURE 5.
Heat shock proteins and Smad1 compete for CHIP binding. A, disruption of the CHIP-Smad1 (DVD) complex by Hsp70-CTD is shown. Some 0.4 mg of recombinant GST-Hsp70-CTD was bound to 0.2 ml glutathione-Sepharose 4B resin (lane 1). The resin was washed five times with 1.0 ml of buffer to remove excess unbound Hsp70 or other contaminants. Then, 0.6 mg of non-tagged CHIP-Smad1 complex was allowed to flow through the resin (lanes 2 and 3). After extensive washing (lane 4), the bound proteins were eluted with 5 mm reduced glutathione (lane 5). All fractions were visualized by SDS-PAGE with Coomassie Blue staining. B, Hsp70/Hsc70-C peptide competes with Smad1 for CHIP binding. The complex of GST-Smad1 and CHIP was loaded to the resin followed by the addition of the chaperone peptide. The column was then washed extensively and eluted with reduced glutathione. Samples were visualized by SDS-PAGE with Coomassie Blue staining. C, heat shock protein antagonizing CHIP-mediated Smad1 ubiquitination is shown. In vitro ubiquitination assays were performed in the absence or presence of increasing amounts of Hsp70/Hsc70-C peptide. IB, immunoblot.
CHIP Fails to Interact with Smad2/3/4
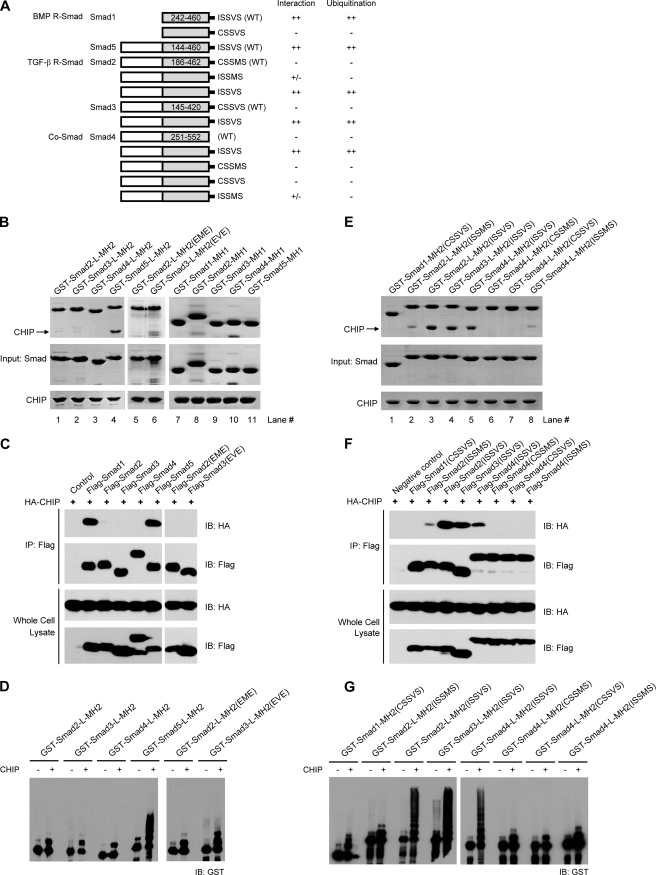
CHIP was previously reported to interact with both BMP and TGF-β R-Smads as well as Co-Smad and further mediate their ubiquitination/degradation (17, 28, 29). We thus carried out in vitro binding assays with recombinant proteins of the linker and MH2 domains of other Smads (Fig. 6A). Unexpectedly, none of the MH2 domains of Smad2/3/4 (TGF-β R-Smads and Co-Smad), not even the pseudophosphorylated Smad2(EME) and Smad3(EVE) mutants, bound to CHIP (Fig. 6B, lanes 1–6). In addition, all MH1 domains of the R- and co-mediator Smads failed to interact with CHIP (Fig. 6B, lanes 7–11). Co-immunoprecipitation assays with full-length Smad proteins confirmed that only Smad1/5, but not Smad2/3/4, can interact with CHIP (Fig. 6C). Consistently, little if any CHIP-mediated polyubiquitination was observed for recombinant MH2 domains of Smad2/3/4 and their pseudophosphorylated mutants, in great contrast with the evident polyubiquitination of CHIP-interactive Smad5-MH2 (Fig. 6D). Therefore, we conclude that CHIP only interacts with and mediates the ubiquitination of Smad1/5 but not Smad2/3/4.
FIGURE 6.
CHIP specifically targets Smad1/5 but not Smad2/3/4. A, shown are schematic diagrams of Smad mutants. The results of the CHIP binding and ubiquitination assays are summarized. B, interactions between the MH1 and MH2 domains of Smad2, -3, -4, and -5 and CHIP. The GST-mediated pulldown assays were carried out using full-length CHIP and various GST-tagged Smad proteins as indicated. C, co-immunoprecipitation (IP) assays detecting the interactions between full-length Smad proteins and CHIP are shown. HEK293T cells were co-transfected with 2 μg of FLAG-Smad1–5 and 1 μg of HA-CHIP plasmids as indicated. After immunoprecipitation with anti-FLAG antibody for Smad proteins, immunoblotting (IB) was carried out using anti-HA and anti-FLAG antibodies for CHIP and Smad, respectively. D, CHIP-mediated ubiquitination of Smad2, -3, -4, and -5 proteins is shown. Pseudophosphorylation of Smad2/3 exhibited little if any effect on both interaction and ubiquitination. E, interactions between the chimeric Smad-MH2 fragments and CHIP analyzed by GST-mediated pulldown assay are shown. F, interactions between full-length Smad proteins and CHIP determined by coimmunoprecipitation analyses are shown. G, CHIP-mediated ubiquitination of chimeric Smad proteins correlated precisely with their interaction abilities.
Because the SXS motif of Smad1 is indispensable for its interaction with CHIP, it is reasonable that Co-Smad Smad4, which lacks the SXS motif unique to R-Smad proteins, failed to associate with CHIP. Indeed, when a C-terminal pentapeptide of ISSVS from BMP R-Smads Smad1/5/8 was added to the C terminus of Smad4, the mutant Smad4-L-MH2(ISSVS) did bind to CHIP (Fig. 6E, lane 5). However, the fact that neither of the TGF-β R-Smads, Smad2/3, exhibited any CHIP affinity suggests that there must be other determinant(s) in addition to the SXS motif. By carefully analyzing the C-terminal sequences, we found that the most prominent difference appears at the fifth position from the C-terminal end, which is a hydrophobic Ile/Met in Smad1/5/8 and heat shock proteins but a polar Cys in Smad2/3 (Fig. 2A). Structurally, the side chain of cysteine is unlike to fulfill the massive hydrophobic interactions observed in the CHIP-Smad1 and CHIP-Hsp complexes (Figs. 3D and 4D). We swapped this residue in Smad1, -2, and -3. Indeed, the Smad3 mutant bearing the Cys to Ile mutation, Smad3-L-MH2(ISSVS), formed a complex with CHIP, whereas the corresponding Smad1 mutant CSSVS did not interact with CHIP (Fig. 6E, lanes 4 and 1). Unexpectedly, the ISSMS mutant of Smad2 barely interacted with CHIP; only when Met-466, the last residue but one, was additionally replaced with valine (the fixed amino acid appears in all other R-Smads and heat shock proteins) did the Smad2 double mutant ISSVS gain reasonable affinity to CHIP (Fig. 6E, lanes 2 and 3). As shown in the left panel of Fig. 3A, the hydrophobic groove on CHIP-TPR for this Val residue is shallow and unlikely to accommodate the bulky side chain of Met-466 in Smad2. We also generated Smad4 mutants with added CSSMS, CSSVS, and ISSMS tails, and only the ISSMS mutant of Smad4-L-MH2 exhibited weak affinity to CHIP as expected (Fig. 6E, lanes 6–8). Moreover, Western blotting analysis verified that the FLAG-Smad proteins/mutants containing the C-terminal ISSVS pentapeptide coimmunoprecipitated with CHIP (Fig. 6F). Importantly, the polyubiquitination levels of these C-terminal mutants were highly correlated with their affinities to CHIP (Fig. 6G). All Smad2/3/4 mutants terminating with the signature sequence ISSVS of BMP R-Smads underwent CHIP-mediated polyubiquitination, whereas those with Cys and/or Met at the fifth and second positions from the C-terminal end displayed little ubiquitin modification. These data together with the Smad1-CHIP structures clearly indicate that the C-terminal hydrophobic residues play a role in the distinct CHIP affinities of R-Smads.
CHIP Disrupts the Functional R-/Co-Smad Complexes
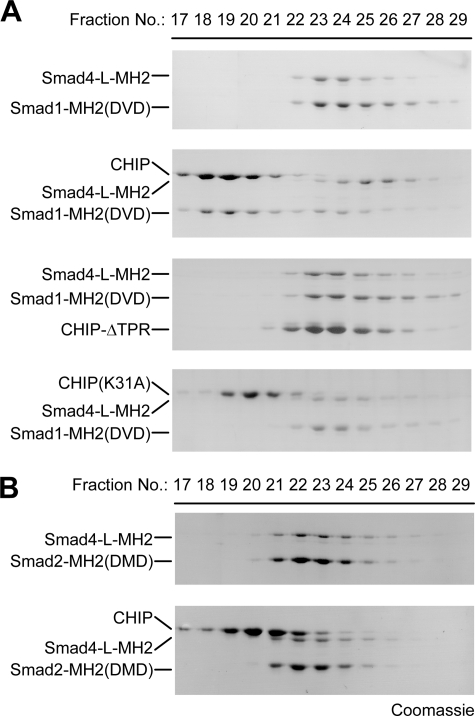
Formation of hetero-complexes between Co-Smad and R-Smads is vital for canonical TGF-β signal transduction, which depends on the specific phosphorylation of the R-Smad SXS motif. We attempted to examine the role of CHIP in R-/Co-Smad complexes as the SXS motif of Smad1/5 is required for both CHIP-Smad and R-Smad-Co-Smad interactions. To address this issue, we prepared and characterized a complex between pseudophosphorylated Smad1-MH2(DVD) and Smad4-L-MH2 and then carried out competition assays between this complex and wild type CHIP or its missense mutants (Fig. 7A). The wild type CHIP did disrupt the Smad1-Smad4 complex and concomitantly formed a binary complex with Smad1-MH2(DVD). On the contrary, neither the CHIP-ΔTPR fragment nor CHIP(K31A) mutant had any impact on the interaction between Smad1 and Smad4. These results demonstrate that an intact TPR domain of CHIP is indispensable for the recruitment of Smad1 from the Smad1-Smad4 complex. The heteromeric complex of Smad2-MH2(DMD) and Smad4-L-MH2 was subjected to a similar competition assay, and CHIP had no effect on the Smad2-Smad4 interaction because of its incapability to recognize Smad2 (Fig. 7B). Taken together, our structural and biochemical results suggest that CHIP competes with Smad4 for the binding to specific R-Smads Smad1/5, disrupts the R-/Co-Smad complex, and further mediates the ubiquitination of Smad1/5 in a chaperone-independent manner.
FIGURE 7.
CHIP disrupts the Smad1-Smad4 complex but not the Smad2-Smad4 complex. A, CHIP disrupts Smad1-Smad4 complex. The wild type CHIP protein or its missense mutants was mixed with the pre-formed complexes of pseudophosphorylated Smad1 and Smad4, and the mixture was analyzed by size exclusion chromatography. B, CHIP fails to disrupt Smad2-Smad4 complex. Gel filtration assay was performed for the mixture of wild type CHIP and the preformed complex of Smad2-MH2(DMD) and Smad4-L-MH2.
DISCUSSION
Because of the important role of TGF-β/BMP signaling in a diverse set of essential cellular processes and the development of various human diseases, understanding the regulatory mechanisms of Smad proteins might provide new therapeutic avenues for human diseases. Here we reported an extensive structural and biochemical study of CHIP-mediated negative regulation of Smad proteins. The CHIP-Smad1 interaction is mediated by the TPR domain of CHIP and the C-terminal sequence of Smad1. Phosphorylation of the Ser residues in the SXS motif can improve the Smad1-CHIP affinity and subsequently promote the polyubiquitination level of Smad1 in vitro, consistent with the cellular evidence that CHIP preferentially mediates the degradation of phosphorylated Smad1 and Smad5 (29). In addition, structural analyses revealed that the last phospho-Ser residue, in particular the covalently bound phosphate and the free carboxylate, contributes a great mass of the hydrophilic interactions between CHIP and Smad (Fig. 3).
The R-Smad proteins mediating the intracellular signal transduction can be classified into two subfamilies, Smad1/5/8 mainly mediating BMP signal and Smad2/3 for TGF-β signaling. These R-Smad proteins are highly conserved within each subfamily but exhibit relative differences between subfamilies. In addition, activities of BMP and TGF-β R-Smads may be regulated via different molecular mechanisms (39, 40). Recently, the ubiquitin ligase Nedd4L was shown to selectively target activated Smad2/3 to destruction and, hence, specifically limit the TGF-β signaling (41). We found that CHIP failed to bind to and ubiquitinate either Co-Smad Smad4 or TGF-β R-Smads Smad2/3 independently of the C-terminal phosphorylation. Therefore, CHIP may be an E3 ligase specifically responsible for BMP R-Smads Smad1/5/8 due to the sequence discrimination at their extreme C termini (Fig. 6). Of a particular note, we did not exclude the possibility of CHIP participating indirectly in the down-regulation of Smad2/3/4. For instance, the transcriptional co-activator SRC-3, a direct target of CHIP-medicated polyubiquitination, was found to enhance Smad2 expression. It is possible that CHIP might down-regulate Smad2 levels through targeting SRC-3 for degradation (27).
The Smad sequence recognized by CHIP consists of a minimum of five amino acids, ΦAX(pS/D/E)ΦB(pS/D/E), where ΦA can be any hydrophobic residue including the bulky Met, and ΦB would be restricted to relatively small hydrophobic amino acids such as Val, Leu, or Ile. This consensus sequence may occur in numerous proteins. However, the structural and biochemical data indicated that this sequence should be located at the extreme C terminus of a protein, which largely limits the number of potential interaction proteins of CHIP. Notably, this C-terminal motif of R-Smads is required not only for CHIP interaction but also for the formation of hetero-complexes between Co-Smad and phosphorylated R-Smads, the central event of TGF-β/BMP signaling. Our data showed that CHIP competes directly with Smad4 for Smad1 binding but fails to disrupt the active complex of Smad2 and Smad4, consistent with its specific recognition of Smad1 (Fig. 7). Although the disruption of R-/Co-Smad heterocomplexes has been used by many other negative regulators, such as I-Smads, MAN1, and Ski/SnoN, to terminate/suppress TGF-β/BMP signal (12), this is to our knowledge the first evidence of a direct competitor for the specific C-terminal SXS motif of R-Smads. Because this negative regulation only requires the TPR domain of CHIP, further investigation on other TPR domain-containing proteins (supplemental Fig. S1) that have been proved or suggested to bind to an extreme C-terminal ‘ΦAX(pS/D/E)ΦB(pS/D/E) motif may lead to the discovery of new regulators of Smad proteins (36).
Unexpectedly, we found that CHIP targets Smad proteins for polyubiquitination in the absence of heat shock proteins, inconsistent with the conventional function of CHIP as a chaperone-dependent E3 ubiquitin ligase. In addition, the same groove on the concave surface of CHIP-TPR accommodates the C-terminal peptides from both Smad1 and molecular chaperons. The structure-based conjecture that Smad and chaperone may compete for CHIP interaction was validated by the competition assay and by the dose-dependent inhibition of CHIP-mediated Smad1 ubiquitination by Hsc70/Hsp70 peptide (Fig. 5). Therefore, in the regulation of TGF-β/BMP signaling, molecular chaperones, rather than assist CHIP-mediated ubiquitination, protect R-Smads from futile degradation. This observation is reminiscent of the chaperone-independent degradation of base excision repair proteins by CHIP (42). However, the base excision repair proteins do not have the C-terminal ΦAX(pS/D/E)ΦB(pS/D/E) motif, suggesting that the base excision repair proteins, unlike Smad1/5 and molecular chaperones, might employ a distinct mode to associate with CHIP.
Our study confirmed the interaction of CHIP with Smad1/5/8, implying a role of CHIP in embryonic development. In a CHIP depletion mouse model, a severe phenotype of early death was observed, as the mouse suffered from an inability of degradation of misfolded proteins and dysfunction of the quality control machinery (43). The role of CHIP in maximal myocardial protection was reported (44), whereas Smad1 also showed a role in protection of cardiomyocytes from ischemia-reperfusion injury (45). However, the physiological relevance in heart disease remains unclear as CHIP regulates degradation of Smad1. We expected that CHIP may also play a role in osteoblast differentiation, which is regulated by Smad1 (46). Indeed, CHIP has been demonstrated to regulate osteoblast differentiation through negative regulation of Runx2, an important factor in response to BMP signaling (47).
In summary, our data reveal the importance of the specific C-terminal ΦAX(pS/D/E)ΦB(pS/D/E) sequence of a protein in the interaction with CHIP-TPR and suggest important roles of CHIP in suppressing specific R-Smad activity. Upon ligand binding, CHIP preferentially binds to the phosphorylated Smad1/5/8, competitively disrupts the functional R-/Co-Smad complexes, and thus down-regulates the intracellular TGF-β/BMP signaling. This effect of CHIP is independent of the E3 ligase activity of the Ubox domain. The CHIP-associated Smad1/5/8 are then targeted to polyubiquitination and subsequent degradation to terminate the TGF-β/BMP signal transduction, for which both the TPR and Ubox domains of CHIP are indispensable. On the other hand, the molecular chaperones antagonize the effects of CHIP by inhibiting the CHIP-mediated R-Smad polyubiquitination and by freeing Smad1/5/8 from CHIP-sequestration. Thus, CHIP suppresses the TGF-β/BMP signaling, whereas the molecular chaperones rescue the TGF-β/BMP signaling in contradiction to the conventional model.
Supplementary Material
Acknowledgments
We thank Jijie Chai for help on data collection and processing and Xueni Li for advice on ubiquitination assay.
This work was supported in part by Ministry of Science and Technology of China Grants 2007CB914400 and 2011CB910803, National Natural Science Foundation of China Grant 31070643, and Tsinghua University Grant 09THZ02235.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 3Q4A, 3Q47, and 3Q49) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BMP
- bone morphogenetic protein
- CHIP
- C terminus of Hsc70-interacting protein
- R-Smad
- receptor-regulated Smads
- Co-Smad
- co-mediator Smad
- TPR
- tetratricopeptide repeat
- MH1
- Mad homology 1
- MH2
- Mad homology 2
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane.
REFERENCES
- 1. Attisano L., Wrana J. L. (2002) Science 296, 1646–1647 [DOI] [PubMed] [Google Scholar]
- 2. Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 3. ten Dijke P., Hill C. S. (2004) Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 4. Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 5. Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pardali E., ten Dijke P. (2009) Front Biosci. 14, 4848–4861 [DOI] [PubMed] [Google Scholar]
- 7. Chen H. B., Shen J., Ip Y. T., Xu L. (2006) Genes Dev. 20, 648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin X., Duan X., Liang Y. Y., Su Y., Wrighton K. H., Long J., Hu M., Davis C. M., Wang J., Brunicardi F. C., Shi Y., Chen Y. G., Meng A., Feng X. H. (2006) Cell 125, 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Knockaert M., Sapkota G., Alarcón C., Massagué J., Brivanlou A. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11940–11945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duan X., Liang Y. Y., Feng X. H., Lin X. (2006) J. Biol. Chem. 281, 36526–36532 [DOI] [PubMed] [Google Scholar]
- 11. Izzi L., Attisano L. (2004) Oncogene 23, 2071–2078 [DOI] [PubMed] [Google Scholar]
- 12. Itoh S., ten Dijke P. (2007) Curr. Opin. Cell Biol. 19, 176–184 [DOI] [PubMed] [Google Scholar]
- 13. Inoue Y., Imamura T. (2008) Cancer Sci. 99, 2107–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 15. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 16. Bonni S., Wang H. R., Causing C. G., Kavsak P., Stroschein S. L., Luo K., Wrana J. L. (2001) Nat. Cell Biol. 3, 587–595 [DOI] [PubMed] [Google Scholar]
- 17. Li L., Xin H., Xu X., Huang M., Zhang X., Chen Y., Zhang S., Fu X. Y., Chang Z. (2004) Mol. Cell. Biol. 24, 856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McDonough H., Patterson C. (2003) Cell Stress Chaperones 8, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murata S., Chiba T., Tanaka K. (2003) Int. J. Biochem. Cell Biol. 35, 572–578 [DOI] [PubMed] [Google Scholar]
- 20. Esser C., Alberti S., Höhfeld J. (2004) Biochim. Biophys. Acta 1695, 171–188 [DOI] [PubMed] [Google Scholar]
- 21. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dickey C. A., Patterson C., Dickson D., Petrucelli L. (2007) Trends Mol. Med. 13, 32–38 [DOI] [PubMed] [Google Scholar]
- 23. Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Esser C., Scheffner M., Höhfeld J. (2005) J. Biol. Chem. 280, 27443–27448 [DOI] [PubMed] [Google Scholar]
- 25. Belova L., Sharma S., Brickley D. R., Nicolarsen J. R., Patterson C., Conzen S. D. (2006) Biochem. J. 400, 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daviau A., Proulx R., Robitaille K., Di Fruscio M., Tanguay R. M., Landry J., Patterson C., Durocher Y., Blouin R. (2006) J. Biol. Chem. 281, 31467–31477 [DOI] [PubMed] [Google Scholar]
- 27. Kajiro M., Hirota R., Nakajima Y., Kawanowa K., So-ma K., Ito I., Yamaguchi Y., Ohie S. H., Kobayashi Y., Seino Y., Kawano M., Kawabe Y., Takei H., Hayashi S., Kurosumi M., Murayama A., Kimura K., Yanagisawa J. (2009) Nat. Cell Biol. 11, 312–319 [DOI] [PubMed] [Google Scholar]
- 28. Xin H., Xu X., Li L., Ning H., Rong Y., Shang Y., Wang Y., Fu X. Y., Chang Z. (2005) J. Biol. Chem. 280, 20842–20850 [DOI] [PubMed] [Google Scholar]
- 29. Li R. F., Shang Y., Liu D., Ren Z. S., Chang Z., Sui S. F. (2007) J. Mol. Biol. 374, 777–790 [DOI] [PubMed] [Google Scholar]
- 30. Otwinowski Z., Minor W. (1997) “Processing of X-ray Diffraction Data Collected in Oscillation Mode,” Methods in Enzymology (Carter C. W., Jr., Sweet R. M. eds) Vol. 276, Macromolecular Crystallography, part A, pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 31. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Shi Y., Hata A., Lo R. S., Massagué J., Pavletich N. P. (1997) Nature 388, 87–93 [DOI] [PubMed] [Google Scholar]
- 35. Wu G., Chen Y. G., Ozdamar B., Gyuricza C. A., Chong P. A., Wrana J. L., Massagué J., Shi Y. (2000) Science 287, 92–97 [DOI] [PubMed] [Google Scholar]
- 36. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 37. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 38. Cliff M. J., Harris R., Barford D., Ladbury J. E., Williams M. A. (2006) Structure 14, 415–426 [DOI] [PubMed] [Google Scholar]
- 39. Miyazawa K., Shinozaki M., Hara T., Furuya T., Miyazono K. (2002) Genes Cells 7, 1191–1204 [DOI] [PubMed] [Google Scholar]
- 40. Xu L. (2006) Biochim. Biophys. Acta 1759, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao S., Alarcón C., Sapkota G., Rahman S., Chen P. Y., Goerner N., Macias M. J., Erdjument-Bromage H., Tempst P., Massagué J. (2009) Mol. Cell 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parsons J. L., Tait P. S., Finch D., Dianova II, Allinson S. L., Dianov G. L. (2008) Mol. Cell 29, 477–487 [DOI] [PubMed] [Google Scholar]
- 43. Sahara N., Murayama M., Mizoroki T., Urushitani M., Imai Y., Takahashi R., Murata S., Tanaka K., Takashima A. (2005) J. Neurochem. 94, 1254–1263 [DOI] [PubMed] [Google Scholar]
- 44. Zhang C., Xu Z., He X. R., Michael L. H., Patterson C. (2005) Am. J. Physiol. Heart Circ. Physiol. 288, H2836–H2842 [DOI] [PubMed] [Google Scholar]
- 45. Masaki M., Izumi M., Oshima Y., Nakaoka Y., Kuroda T., Kimura R., Sugiyama S., Terai K., Kitakaze M., Yamauchi-Takihara K., Kawase I., Hirota H. (2005) Circulation 111, 2752–2759 [DOI] [PubMed] [Google Scholar]
- 46. Yamamoto N., Akiyama S., Katagiri T., Namiki M., Kurokawa T., Suda T. (1997) Biochem. Biophys. Res. Commun. 238, 574–580 [DOI] [PubMed] [Google Scholar]
- 47. Li X., Huang M., Zheng H., Wang Y., Ren F., Shang Y., Zhai Y., Irwin D. M., Shi Y., Chen D., Chang Z. (2008) J. Cell Biol. 181, 959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.