Abstract
Malaria during pregnancy is a major health problem for African women. The disease is caused by Plasmodium falciparum malaria parasites, which accumulate in the placenta by adhering to chondroitin sulfate A (CSA). The interaction between infected erythrocytes and the placental receptor is mediated by a parasite expressed protein named VAR2CSA. A vaccine protecting pregnant women against placental malaria should induce antibodies inhibiting the interaction between VAR2CSA and CSA. Much effort has been put into defining the part of the 350 kDa VAR2CSA protein that is responsible for binding. It has been shown that full-length recombinant VAR2CSA binds specifically to CSA with high affinity, however to date no sub-fragment of VAR2CSA has been shown to interact with CSA with similar affinity or specificity. In this study, we used a biosensor technology to examine the binding properties of a panel of truncated VAR2CSA proteins. The experiments indicate that the core of the CSA-binding site is situated in three domains, DBL2X-CIDRPAM and a flanking domain, located in the N-terminal part of VAR2CSA. Furthermore, recombinant VAR2CSA subfragments containing this region elicit antibodies with high parasite adhesion blocking activity in animal immunization experiments.
Keywords: Adhesion, Carbohydrate-binding Protein, Chondroitin Sulfate, Glycosaminoglycan, Parasitology, Malaria, PfEMP1, Pregnancy-associated Malaria
Introduction
Carbohydrates, or glycans, are highly diverse in structure and biological function and are expressed by virtually all mammalian cells. Many microorganisms, i.e. viruses, bacteria, and protozoa, have evolved mechanisms to establish infection by interacting with glycans on the host cell surface (1), and the Plasmodium parasites causing malaria are no exception. Of the different Plasmodium species that can infect humans, Plasmodium falciparum is by far the most virulent form and was the cause of 89% of all deaths due to malaria infection in the African region in 2008 (the World Malaria Report 2009, WHO).
A number of important potential P. falciparum vaccine targets have been defined as glycan-binding proteins or lectins, e.g. circumsporozoite protein, which interacts with highly sulfated heparan sulfate proteoglycans (HSPG)3 during sporozoite invasion of the liver, and the EBA-175 surface antigen that mediates merozoite invasion of erythrocytes through the interaction with sialic acid on glycophorin A (2). In addition to these proteins, VAR2CSA, a unique member of the P. falciparum erythrocyte membrane protein 1 (PfEMP1) family has been characterized as a lectin with binding preference for a low-sulfated form of chondroitin sulfate A (CSA) anchored to proteoglycans (CSPG) in placental tissue (3–7). VAR2CSA, is a large multidomain antigen expressed on the surface of P. falciparum-infected erythrocytes (IEs) where it mediates sequestration of the IEs to the placenta (8). This placental sequestration causes placental malaria (PM), a major cause of mother and offspring morbidity such as severe maternal anemia, low birth weight, and stillbirth (9).
Pregnant women and their unborn children are the victims of this severe disease, and it should be feasible to protect them by a vaccine. This is based on the fact that immunity to PM in malaria-endemic regions is acquired as a function of parity (10–12) and the main antigen-mediating sequestration, VAR2CSA, has been found in all P. falciparum genomes studied. When looking at PfEMP1 sequence variation VAR2CSA appears as an unusually conserved PfEMP1 protein with as high as 75–83% amino acid identity between variants (13). The large molecular weight of VAR2CSA makes production of full-length recombinant protein for vaccine use difficult. The ideal scenario for vaccine development is to define a region of the VAR2CSA, which induces antibodies that can abrogate placental adhesion of the full repertoire of genetically different P. falciparum parasites. The consequences of vaccinating to induce VAR2CSA-specific antibodies that are opsonising but not able to block parasite sequestration are not known but could potentially worsen the parasite-induced inflammation in the placenta.
Considering the size and the complex structure it is essential to define smaller regions of VAR2CSA that can be included in a vaccine, the obvious approach being to define the glycan-binding site. However, this has not been straightforward and the molecular mechanism underlying the interaction between VAR2CSA and CSA remains unresolved. This is in part due to the difficulties in analyzing protein-glycan binding in vitro, which easily gives rise to false positive identification of nonspecific glycan-binding proteins because of the large impact of low affinity charge-charge interactions (14).
To date, vaccine development has relied on the production of a large panel of various recombinant VAR2CSA-derived protein fragments and analysis of their capacity to induce antibodies in animals that can block adhesion of IEs to CSA in vitro. This trial and error approach has generated several potential vaccine antigens (15–17) although the capacity of these proteins to specifically bind CSA is questionable (18). Nevertheless, understanding the mode of CSA binding of VAR2CSA is likely to be essential for the optimal design of a vaccine against PM.
An important breakthrough in this respect was recently made when the full-length ectodomain of VAR2CSA was successfully expressed as a recombinant protein and for the first time high affinity binding, within the nanomolar range, was attained when binding this complete ectodomain of VAR2CSA to CSA (6, 7). Based on a few truncated proteins and a compact low-resolution structure of VAR2CSA, Srivastava et al. (7) proposed that the entire ectodomain is required for a specific high affinity CSA interaction. This would explain the reported observations that no single recombinant VAR2CSA domain binds with high affinity to CSA (18, 19).
In this study we have systematically analyzed recombinant fragments of VAR2CSA for affinity to CSA and defined two overlapping protein constructs that possess the same binding properties as the full-length VAR2CSA ectodomain. Specific binding activity locates to the four N-terminal domains, DBL1X-ID1, DBL2X, CIDRPAM, and DBL3X. A major part of the interaction seems to be conferred by proteins encompassing the DBL2X-CIDRPAM region, however an additional domain is required to obtain high affinity binding. Importantly animal-induced antibodies against a recombinant protein encoding the entire binding region effectively block adhesion of IEs to CSA.
EXPERIMENTAL PROCEDURES
P. falciparum Cultures
Parasite cultures of the FCR3 strain were grown as previously described (20). In brief, parasites were maintained in culture using 5% hematocrit of blood (human blood group 0+) in parasite medium consisting of RPMI 1640 supplemented with 25 mm sodium bicarbonate (Sigma-Aldrich), 0.125 μg/ml gentamicin, 0.125 μg/ml Albumax II (Invitrogen), and 2% normal human serum. Parasites were repeatedly panned on BeWo cells to select for VAR2CSA expression as described (21). All parasite isolates were tested mycoplasma negative and were regularly genotyped using nested GLURP (glutamate-rich protein) and MSP-2 (merozoite surface protein 2) primers in a single PCR step.
Cloning and Protein Expression
Subgenetic fragments were amplified from the codon-optimized FCR3 var2csa gene (GenBankTM Accession No. GU249598) or FCR3 genome DNA and cloned into the baculovirus vector pAcGP67-A (BD Biosciences) modified to contain a V5 epitope upstream of a His tag at the C-terminal end (Table 1). Linearized Bakpak6 Baculovirus DNA (BD Biosciences) was co-transfected with pAcGP67-A into Sf9 insect cells for the generation of recombinant virus particles. High-Five cells grown in suspension in 400 ml serum-free medium (10486, GIBCO) were infected with 10 ml of the second amplification of recombinant virus particles when the cell density had reached 0.9–1 × 106 cells/ml. The culture supernatant containing the secreted recombinant protein was harvested 26 h after virus infection and filtered (0.2 μm) before dialysis.
TABLE 1.
Primers for cloning of var2csa fragments that were expressed as proteins and analysed for receptor binding activity
Constructs were cloned from the codon-optimized FCR3 var2csa gene (GenBankTM accession no. GU249598) into the pAcGP67-A vector (BD Biosciences) using either EcoRI/NotI or BamHI/NotI restriction sites. FV2, full-length ectodomain of VAR2CSA without N-terminal segment; NTS, N-terminal segment; DBL, Duffy binding-like domain; ID, interdomain; CIDRPAM, cysteine-rich interdomain region specific for VAR2CSA and pregnancy-associated malaria (PAM).
| PROTEIN | Forward primer | Reverse primer |
|---|---|---|
| FV2 | CCACAGCGATAGCGGCAAG | CGAAGGGCTCGTCGGCCTGGGG |
| NTS-FV2 | CATGGACAGCACCAGCACA | CCTGGGGTGGCAGATCGTC |
| NTS-DBL4ϵ | ATGGACAGCACCAGCACA | TGTCCAGGGGGTACAGCTT |
| NTS-DBL3X | ATGGACAGCACCAGCACA | CGGCATTGTTGTACTTATA |
| NTS-DBL2Xa | ATGGATAGTACAAGCACT | ATCCTATATCAATTAAGTG |
| ID1-DBL4ϵ | CTGAGCTTCATCCTGAAC | TGTCCAGGGGGTACAGCTT |
| ID1-DBL3X | CTGAGCTTCATCCTGAAC | CGGCATTGTTGTACTTATA |
| DBL2X-DBL4ϵ | CTGACCAACGGCTACAAG | TGTCCAGGGGGTACAGCTT |
| DBL2X-DBL3Xb | CTGACCAACGGCTACAAG | CGGCATTGTTGTACTTATA |
| DBL3Xa | ACCAATATTAATAAAAGTGAA | CAGCATTATTATATTTGTA |
| DBL2Xa | TTAACGAATGGTTATAAATGC | ATCCTATATCAATTAAGTG |
| DBL1X-CIDRPAMc | CACAGCGATAGCGGCAAG | TCTTGTTGATATTGGTGTCGGT |
| DBL2X-CIDRPAM | CTGACCAACGGCTACAAG | TCTTGTTGATATTGGTGTCGGT |
a Genome DNA from the FCR3 parasite was used as cloning template.
b Could not be expressed as a soluble protein.
c Previously known as ID2, but is called CIDRPAM according to the updated nomenclature recently described (37).
Protein Purification and SDS-PAGE
Filtrated cell culture supernatant was dialyzed using an ÄKTA cross-flow (GE Healthcare) in 10 mm NaH2PO4 (pH 7.4) and 500 mm NaCl, yielding a final volume of 45 ml. The retentate was filtered (0.2 μm) and imidazole (pH 7.4, Sigma-Aldrich) was added to the sample to give a final imidazole concentration of 15 mm before loading onto a 1-ml HisSelect column (H8286, Sigma-Aldrich). The bound protein was eluted with 10 mm NaH2PO4 (pH 7.4), 500 mm NaCl, and 500 mm imidazole. When required, protein samples were subsequently purified by size-exclusion chromatography on a HiLoad 16/60 SuperdexTM 200 column (GE Healthcare) using a buffer of 20 mm Tris (pH 8.0) and 200 mm NaCl, to obtain monomers for the quartz crystal microbalance measurements. Protein purity and formation of disulfide bridges were verified by SDS-PAGE using NuPage Novex Tris-acetate mini gels (Invitrogen) and NuPage Tris-acetate SDS running buffer. A high molecular weight protein marker (LC5699, Invitrogen) was used as a standard. Around 1–2 μg of protein was mixed with SDS loading dye, with and without reducing agent (50 mm DTT), and heated at 75 °C for 10 min prior to loading onto the gel. Gels were run at 140 V for 1 h and 30 min and finally stained using BioSafeTM Coomassie (Bio-Rad).
Animal Immunizations and IgG Purification
All procedures regarding animal immunizations complied with European and national regulations. Wistar rats received 30 μg of recombinant protein in Freund's complete adjuvant, and were boosted three times with 15 μg of protein in Freund's incomplete adjuvant at 3-week intervals. Antibodies against the recombinant proteins were generated in rats as described in Nielsen et al. (15). Immune sera from all immunizations were collected 8 days after the final boost and used for IgG purification. Recombinant GammaBindTM G type 2 coupled to SepharoseTM 4B (GE Healthcare) was used to purify IgG according to the manufacturer's instructions. In brief, 0.5–1 ml of filtered (0.2 μm) rat serum was diluted 1:1 in 20 mm NaH2PO4 (pH 7.0) and manually passed through a column packed with Sepharose (300 μl of Sepharose per ml of serum). The same serum was passed through the column ten times and then washed with 20 mm NaH2PO4 (pH 7.0) followed by a wash with 100 mm citric acid (pH 4.0). Finally, bound IgG was eluted with 9 ml of 100 mm citric acid (pH 2.7) and collected in tubes with 3.6 ml of 1 m Tris (pH 9.0). Purified IgG was concentrated and stored in aliquots at −80 °C.
Quartz Crystal Microbalance Assay
CSPG from bovine articular cartilage (decorin) was used as a source of CSA. Importantly, IE adhesion to decorin can be abrogated by soluble CSA as well as by chondroitinase treatment of the decorin. This CSPG has either a CSA chain or a dermatan sulfate chain attached to the protein core. By using a CSA-specific antibody, it was determined that the decorin used carried a CSA side chain in addition we confirmed that VAR2CSA expressing parasites could not adhere to dermatan sulfate (data not shown). All biosensor measurements were performed using an Attana A100 (Attana AB). Gold-plated, 10 MHz, AT-cut quartz crystals which were pretreated to generate a polystyrene coating were purchased from Attana AB. The surface was housed in a cylindrical flow chamber with a volume of 1.25 μl. All buffers and solutions were sterile filtered (0.2 μm) before use. A solution of 100 μg/ml CSPG (D8428, Sigma-Aldrich), or 100 μg/ml HSPG (H4777, Sigma-Aldrich) in PBS was allowed to adsorb to the surface at room temperature for 30 min. Following this, the surface was washed and then blocked with PBS containing 0.1% Ig-free BSA (BSA-50, Rockland) at room temperature for 30 min. The assays were performed at a flow rate of 25 μl/min at 25 °C, and with PBS as running buffer. After the baseline stabilized, buffer injections were performed to show that the base line was unaffected by the injection process. The data were processed to line up the curves according to injection time and the curves were fitted to either a simple 1:1 binding model allowing for mass transport limitations using software package Scrubber 2 (BioLogic Software Pty Ltd, Campbell, Australia), or a heterogeneity model using ClampXP (Tom Morton and David Myszka, version 3.50) if a simple 1:1 model fit gave an error of more than 10% of the parameter value. kon and koff were given by the curve fitting and the KD was calculated by dividing the koff by the kon. The software use the Levenberg-Marquardt method for nonlinear sum of squares error minimization.
The data were processed and curves fitted to either a simple 1:1 binding model using software package Scrubber 2 (BioLogic Software Pty Ltd, Campbell, Australia), or a heterogeneity model using ClampXP (Tom Morton and David Myszka, version 3.50). This software uses the Levenberg-Marquardt method for nonlinear sum of squares error minimization.
Solid Phase Binding Assay
To remove the CSA chains from the protein core, CSPG (40 μg) was digested with 25.6 mU chondroitinase ABC (C2905, Sigma-Aldrich) in enzyme buffer (50 mm Tris, 50 mm sodium acetate, pH 8.0) in a total volume of 200 μl. The reaction was incubated overnight at 37 °C with gentle rotation. Digested CSPG was subsequently dialyzed against PBS. The final product was analyzed by ELISA using a mAb (MAB2030, Chemicon) targeting chondroitinase ABC-digested CSA and by SDS-PAGE as described above, to verify that the CSA chains had been removed and that the protein core was intact after enzyme treatment (data not shown).
Falcon microtiter plates (351172, BD Biosciences) were pre-incubated with either 3 μg/ml CSPG, 3 μg/ml enzyme treated CSPG or 3 μg/ml HSPG for 2 h at 37 °C. Following coating with proteoglycan, the wells were blocked with TSM binding buffer (20 mm Tris, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 0.05% Tween-20, and 1% BSA, pH 7.4 at 25 °C) for 2 h at 37 °C. A 2-fold dilution series (1.56–100 nm) of each protein was prepared in TSM binding buffer, added in triplicates to the wells and incubated for 1 h at 37 °C with gentle shaking. After washing three times in TSM washing buffer (20 mm Tris, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 0.05% Tween-20, pH 7.4 at 25 °C), an anti-V5-HRP antibody (R96125, Invitrogen) diluted 1:3000 in binding buffer was added to each well and incubated for 1 h at 37 °C with gentle shaking. The assay was finalized by washing three times and developed using 100 μl per well of o-phenylenediamine substrate (DAKO) for 15 min. Absorbance was measured at 490 nm after quenching the reaction with 100 μl of 2.5 m H2SO4.
Flow Cytometry
Antibody binding to the native VAR2CSA protein expressed on the surface of FCR3 IEs was measured by flow-cytometry (FCM). Briefly, the parasite culture was enriched for late trophozoite and schizont stage IEs in a strong magnetic field (MACS, Miltenyi). Aliquots of 2 × 105 IEs in a total volume of 100 μl of PBS with 2% FCS were labeled by antibodies in a final concentration of 0.5 mg/ml purified IgG. The cells were washed three times in a total volume of 200 μl of PBS with 2% FCS and subsequently exposed to a final concentration of 2 μg/ml ethidium bromide and a 1:100 dilution of FITC labeled secondary antibody specific for rat IgG (62-9511, Invitrogen). As a negative control, IEs were incubated both with IgG from rats immunized with an irrelevant antigen (CIDR1 from VAR4 3D7) and with secondary antibodies alone. Data from 5000 IEs (ethidium bromide positive) was collected on a FC500 flow cytometer (Beckman Coulter). The median FITC fluorescence intensity was determined using Winlist Software (Verity Software House).
Parasite Binding Assays
To analyze the capacity of IgG to inhibit parasite binding to CSA we used a 96- well plate format and a robot-standardized washing method. Briefly, tritium-labeled late trophozoite and schizont stage IEs were MACS purified (Miltenyi). A total of 2 × 105 IEs in 100 μl were added in triplicates to wells coated with 2 μg/ml CSPG. Following incubation with different concentrations of IgG for 90 min at 37 °C, unbound IEs were washed away by resuspension performed by a pipetting robot (Beckman Coulter). The proportion of adhering IEs was determined by liquid scintillation counting on a Topcount NXT (Perkin-Elmer). The CSA specificity of IE binding to CSPG was verified by abrogating the binding by 100 μg/ml soluble CSA (Sigma-Aldrich) and by chondroitinase ABC treatments (Sigma-Aldrich) (data not shown).
RESULTS
Production of Recombinant Proteins
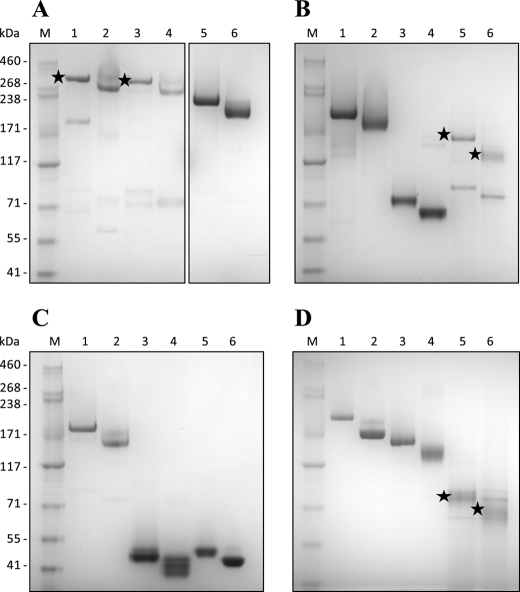
All constructs in this study were expressed in baculovirus-infected insect cells as soluble proteins secreted into the cell culture supernatant (Fig. 1). All proteins formed the expected intra-molecular disulfide bridges, which were confirmed by the difference in mobility when comparing reduced and non-reduced protein samples by SDS-PAGE (Fig. 1). Several proteins formed high-molecular complexes detected by non-reduced SDS-PAGE, probably due to the formation of inter-molecular disulfide bonds between unpaired cysteines because these complexes were readily reduced to monomeric protein using DTT (data not shown). Thus to obtain monomeric proteins most recombinant constructs were further purified by size-exclusion chromatography prior to receptor binding analysis. Only the main peak fraction from the size-exclusion chromatography was used for receptor binding assays, however when analyzing these peak fractions by SDS-PAGE it was noted that small molecular bands appeared for some proteins (Fig. 1). This could be degradation products following the size-exclusion chromatography or artifacts from the SDS-PAGE.
FIGURE 1.
Coomassie-stained SDS-PAGE of recombinant VAR2CSA proteins. All constructs were produced in baculovirus-infected insect cells and purified as soluble proteins on a HisSelect column. 1–2 μg of protein was mixed with SDS loading dye, with and without reducing agent (50 mm DTT). A, lane 1: reduced NTS-FV2; lane 2: non-reduced NTS-FV2; lane 3: reduced FV2; lane 4: non-reduced FV2; lane 5: reduced NTS-DBL4ϵ; lane 6: non-reduced NTS-DBL4ϵ. B, lane 1: reduced NTS-DBL3X; lane 2: non-reduced NTS-DBL3X; lane 3: reduced NTS-DBL2X; lane 4: non-reduced NTS-DBL2X; lane 5: reduced ID1-DBL3X; lane 6: non-reduced ID1-DBL3X. C, lane 1: reduced DBL2X-DBL4ϵ; lane 2: non-reduced DBL2X-DBL4ϵ; lane 3: reduced DBL2X; lane 4: non-reduced DBL2X; lane 5: reduced DBL3X; lane 6: non-reduced DBL3X. D, lane 1: reduced ID1-DBL4ϵ; lane 2: non-reduced ID1-DBL4ϵ; lane 3: reduced DBL1X-CIDRPAM; lane 4: non-reduced DBL1X-CIDRPAM; lane 5: reduced DBL2X-CIDRPAM; lane 6: non-reduced DBL2X-CIDRPAM. M, protein marker. Black stars indicate correct protein size when multiple bands are visible.
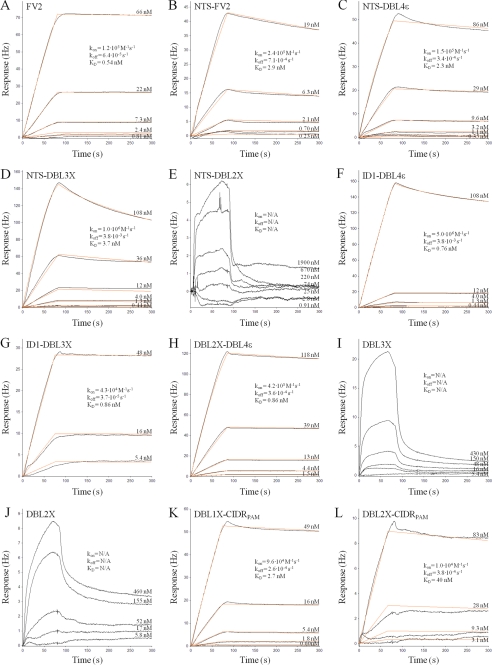
Kinetic Analysis of VAR2CSA Proteins Binding to CSPG
The full-length ectodomain of VAR2CSA from two genetically different parasite lines (FCR3 and 3D7) have previously been shown to bind specifically to CSPG with nanomolar affinity (6, 7). These affinity measurements were obtained using a surface plasmon resonance technology (Biacore). Here we analyzed the full-length VAR2CSA, with (NTS-FV2) and without (FV2) the N-terminal segment (NTS) as well as truncated VAR2CSA proteins, for affinity to CSPG using a quartz crystal microbalance biosensor (Attana A100) with the aim of mapping the minimal binding region of VAR2CSA from FCR3. Association and dissociation data for each protein was collected at different protein concentrations to allow an accurate estimation of the binding affinity (dissociation equilibrium constant, KD). Sensorgrams and corresponding association rate constant (kon), dissociation rate constant (koff) and KD values for each protein are shown in Fig. 2. The measurements obtained with the Attana biosensor confirm the previously published results (6), i.e. full-length VAR2CSA binds to CSPG with nano-molar affinity, and it is apparent that the absence or presence of the NTS region does not significantly change the affinity to CSPG (KD is 0.54 nm for FV2 and 2.9 nm for NTS-FV2, Fig. 2, A and B, respectively). By systematic truncation of the VAR2CSA ectodomain we find that the functional CSA-binding site of VAR2CSA is located in the N-terminal part of the protein and the core of the binding region consists of two domains, DBL2X and CIDRPAM (Fig. 2 and Table 2). This region binds with relatively high affinity to CSPG (KD of 40 nm) and the data fitted to a 1:1 binding model (Fig. 2L). Although DBL2X-CIDRPAM alone binds to CSPG, the interaction with CSPG is reinforced by a flanking domain in either end of the construct, and the proteins DBL1X-CIDRPAM and ID1-DBL3X bind to CSPG with a KD of 2.7 nm and 0.86 nm, respectively (Fig. 2, K and G), similar to that observed for NTS-FV2 and FV2. The DBL3X domain alone binds weakly and heterogeneously to CSPG (Fig. 2I), consistent with results obtained in a previous study (19). Likewise, binding of DBL2X to CSPG is of weak affinity and heterogeneous consistent with previously published (18) (Fig. 2J). The NTS-DBL2X protein does not interact with CSPG (Fig. 2E) even though this construct includes DBL2X, which binds weakly as a single domain, most likely because the DBL2X residues mediating the interaction to CSPG are not exposed in this protein. The CIDRPAM domain and the DBL2X-DBL3X protein were not analyzed because of difficulties in producing these proteins.
FIGURE 2.
Kinetic analysis of VAR2CSA proteins binding to CSPG. A quartz crystal microbalance biosensor (Attana A100) was used for the kinetic analyses. Sensorgrams show VAR2CSA fragments binding to immobilized CSPG as a response (in Hertz) as a function of time (in seconds). Both the association and dissociation phase are shown. Black lines represent data, and red lines represent fitted curves attained by a 1:1 binding model. The concentrations of VAR2CSA proteins for individual curves are shown, and the insets list kon, koff, and the calculated KD. The protein tested is indicated in the upper left corner of each sensorgram.
TABLE 2.
CSPG binding analysis of recombinant VAR2CSA proteins
The kinetic analyses were performed using a quartz crystal microbalance biosensor (Attana A100). Estimated KD values are based on a 1:1 binding model (see Fig. 2 for fitted binding curves) for each protein interacting with CSPG.
a Kinetic data could not be obtained due to lack of binding to CSPG.
b Both DBL2X and DBL3X bind heterogeneously and with weak affinity to CSPG, therefore the binding strength could not be satisfactory calculated.
c Binding of DBL2X-CIDRPAM is not specific for CSPG, and binding to HSPG is of similar affinity.
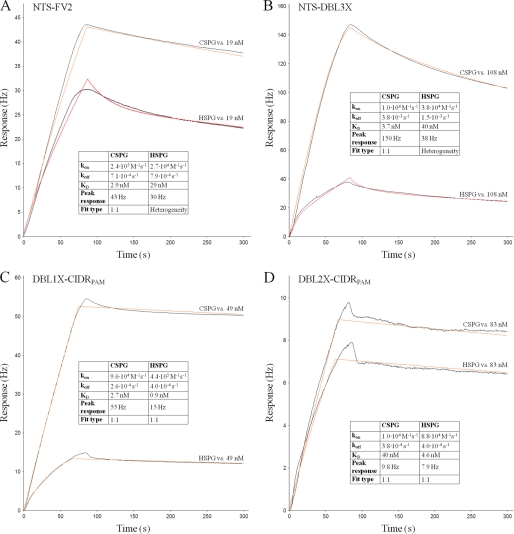
Analysis of VAR2CSA Proteins Binding to HSPG
Placental IEs seem to selectively adhere to the CSA chains of CSPG and not to other glycosaminoglycans (4, 22), and it has previously been shown that FCR3 IEs expressing VAR2CSA on the cell surface do not bind to HSPG (18). To assess whether this receptor specificity also applies for recombinant full-length VAR2CSA and the minimal CSA binding region defined in the N-terminal, a number of selected proteins were analyzed for binding to HSPG by both quartz crystal microbalance measurements (Fig. 3) and by a solid phase binding assay (Fig. 4). High affinity binding of full-length VAR2CSA (NTS-FV2) and truncated VAR2CSA proteins was detected to HSPG. However, for several recombinant proteins it was not possible to fit a 1:1 binding model to the HSPG affinity curves and it is apparent that the peak response of HSPG binding is considerably lower than the CSPG response, indicating that only a fraction of the VAR2CSA proteins interact with HSPG (Fig. 3). Affinity measurements using VAR2CSA proteins not purified by size-exclusion chromatography show that aggregated protein has a much higher tendency to bind to HSPG (data not shown). The peak response for the core DBL2X-CIDRPAM was low and at a similar level for both CSPG and HSPG binding (Fig. 3D). This lack of specificity of DBL2X-CIDRPAM was also confirmed in the solid phase binding assay (Fig. 4E).
FIGURE 3.
Comparing VAR2CSA binding to CSPG and HSPG using a biosensor. A quartz crystal microbalance biosensor (Attana A100) was used for the kinetic analyses. Sensorgrams show VAR2CSA fragments binding either to immobilized CSPG or immobilized HSPG. The graphs show response (in Hertz) as a function of time (in seconds). Both the association and dissociation phase are shown. Black lines represent data, and red lines represent fitted curves attained by either a 1:1 binding model or a heterogeneity model. Insets show kon and koff as well as KD calculated from these. Furthermore, the insets show the maximum response obtained with the given concentration (peak response) and the type of fit used. The protein in flow is listed in the upper left corner of each sensorgram.
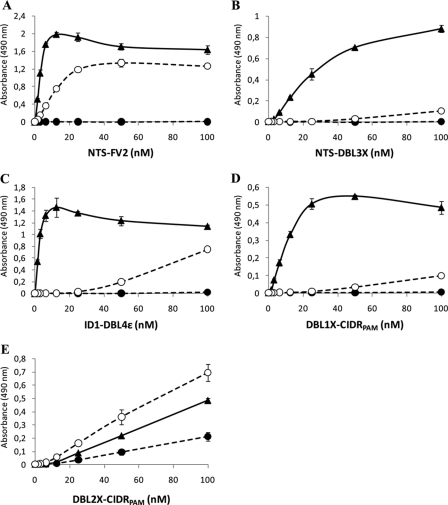
FIGURE 4.
Comparing VAR2CSA binding to CSPG and HSPG using a solid phase binding assay. Microtiter plates were coated with either 3 μg/ml CSPG (▴), 3 μg/ml enzyme treated CSPG (●) or 3 μg/ml HSPG (○). A 2-fold dilution series (1.56–100 nm) of each VAR2CSA protein was added to the wells, and an HRP-conjugated antibody targeting the V5-tag of the proteins was used to detect protein-glycan binding by measuring absorbance at 490 nm. A, NTS-FV2. B, NTS-DBL3X. C, ID1-DBL4ϵ. D, DBL1X-CIDRPAM. E, DBL2X-CIDRPAM. Error bars indicate the standard deviation of triplicate measurements.
Overall, the solid phase binding experiments corroborate the kinetic data by showing high CSPG specificity for all VAR2CSA proteins containing the CSA binding core DBL2X-CIDRPAM flanked by either the DBL1X in the N-terminal end or the DBL3X domain in the C-terminal end (Fig. 4). Limited amounts of the ID1-DBL4ϵ protein restricted the specificity analysis to the solid phase binding assay. Similarly, there was not enough protein of ID1-DBL3X to analyze HSPG binding. None of the proteins tested appeared to bind the protein core of CSPG, as demonstrated by their lack of binding to chondroitinase ABC-treated CSPG (Fig. 4).
Analysis of VAR2CSA-specific Antibodies for Inhibition of Parasite Adhesion to CSPG
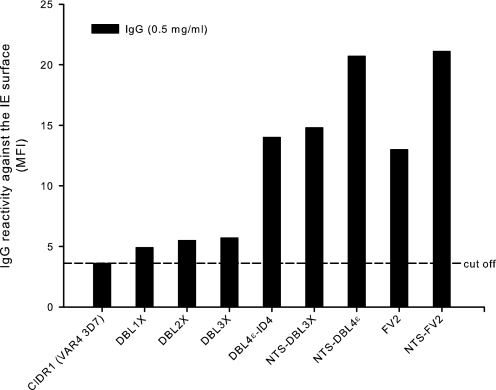
From the point of view of vaccine development it is important to analyze the ability of induced VAR2CSA-specific antibodies to block adhesion of IEs to CSA. Furthermore, such data may provide additional information about the receptor binding site. IgG induced by VAR2CSA constructs were analyzed by FCM for reactivity with native VAR2CSA protein on the surface of homologous FCR3 IEs. All the VAR2CSA-specific IgG purifications were positive in the FCM analysis although to a varying degree (Fig. 5). As expected, the multidomain proteins clearly induced a higher FCM antibody titer.
FIGURE 5.
Levels of anti-VAR2CSA IgG reactivity with the native protein after immunizations with recombinant proteins. Bars indicate the relative reactivity against VAR2CSA expressed on the surface of erythrocytes infected with the parasite line FCR3 (IEs). The IEs were labeled by purified IgG (0.5 mg/ml) from rats immunized with recombinant VAR2CSA proteins. IgG specific for the CIDR1 domain from the VAR4 3D7 protein was used as a negative control. The mean fluorescence intensity (MFI) was recorded from 5000 IEs. The assay was performed twice with similar results.
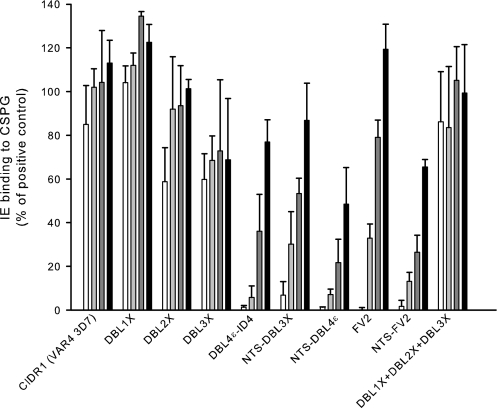
The purified IgG was subsequently assessed for the ability to inhibit adhesion of FCR3 IEs to CSPG in an inhibition binding assay. Each IgG was analyzed in a dilution series as follows: 0.5, 0.1, 0.02, and 0.004 mg/ml (Fig. 6). As previously shown antibodies specific for DBL4ϵ-ID4 (15), which is our leading vaccine candidate, and full-length VAR2CSA including the NTS region (NTS-FV2) (6) are both highly inhibitory (complete inhibition at an IgG concentration of 0.5 mg/ml and ∼90% inhibition at 0.1 mg/ml). In line with this, antibodies against the five domain construct NTS-DBL4ϵ inhibit parasite adhesion to CSPG with the same efficiency as the DBL4ϵ-ID4-specific IgG. Interestingly, antibodies specific for the NTS-DBL3X construct are also potent inhibitors (>90% inhibition at 0.5 mg/ml) despite the finding that IgG induced by the individual domains DBL1X, DBL2X, and DBL3X do not exhibit any inhibitory activity when pooled.
FIGURE 6.
Inhibition of parasite binding to CSPG by VAR2CSA-specific IgG induced by recombinant proteins. Bars indicate FCR3 P. falciparum-infected erythrocytes (IEs) binding to CSPG, after incubation with VAR2CSA-specific IgG and washing, as a percentage of binding by the positive control (binding in cell medium alone). Error bars indicate the standard deviation of triplicate measurements. The assay was performed with IgG purified from serum pools of rats immunized with the indicated recombinant proteins. IgG concentrations used: 0.5 mg/ml (white bars), 0.1 mg/ml (light gray), 0.02 (dark gray), and 0.004 mg/ml (black). IgG specific for the CIDR1domain from the VAR4 3D7 protein was used as a negative control. The assay was performed three times with similar results.
Although antibodies against the FV2 construct (lacking the NTS region) completely abolish parasite binding at 0.5 mg/ml of IgG, these are not as inhibitory as the NTS-FV2 antibodies at lower IgG concentrations. This observation is in some respects surprising since the FV2 protein binds, if not better than, at least as strongly as the NTS-FV2 to CSPG. This implies a proper folding of these two proteins thus displaying the same epitopes. However, it is possible that this discrepancy is due to variations in the animal immunizations or IgG preparation because the FCM results also show a difference in parasite surface reactivity between these two full-length constructs. Adhesion of the FCR3 IEs to CSPG was unaffected by IgG specific for the control protein (CIDR1 from VAR4 3D7).
DISCUSSION
Specific accumulation of P. falciparum-infected erythrocytes in the developing placenta causes the severe malaria syndrome named placental malaria. The main placental receptor for parasite binding are the CSA chains of CSPG (22).The unique sulfation pattern of the placental CSA (4, 5) probably explains the exclusively placental trophism. The VAR2CSA protein is the major parasite-encoded antigen associated with placental parasite adhesion and is the leading PM vaccine candidate. Several different regions of VAR2CSA have been proposed as responsible for the binding to CSA (23, 24), and it was recently suggested that the entire ectodomain is required for a high affinity interaction (7). In this study, we aimed to define the exact minimum CSA-binding region of the VAR2CSA protein.
Proteins of the PfEMP1 family consist of several highly cysteine-rich domains, which form a complex structure held together by a number of intramolecular disulfide bridges. Because of the many disulfide bonds, the production of these molecules as correctly folded protein has been, and remains, a challenge. VAR2CSA is the only PfEMP1 molecule in which the entire extracellular region has been successfully produced as a recombinant protein, and this has only been done in an insect cell expression system (6) and in HEK293 cells (7). In this study, all VAR2CSA proteins were expressed in baculovirus-infected insect cells and successful formation of intramolecular disulfide bridges was confirmed by reduced and non-reduced SDS-PAGE.
Our protein-glycan binding results provide compelling evidence that the receptor binding site of VAR2CSA from the FCR3 parasite lies within the N-terminal region, i.e. DBL1X-ID1-DBL2X-CIDRPAM-DBL3X, within which the binding core consists of DBL2X-CIDRPAM (summarized in Table 2). The DBL2X-CIDRPAM protein does not possess all the elements required for a specific CSA interaction of high affinity, but when this core assembles with either the DBL1X domain in the N-terminal end or the DBL3X domain in the C-terminal end, the CSA binding strength is increased to the level of the full-length protein and the specificity seems to be retained. It is possible that these flanking domains have a stabilizing effect on the binding core without directly participating in the CSA binding. Alternatively, DBL1X and DBL3X exhibit residues that are directly involved in the CSA binding, which is enhanced by the presence of these domains. We have previously shown that the DBL2X single domain does bind to some extend to CSA in a solid phase binding assay but with low specificity (18). This is supported by the present data showing that the affinity of the DBL2X protein to CSA is low.
The CSA binding properties of the DBL3X domain has been extensively studied (19, 25–27), but these results have not conformed to the expected specificity and affinity of the interaction between VAR2CSA and CSA. Although the DBL3X domain alone binds with weak affinity and not specifically to CSA, this domain has been considered to be part of the true CSA-binding site. Thus, it was an unexpected discovery that DBL3X is not necessary for achieving specific high affinity binding in vitro.
Many of the tested proteins also showed some affinity to HSPG. However when looking at the raw data in the sensorgrams it is apparent that the peak response is much lower than the CSPG binding, indicating that only a subfraction of the proteins binds to HSPG. This prompts the use of homogenous proteins as well as direct comparative analyses as shown here.
On the basis of our results it seems likely that all four N-terminal domains (DBL1X-ID1-DBL2X-CIDRPAM-DBL3X) form a binding cleft or pocket in the native VAR2CSA protein, which enables specific CSA interactions of high affinity. This kind of multivalent binding is rather typical for protein-glycan interactions, although there are exceptions. Monovalent protein-glycan binding tends to be of relatively low affinity (micro-molar range) and high avidity is therefore often required for the protein-glycan interaction to achieve specificity and function in vivo (1).
The full-length ectodomain of VAR2CSA from the 3D7 parasite binds with similar affinity to CSA as the FCR3 variant analyzed here (7). It has been shown that the overall structure of VAR2CSA from 3D7 is quite compact, which supports the idea that the binding site resides in multiple rather than single domains (7). From a broader perspective, it is interesting that the receptor binding region of VAR2CSA from the FCR3 isolate lies within the N-terminal part of the protein, which corresponds to the conserved domain composition of other PfEMP1 molecules (28). Several domains within this N-terminal region have been implicated in receptor binding activities, e.g. CD36 binding of CIDR1 domains (29–31), ICAM-1 binding of DBLβ-C2 domains (32–34) and binding of the DBL1α domain to HSPG (35).
It is tempting to speculate that all PfEMP1 molecules share a fundamentally similar mode of binding such that the receptor/ligand contact zone is always in the semi-conserved N-terminal part, although the number of domains necessary for high affinity and specificity may vary. However, considerably more structural knowledge about the diverse ectodomains and binding sites of PfEMP1 proteins are needed to substantiate this proposal.
The inhibitory function of the VAR2CSA-specific antibodies generated for this study support the location of the CSA-binding site in the N-terminal part since antibodies induced against the NTS-DBL3X protein efficiently abrogate parasite adhesion. The NTS-DBL3X construct binds specifically and with nanomolar affinity to CSPG, which suggests that it displays an essentially fully functional receptor binding site and that epitopes within and around this site are the targets of the inhibitory antibodies. By contrast, the individual domains DBL2X and DBL3X both bind poorly to CSPG, probably because many residues necessary for a strong interaction are not present in a single domain. This would explain why the antigen epitopes necessary for inducing adhesion-blocking antibodies, which are probably conformation dependent and contributed by separate domains, are absent from these constructs. Supporting this view, pooled antibodies raised against individual domains (DBL1X, DBL2X, and DBL3X) were not able to inhibit the interaction between VAR2CSA and CSPG. One additional possible explanation for the lack of adhesion inhibition demonstrated by pooled IgG could be the absence of antibodies specific for the CIDRPAM domain, which seems to be an essential region of the CSA-binding site.
Significantly, our current leading PM vaccine candidate, which is based on the DBL4ϵ domain including a small interdomain region (ID4) from the FCR3 parasite, is not required for a specific CSA interaction of high affinity in vitro, although this protein induces inhibitory antibodies that effectively block parasite adhesion to CSA (15). The mechanism behind the binding inhibition induced by immunization with this domain is not known, but it could be explained by steric hindrance since the DBL4ϵ domain has been shown by small angle x-ray scattering (SAXS) analysis to be in close proximity to the CSA binding structure of VAR2CSA.4 These DBL4ϵ-ID4 antibodies have recently proven to be capable of inhibiting the CSA binding of a panel of P. falciparum parasites isolated from the placenta of delivering mothers in both Tanzania and Benin (36). Cross-inhibition is an essential property of any candidate for PM vaccine development and by targeting the exact CSA-binding site it might be possible to induce an even more potent immune response than has been achieved with the DBL4ϵ-ID4 construct. In addition, such a directed antibody response against the actual receptor binding site would be much more difficult for the parasite to evade by mutation, following introduction of an inhibitory PM vaccine. Possibly the DBL4 domain could be fused to the DBL2X-CIDRPAM to make a stable chimeric protein, both presenting the minimal binding region as well as the DBL4 epitopes that has been shown to induce inhibitory antibodies.
Acknowledgments
We thank Besim Berisha, Khalid Pardes, Nahla Chehabi, and Jonas Fjelbye Hansen for excellent technical assistance.
This work was funded in part by the Danish Council for Independent Research/Medical Sciences (FSS) Grant 271-07-0696, the Seventh Framework Programme ([FP7/2007-2013]) under Grant Agreements 201222 (PreMalStruct) and 200889 (STOPPAM), and the Bill and Melinda Gates Foundation, Grant 47029 (PMI2). This study also received Proof-of-Concept Funding from VTU, Grant 07-017766, support from University of Copenhagen Program of Excellence (Membrane Topology and Quaternary Structure of Key Parasite Proteins Involved in P. falciparum Malaria Pathogenesis and Immunity), and a Grant for a Visiting Professorship from the Danish National Research Foundation (to D. E. A.).
S. Christoffersen, A. E. Langkilde, L. Turner, T. Lavstsen, B. Berisha, K. E. Jensen, M. A. Nielsen T. G. Theander, S. Larsen, and A. Salanti, manuscript in preparation.
- HSPG
- heparan sulfate proteoglycan
- CSA
- chondroitin sulfate A
- PfEMP1
- P. falciparum erythrocyte membrane protein 1
- CIDR
- cysteine-rich interdomain region
- CSPG
- chondroitin sulfate proteoglycan
- DBL
- Duffy binding-like domain
- FCM
- flow cytometry
- FV2
- full-length ectodomain of the VAR2CSA protein without N-terminal segment
- GLURP
- glutamate-rich protein
- ID
- interdomain
- IE
- P. falciparum-infected erythrocyte
- MSP-2
- merozoite surface protein 2
- NTS
- N-terminal segment
- PAM
- pregnancy-associated malaria
- PM
- placental malaria.
REFERENCES
- 1. Edited by Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 2. Brown A., Higgins M. K. (2010) Curr. Opin. Struct. Biol. 20, 560–566 [DOI] [PubMed] [Google Scholar]
- 3. Salanti A., Staalsoe T., Lavstsen T., Jensen A. T., Sowa M. P., Arnot D. E., Hviid L., Theander T. G. (2003) Mol. Microbiol. 49, 179–191 [DOI] [PubMed] [Google Scholar]
- 4. Achur R. N., Valiyaveettil M., Alkhalil A., Ockenhouse C. F., Gowda D. C. (2000) J. Biol. Chem. 275, 40344–40356 [DOI] [PubMed] [Google Scholar]
- 5. Alkhalil A., Achur R. N., Valiyaveettil M., Ockenhouse C. F., Gowda D. C. (2000) J. Biol. Chem. 275, 40357–40364 [DOI] [PubMed] [Google Scholar]
- 6. Khunrae P., Dahlbäck M., Nielsen M. A., Andersen G., Ditlev S. B., Resende M., Pinto V. V., Theander T. G., Higgins M. K., Salanti A. (2010) J. Mol. Biol. 397, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srivastava A., Gangnard S., Round A., Dechavanne S., Juillerat A., Raynal B., Faure G., Baron B., Ramboarina S., Singh S. K., Belrhali H., England P., Lewit-Bentley A., Scherf A., Bentley G. A., Gamain B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4884–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salanti A., Dahlbäck M., Turner L., Nielsen M. A., Barfod L., Magistrado P., Jensen A. T., Lavstsen T., Ofori M. F., Marsh K., Hviid L., Theander T. G. (2004) J. Exp. Med. 200, 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brabin B. J., Romagosa C., Abdelgalil S., Menéndez C., Verhoeff F. H., McGready R., Fletcher K. A., Owens S., d'Alessandro U., Nosten F., Fischer P. R., Ordi J. (2004) Placenta 25, 359–378 [DOI] [PubMed] [Google Scholar]
- 10. Fried M., Nosten F., Brockman A., Brabin B. J., Duffy P. E. (1998) Nature 395, 851–852 [DOI] [PubMed] [Google Scholar]
- 11. Ricke C. H., Staalsoe T., Koram K., Akanmori B. D., Riley E. M., Theander T. G., Hviid L. (2000) J. Immunol. 165, 3309–3316 [DOI] [PubMed] [Google Scholar]
- 12. Staalsoe T., Megnekou R., Fievét N., Ricke C. H., Zornig H. D., Leke R., Taylor D. W., Deloron P., Hviid L. (2001) J. Infect. Dis. 184, 618–626 [DOI] [PubMed] [Google Scholar]
- 13. Bockhorst J., Lu F., Janes J. H., Keebler J., Gamain B., Awadalla P., Su X. Z., Samudrala R., Jojic N., Smith J. D. (2007) Mol. Biochem. Parasitol. 155, 103–112 [DOI] [PubMed] [Google Scholar]
- 14. Dahlbäck M., Nielsen M. A., Salanti A. (2010) Trends Parasitol. 5, 230–235 [DOI] [PubMed] [Google Scholar]
- 15. Nielsen M. A., Pinto V. V., Resende M., Dahlbäck M., Ditlev S. B., Theander T. G., Salanti A. (2009) Infect. Immun. 77, 2482–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salanti A., Resende M., Ditlev S. B., Pinto V. V., Dahlbäck M., Andersen G., Manczak T., Theander T. G., Nielsen M. A. (2010) Malar. J. 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernandez P., Kviebig N. K., Dechavanne S., Lépolard C., Gysin J., Scherf A., Gamain B. (2008) Malar. J. 7, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Resende M., Ditlev S. B., Nielsen M. A., Bodevin S., Bruun S., Pinto V. V., Clausen H., Turner L., Theander T. G., Salanti A., Dahlbäck M. (2009) Int. J. Parasitol. 39, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 19. Khunrae P., Philip J. M., Bull D. R., Higgins M. K. (2009) J. Mol. Biol. 393, 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen M. A., Resende M., Alifrangis M., Turner L., Hviid L., Theander T. G., Salanti A. (2007) Exp. Parasitol. 117, 1–8 [DOI] [PubMed] [Google Scholar]
- 21. Haase R. N., Megnekou R., Lundquist M., Ofori M. F., Hviid L., Staalsoe T. (2006) Infect. Immun. 74, 3035–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fried M., Duffy P. E. (1996) Science 272, 1502–1504 [DOI] [PubMed] [Google Scholar]
- 23. Gamain B., Trimnell A. R., Scheidig C., Scherf A., Miller L. H., Smith J. D. (2005) J. Infect. Dis. 191, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 24. Avril M., Gamain B., Lépolard C., Viaud N., Scherf A., Gysin J. (2006) Microbes Infect. 8, 2863–2871 [DOI] [PubMed] [Google Scholar]
- 25. Higgins M. K. (2008) J. Biol. Chem. 283, 21842–21846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh K., Gittis A. G., Nguyen P., Gowda D. C., Miller L. H., Garboczi D. N. (2008) Nat. Struct. Mol. Biol. 15, 932–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh K., Gitti R. K., Diouf A., Zhou H., Gowda D. C., Miura K., Ostazeski S. A., Fairhurst R. M., Garboczi D. N., Long C. A. (2010) J. Biol. Chem. 285, 24855–24862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A., Wellems T. E. (1995) Cell 82, 89–100 [DOI] [PubMed] [Google Scholar]
- 29. Smith J. D., Kyes S., Craig A. G., Fagan T., Hudson-Taylor D., Miller L. H., Baruch D. I., Newbold C. I. (1998) Mol. Biochem. Parasitol. 97, 133–148 [DOI] [PubMed] [Google Scholar]
- 30. Chen Q., Heddini A., Barragan A., Fernandez V., Pearce S. F., Wahlgren M. (2000) J. Exp. Med. 192, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson B. A., Welch T. L., Smith J. D. (2003) Mol. Microbiol. 47, 1265–1278 [DOI] [PubMed] [Google Scholar]
- 32. Smith J. D., Subramanian G., Gamain B., Baruch D. I., Miller L. H. (2000) Mol. Biochem. Parasitol. 110, 293–310 [DOI] [PubMed] [Google Scholar]
- 33. Howell D. P., Levin E. A., Springer A. L., Kraemer S. M., Phippard D. J., Schief W. R., Smith J. D. (2008) Mol. Microbiol. 67, 78–87 [DOI] [PubMed] [Google Scholar]
- 34. Oleinikov A. V., Amos E., Frye I. T., Rossnagle E., Mutabingwa T. K., Fried M., Duffy P. E. (2009) PLoS. Pathog. 5, e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vogt A. M., Barragan A., Chen Q., Kironde F., Spillmann D., Wahlgren M. (2003) Blood 101, 2405–2411 [DOI] [PubMed] [Google Scholar]
- 36. Magistrado P. A., Minja D., Doritchamou J., Ndam N. T., John D., Schmiegelow C., Massougbodji A., Dahlbäck M., Ditlev S. B., Pinto V. V., Resende M., Lusingu J., Theander T. G., Salanti A., Nielsen M. A. (2011) Vaccine 29, 437–443 [DOI] [PubMed] [Google Scholar]
- 37. Rask T. S., Hansen D. A., Theander T. G., Gorm P. A., Lavstsen T. (2010) PLoS. Comput. Biol. 6, e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]