Abstract
Nitro-fatty acids (NO2-FA) are electrophilic signaling mediators formed by reactions of nitric oxide and nitrite. NO2-FA exert anti-inflammatory signaling actions through post-translational protein modifications. We report that nitro-oleic acid (OA-NO2) stimulates proMMP-7 and proMMP-9 proteolytic activity via adduction of the conserved cysteine switch domain thiolate. Biotin-labeled OA-NO2 showed this adduction occurs preferentially with latent forms of MMP, confirming a role for thiol alkylation by OA-NO2 in MMP activation. In addition to regulating pro-MMP activation, MMP expression was modulated by OA-NO2 via activation of peroxisome proliferator-activated receptor-γ. MMP-9 transcription was decreased in phorbol 12-myristate 13-acetate-stimulated THP-1 macrophages to an extent similar to that induced by the peroxisome proliferator-activated receptor-γ agonist Rosiglitazone. This was affirmed using a murine model of atherosclerosis, ApoE−/− mice, where in vivo OA-NO2 administration suppressed MMP expression in atherosclerotic lesions. These findings reveal that electrophilic fatty acid derivatives can serve as effectors during inflammation, first by activating pro-MMP proteolytic activity via alkylation of the cysteine switch domain, and then by transcriptionally inhibiting MMP expression, thereby limiting the further progression of inflammatory processes.
Keywords: Atherosclerosis, Inflammation, Matrix Metalloproteinase, Post Translational Modification, PPAR, Electrophiles, Nitro-fatty Acids, Nitroalkenes, Reactive Oxygen Species (ROS)
Introduction
Nitro-fatty acids, products of the reaction of nitric oxide (NO), secondary oxides of nitrogen, and unsaturated fatty acids, act as anti-inflammatory signaling mediators (1, 2). Nitroalkene derivatives of fatty acids, including nitro-linoleic acid (LNO2)2 and nitro-oleic acid (OA-NO2), are electrophilic and exert signaling actions via peroxisome proliferator-activated receptor-γ (PPARγ)-dependent and -independent mechanisms (3). NO2-FA also modulate inflammation by inhibiting nuclear factor κB (NF-κB)-dependent gene expression (4), suppression of the proinflammatory signal transducer and activator of transcription-1 (STAT-1) (5), and induction of heat shock factor and nuclear factor-erythroid 2-related factor 2/Kelch-like ECH-associating protein 1 (Keap1/Nrf2)-dependent phase II gene expression (6).
A broad range of enzymatically and non-enzymatically modified fatty acids mediate different stages of inflammation, ranging from cytokine secretion and cell recruitment to the resolution of inflammation (7). In particular, electrophilic fatty acids stemming from oxidative inflammatory reactions (i.e. 15-deoxy-Δ12,14-prostaglandin J2, 4-hydroxynonenal, 4-oxononenal, acrolein, and NO2-FA) undergo Michael addition reaction with nucleophilic amino acids (i.e. cysteine and histidine) located in electrophile-sensitive protein domains (8). The sites, rates, and reversibility of reaction of these different species are strongly influenced by the electron withdrawing group. In this regard, the rate constant for the reversible Michael addition of cysteine and glutathione (GSH) by NO2-FA is 2 orders of magnitude greater than for α,β-unsaturated carbonyl fatty acids including 4-hydroxynonenal, 4-oxononenal, prostaglandin A2, and 15-deoxy-Δ12,14-prostaglandin J2 (9). The facile reaction of NO2-FA with susceptible thiols of key transcriptional regulatory proteins, including the p65 subunit of NF-κB (10), PPARγ (11), and Keap1/Nrf2 (12). This supports that nitroalkenes are redox reaction-derived byproducts that link both gene expression and cell function with metabolic and inflammatory status (13). It is expected that electrophile-induced cytoprotective versus toxic reactions will be dependent on multiple factors, including electrophile concentration, molecular target specificity, reaction reversibility, metabolism, and excretion (14, 15).
Matrix metalloproteinases, a class of proteolytic enzymes under strict transcriptional and post-translational regulation, are composed of 24 members in mammals. MMPs are responsible for the degradation and turnover of extracellular matrix components. MMPs all share a common structure that consists of a signal peptide, the pro-peptide domain, and a catalytic domain containing a Zn2+ ion at the active site (16). In general, MMPs are secreted as zymogens and are maintained in a latent state until activated. Latency is maintained by a covalent thiol-Zn2+ interaction between a conserved cysteine in the pro-peptide domain and a Zn2+ ion complexed in the catalytic domain. Activation of pro-MMP occurs upon the dissociation of the Cys-S-Zn2+ bond or by proteolytic cleavage of the pro-domain (17). Dysregulation of MMP activity is linked with the initiation and development of numerous pathological processes such as atherosclerosis, cardiac remodeling, emphysema, and metastatic cancers (18).
Byproducts of inflammatory reactions play a role during MMP activation in vivo. In this context, compelling evidence supports that a product of neutrophil myeloperoxidase, hypochlorous acid, induces in vitro activation of proMMP-7 by Cys-SH oxidation to sulfinic acid (Cys-SO2H) (19). Also, the product of superoxide and NO reaction, peroxynitrite (ONOO−), induces oxidation and glutathiolation (Cys-S-SG) of the MMP cysteine switch thiol (20). In contrast, MMP activation by NO-mediated Cys-S-nitrosation remains controversial, because of the broad range of concentrations and chemical natures of different NO donors being used (21, 22).
During the onset of inflammation, immune cell access to sites of injury and infection is facilitated by MMP activity. Uncontrolled or prolonged exposure to MMP activity results in tissue damage, remodeling, poor resolution of inflammation, and even progression of disease toward more chronic states. For example, atheroma formation in blood vessels is associated with, neointimal proliferation and infiltration of macrophages (23). Plaque stability is critically related to the action of MMPs, as the hydrolysis of extracellular matrix leads to plaque instability, plaque rupture, and vessel occlusion (24).
It remains unclear as to what regulates the beneficial versus deleterious actions of MMPs toward atherosclerotic plaque. Studies of MMP-9−/− mice have revealed opposing results on plaque size and stability (25, 26). Therefore, the regulation of MMP expression by macrophages and control of the inflammatory process seem to be crucial aspects for the prevention of plaque rupture. In this regard, in vivo administration of OA-NO2 inhibits the formation of fatty streaks and stabilizes atheromatous plaque in ApoE−/− mice fed a high-fat diet, a model of atherosclerosis (27). Notably, tissue and plasma levels of NO2-FA can be increased by inflammatory and ischemic processes (10, 28, 29), suggesting that downstream signaling actions may also be of relevance. For these reasons, we examined the impact of an exemplary nitroalkenyl fatty acid, OA-NO2, on the activity and transcriptional regulation of MMPs using MMP-7 and MMP-9 as enzymatic and biological models.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Sigma, if not stated otherwise. Recombinant human proMMP-7, proMMP-9, MMP-2/MMP-9 substrate II, and MMP-7 fluorescent Mca peptide (7-methoxycoumarin-4-acetyl-Pro-Leu-Gly-Leu-β-(2,4-dinitrophenylamino)Ala-Ala-Arg-NH2) were from Calbiochem (La Jolla, CA). Polyclonal MMP-9 antibody was obtained from Sigma and streptavidin-conjugated HRP from Pierce. Nitro-linoleic acid (9-, 10-, 12-, or 13- octadeca 9,12-cis, cis dienoic acids; LNO2) and nitro-oleic acid (9- or 10- octadeca 9-cis enoic acids; OA-NO2) were prepared as previously described (30, 31). OA-NO2-biotin and OA-biotin were prepared as described (4). The human macrophage THP-1 cell line was obtained from the American Type Culture Collection (ATCC). Cells were maintained in DMEM, 10% fetal serum bovine (FBS), and 100 units/ml of penicillin and streptomycin at 37 °C, 5% CO2.
MMP-9 Activity Determination
Pro-MMP-9 activation was determined by following the proteolytic product of the MMP synthetic peptide substrate by mass spectrometry. Pro-MMP-9 was incubated with various concentrations of OA-NO2 (1–20 μm), oleic acid (OA, 1–20 μm), and methanol (vehicle control) in 50 mm sodium phosphate buffer, pH 7.5, for 45 min at 23 °C. Then, 30 μmol of the synthetic substrate was added and, at the indicated time points, the reaction was terminated by acidification with formic acid (1% final concentration) and proteolytic products were quantified by LC-ESI-MS. Mass spectrometric analysis and quantification were performed using a hybrid triple-quadrupole linear ion trap mass spectrometer (4000 Q trap; Applied Biosystems) coupled to a Shimadzu HPLC system. Peptides were separated on a reverse-phase column (Zorbax SB300-C18, 100 × 0.5 mm, 3.5 μm) using mobile phase: solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Elution was carried out with a linear gradient of solvent B (2–100% in 7 min). MS analysis in the positive ion mode showed an ion corresponding to the MMP peptide substrate m/z 656.5, and the MMP cleavage product ion with an m/z 369.5 resulting from the activation of proMMP-9 with p-aminophenylmercuric acetate, a known activator of latent MMP-9. Quantification of substrate consumption and product formation was done using the multiple reaction monitoring scan mode by recording peak areas using an external standard curves. The following multiple reaction monitoring transitions were used for substrate consumption (m/z 656.5/369.5) and product formation (m/z 369.5/239.1).
ProMMP-7 Activity
Proteolytic activity of proMMP-7 was measured by fluorescence analysis using the synthetic peptide, 7-methoxycoumarin-4-acetyl-Pro-Leu-Gly-Leu-β-(2,4-dinitrophenylamino)Ala-Ala-Arg-NH2 (Mca peptide), which under proteolytic cleavage release the fluorescence domain. Briefly, the proMMP-7 sample was incubated with NO2-FA as described above with proMMP-9. Then, aliquots of proMMP-7 (30 ng) were transferred to a 96-well microtiter plate containing 3 μm substrate (Mca) in 500 μl of reaction buffer (50 mm Tris-HCl, pH 7.4, 0.2 m NaCl, 10 mm CaCl2, and 0.02% NaN3). Fluorescence was acquired using an excitation wavelength of λex = 328 nm and emission λem = 392 nm in a SpectraMax M2 microplate reader (Molecular Devices). Control reactions were performed to evaluate the effects of the reaction mixture components on fluorescence yields.
Gelatin Zymography
MMP-9 activity was assessed in conditioned media from THP-1 cells incubated with OA-NO2, OA, or preincubated with the PPARγ antagonist GW9662. Samples were prepared in non-reducing buffer and loaded on 4–12% polyacrylamide gradient gels containing 0.1% gelatin. After electrophoresis, the gels were washed with 2.5% Triton X-100 and incubated for 18 h in activation buffer containing 10 mm Tris-HCl, pH 7.4, 100 mm NaCl, and 10 mm CaCl2. Then, gels were stained in 0.25% Coomassie Brilliant Blue G-250 solution. Proteolytic activity was determined as proteolysed areas of gelatin against the blue background staining.
Mass Spectrometry
Native proMMP-7 (10 μg) was digested before and after OA-NO2 and OA treatment with sequencing grade trypsin (1:50) in 50 mm phosphate buffer, pH 7.4, at 37 °C for 16 h. ESI-LC-MS analysis of the peptide digest was performed in the positive ion mode with a linear ion trap mass spectrometer (LTQ-XL, Thermo Electron Corp., San Jose, CA). Peptides were separated in a reverse-phase column (Grace Vydac, 5 μm, 2.1 × 150 mm, 300 Å) with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) at a flow rate of 0.2 ml/min. Peptides were eluted with a linear gradient of solvent B (5–60% in 70 min). The electrospray voltage was set at 5 kV and the capillary temperature was 260 °C.
Western Blotting
ProMMP-9 and MMP-9 treated with OA-NO2-biotin or OA-biotin were separated on 10% SDS-polyacrylamide gels (CriterionTM XT Precast Gels, Bio-Rad) and transferred to nitrocellulose membranes at 90 V for 1 h at 4 °C. Membranes were blocked with 10% nonfat milk in Tris-buffered saline, 0.05% Tween 20 (TTBS) for 1 h at room temperature before incubation with a 1:1000 dilution of streptavidin-horseradish peroxidase (HRP) conjugate (Pierce) in TTBS, 1% nonfat milk for 1 h at RT. Then, membranes were washed in TTBS and developed with chemiluminescent substrate (GE Healthcare). To ensure even protein loading, membranes were stripped and re-probed with anti-MMP-9 antibody, which recognizes pro- and active forms.
Quantitative Real Time PCR
THP-1 cells (90% of confluent) were treated with OA-NO2 (1–2.5 μm), Rosiglitazone (1–2.5 μm), and methanol (vehicle control) for 1 h before stimulation with 0.1 μm PMA during 4, 9, and 16 h. Then, cells were harvested for RNA extraction. Total RNA was extracted by TRIzol® (Invitrogen) according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed in a 25-μl reaction using iScriptTM cDNA synthesis kit (Bio-Rad). Samples were analyzed by TaqMan fast universal PCR master mixture using gene expression primers for MMP-9 (Hs00234579_m1) and actin as a housekeeping gene. Samples were run in triplicate in the StepOne detection system (Applied Biosystems).
Murine Atherosclerosis Studies
Male ApoE−/− mice were purchased from The Jackson Laboratory. At 8 weeks age, mice were placed on an atherogenic diet for 12 weeks (21% fat and 1.25% cholesterol, Harlan Teklan). Concurrently, mice were implanted with a subcutaneous osmotic mini-pump (Alzet®, Durect Corp., Cupertino, CA), which delivered OA-NO2 (8 mg/kg/d), OA (8 mg/kg/d, Nu-Chek Inc.), or vehicle control (polyethylene glycol/ethanol). At the termination of treatment, mice were sacrificed and aortic roots dissected and embedded in OCT medium for posterior sectioning. Tissue was fixed before staining and atherosclerotic lesions were detected by Oil Red O staining. Immunofluorescence of aortic root sections was performed with an anti-MMP-9 antibody (Sigma). All animal studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (approval 0709432).
RESULTS
OA-NO2 Activation of ProMMP-9 and ProMMP-7
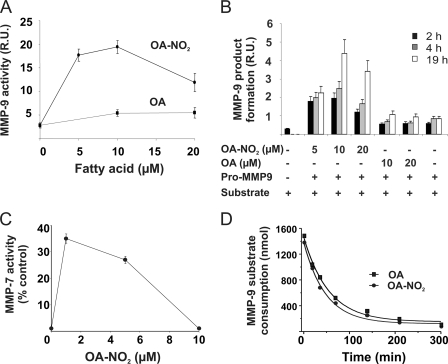
To evaluate whether OA-NO2 disrupts Cys-Zn2+ interactions to activate proMMP-9, the pro-enzyme was treated with OA-NO2 and extents of proteolytic product generation from an added synthetic peptide substrate was monitored by mass spectrometry (Fig. 1). Incubation with OA-NO2 (1–10 μm) for 30 min at 37 °C induced a 3-fold increase in the activation of proMMP-9, whereas both vehicle control (not shown) and OA had no effect (Fig. 1A). Activation of proMMP-9 was both time- and dose-dependent, with pro-enzyme not being activated by native OA (Fig. 1B). In addition, the activation of proMMP-9 by OA-NO2 presented a biphasic response profile, showing activation at concentrations lower than 10 μm and reduction of activity at higher and less biological concentrations (>10 μm) (Fig. 1, A and B). ProMMP-7 activation by OA-NO2 was also dose-dependent and unaffected by native OA (Fig. 1C), with proMMP-7 slightly more sensitive to activation by OA-NO2 than proMMP-9 (Fig. 1A, C). MMP-9 activity was not affected by 10 μm OA-NO2 and its native fatty acid counterpart OA (Fig. 1D).
FIGURE 1.
OA-NO2 induces activation of proMMP-9 and proMMP-7. Pro-MMPs were treated with either OA-NO2 or OA (1–20 μm) in 50 mm phosphate buffer, pH 7.5, for 30 min at RT. Aliquots were removed and proteolytic activity was determined by LC-ESI-MS using a synthetic peptide as MMP substrate. A, proMMP-9 activation by OA-NO2. B, OA-NO2 time and dose dependence of proMMP-9 activation. C, proMMP-7 activation by OA-NO2. D, OA-NO2 (10 μm) does not affect MMP-9 activity. Results are the mean ± S.D. of three independent determinations.
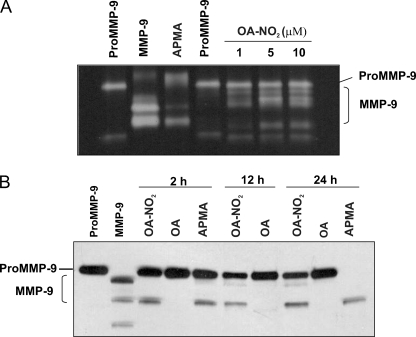
To verify that OA-NO2 activated MMP-9 by inducing cleavage of the pro-domain, recombinant proMMP-9 was incubated with OA-NO2 and evaluated by both gelatin zymography and Western blot analysis. Zymography gels (Fig. 2A) showed that OA-NO2 treatment induces, in a dose-dependent fashion (1–10 μm), conversion of the zymogen (92 kDa) to the active form of MMP-9, as revealed by an emerging band showing both proteolytic activity and a lower molecular mass (82 kDa). In separate experiments over time (2, 12, and 24 h), treatment with OA-NO2 gave similar results, confirming conversion to the active MMP-9 form, as compared with the MMP activator p-aminophenylmercuric acetate. Treatment with native OA did not induce cleavage of proMMP-9 (Fig. 2B).
FIGURE 2.
OA-NO2 induces activation of proMMP-9. A, recombinant proMMP-9 treated with OA-NO2 is activated in a dose-dependent fashion (1–10 μm). Conversion of proMMP-9 (92 kDa) to the low molecular mass active form (82 kDa) is demonstrated by gelatin zymography. B, Western blot of proMMP-9 activated by OA-NO2. One representative experiment of three is shown.
OA-NO2 Adduction of ProMMP-9
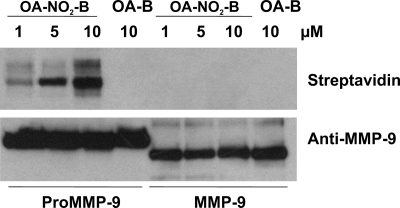
The activation of proMMP-9 was further evaluated with a biotin-OA-NO2 derivative. MMP-9 and proMMP-9 (0.2 μg) were treated with either biotin-OA-NO2 or the corresponding native fatty acid control, biotin-OA, and the proteins were analyzed by Western blotting (Fig. 3). Formation of the proMMP-9-OANO2-biotin adduct was increased in a dose-dependent manner (1–10 μm) (Fig. 3). In contrast, the active form of MMP-9, which lacks the 10-kDa amino-terminal pro-domain, was not alkylated by biotin-OA-NO2 at concentrations up to 10 μm. Non-electrophilic OA-biotin was used as a control for nonspecific interactions (Fig. 3). These results reveal that biotin-OA-NO2 adduction to proMMP-9 occurs in the pro-domain region, which contains the critical thiol group involved in the activation of MMPs (Cys-70 and Cys-100 for proMMP-7 and proMMP-9, respectively).
FIGURE 3.
OA-NO2 induces nitroalkylation of the proMMP-9 cysteine switch domain. ProMMP-9 and MMP-9 (200 ng) were incubated with various concentrations of biotinylated OA-NO2 (1, 5, and 10 μm) in 50 mm phosphate buffer, pH 7.5, at room temperature (23 °C) for 30 min. Pro-MMP-9 and MMP-9 controls were incubated with 10 μm biotinylated native oleic acid (OA-B). Samples were subjected to SDS-PAGE (10% acrylamide gel) under non-reducing conditions. Proteins were transfered and immunoblotted with streptavidin-HRP or polyclonal anti-MMP-9 antibody, which demonstrates an equal loading of proMMP-9 (92 kDa) and the active form of MMP-9 (82 kDa). This result is representative of three independent experiments.
Mass Spectrometric Analysis of ProMMP-7
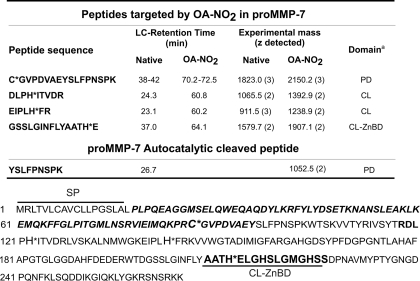
To define specific sites of OA-NO2 adduction, proMMP-7 was trypsin digested and sequenced by mass spectrometry. ProMMP-7 was selected for analysis due to its lower molecular mass (29 kDa) and the fact that it contains only 1 cysteine (in the prodomain) and 8 histidine residues. Untreated proMMP-7 (5 μg) tryptic digests were analyzed by ESI-LC-MS, resulting in 65% peptide coverage of the primary sequence (not shown). When digests of OA-NO2-treated proMMP-7 were analyzed, four OA-NO2-adducted peptides were identified (Table 1). These peptides showed both the addition of m/z 327.3 upon OA-NO2 reaction (the mass of OA-NO2) and a shift in retention time compared with the non-modified peptides, due to an increased hydrophobicity induced by OA-NO2 adduction. One of the four modified peptides encompassed residues 70–86 that contain the cysteine switch domain coordinating the Zn2+ ion in the catalytic domain. ESI-LC-MS analysis also showed a new peak in the treated sample as the doubly charged ion m/z 1075.4; [C(OA-NO2)GVPDVAEYSLFPNSPK]2+ having a theoretical mass of m/z of 1075.8 (Table 1 and supplemental Fig. S1A). The unmodified peptide was detected as a doubly charged ion of m/z 912.0 [CGVPDVAEYSLFPNSPK]2+; which was present in low abundance in OA-NO2-treated samples (supplemental Fig. S1A). The mass difference between the non-modified and the modified peptide was m/z 163.4 atomic mass units, corresponding to the mass of OA-NO2 in a doubly charged peptide (supplemental Fig. S1A). To verify these results, MS/MS analysis of ions m/z 912.0 and 1075.4 was performed and showed a sequence of b and y ions that match the expected ion products from the cysteine switch domain peptide (supplemental Fig. S1B). These results indicate that the thiol group of the pro-domain is the principal target for covalent adduction by OA-NO2, and supports that lipid electrophiles can play a role in the activation of MMPs.
TABLE 1.
LC-MS analysis of proMMP-7 cysteine and histidine adduction by OA-NO2
Tryptic peptides having OA-NO2 adducts displayed an increase in HPLC retention time. The proMMP-7 amino acid sequence is shown below. SP, signal peptide; italic bold letters indicate propeptide domain, PD; CL, catalytic domain and CL-ZnBD, CL zinc binding domain.
Activation of pro-MMP involves the disruption of the thiol-Zn2+ interactions that in turn trigger an autoproteolytic cleavage of the prodomain. MS analysis of the peptide digest of OA-NO2-treated proMMP-7 identified a doubly charged ion of m/z 526.7 [YSLFPNSPK]2+, which corresponds to the autoproteolytic product of the prodomain peptide, CGVPDVAE77Y78SLFPNSPK (supplemental Fig. S1C). Analysis of unmodified proMMP-7 did not detect this peptide ion. Failure to detect the complementary ion CGVPDVAE+ (m/z 1117.3) in both treatments may have been the result of poor ionization.
Regulation of MMP-9 Gene Expression by OA-NO2
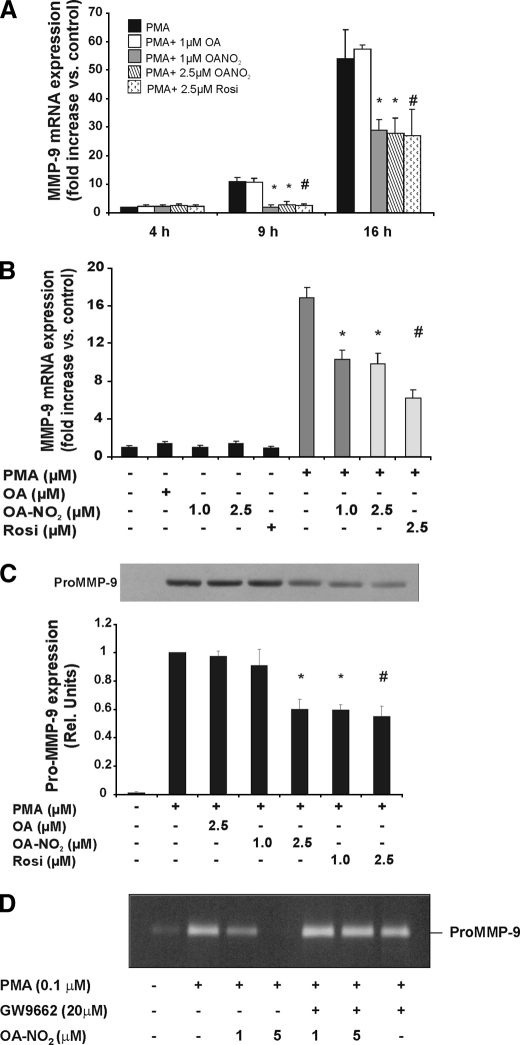
To determine whether OA-NO2 modulates MMP-9 transcription, THP-1 macrophages were stimulated with 0.1 μm PMA after OA-NO2 treatment. Upon activation, MMP-9 mRNA transcription was assessed at 4, 9, and 16 h by quantitative real time PCR. PMA induced 10 ± 1.5- and 65 ± 4.3-fold increases in mRNA levels at 9 and 16 h, respectively (Fig. 4A). OA-NO2 (1–2.5 μm) and Rosiglitazone (2.5 μm), both potent PPARγ ligands (11), markedly suppressed PMA induction to 2 ± 0.2- and 1.8 ± 0.3-fold increases, respectively, after 9 h of incubation. Comparable results were obtained after 16 h of treatment, with OA-NO2 and Rosiglitazone reducing PMA induction of MMP-9 mRNA transcription to 31.3 ± 4.1- and 35 ± 3.8-fold increases. Only slight effects were observed at 4 h after PPARγ ligand addition (Fig. 4A). No change in MMP-9 mRNA expression was induced by OA-NO2 or Rosiglitazone in the absence of cell differentiation by PMA (Fig. 4B).
FIGURE 4.
PMA-induced MMP-9 expression is down-regulated by OA-NO2. THP-1 cells were preincubated with the indicated concentrations of OA-NO2 and Rosiglitazone for 1 h prior to addition of PMA. A, PMA induced MMP-9 mRNA expression is significantly decreased by OA-NO2 (1.0–2.5 μm) and Rosiglitazone (2.5 μm) at 9 and 16 h compared with PMA treatment. Data are presented as mean ± S.E. of three independent experiments (OA-NO2 (1.0–2.5 μm), *, p < 0.05; and Rosiglitazone (2.5 μm), #, p < 0.05 compared with PMA, analysis of variance). B, OA-NO2 did not alter MMP-9 mRNA levels in cells in the absence of PMA stimulation but significantly reduced its expression under PMA stimulation. Data are presented as mean ± S.E. of three independent experiments (OA-NO2 (1.0–2.5 μm), *, p < 0.05; and Rosiglitazone (2.5 μm), #, p < 0.05 compared with PMA induction, analysis of variance). C, MMP-9 protein expression is decreased in the medium of THP-1 cells exposed to OA-NO2 previous to PMA stimulation. Results are representative of three independent experiments. D, PPARγ antagonist GW9662 blocks MMP-9 down-regulation by OA-NO2. Gel zymography of the THP-1 cells supernatant incubated with OA-NO2 (1–5 μm) in the presence or absence of PPARγ inhibitor, GW9662, significantly inhibited down-regulation of MMP-9. These results are representative of three independent experiments.
To evaluate whether a change in transcription affected MMP-9 protein secretion, extracellular levels of MMP-9 were assessed. THP-1 cells in serum-free medium were treated for 1 h with OA-NO2 (0–2.5 μm) or Rosiglitazone (0–2.5 μm) before PMA stimulation and then secretion into the medium was monitored by Western blotting. MMP-9 was only detectable in cell medium upon PMA stimulation (Fig. 4C). Both Rosiglitazone and OA-NO2 significantly decreased extracellular MMP-9 secretion, although the effect of NO2-FA was less pronounced. Trypan blue exclusion staining for cell viability did not show any difference between treatments (not shown).
Gelatin zymography of THP-1-conditioned medium confirmed the inhibition of MMP-9 expression by OA-NO2 (1–5 μm). Notably, when cells were incubated with the PPARγ receptor antagonist GW9662 (20 μm), OA-NO2-induced inhibition of MMP-9 expression was abrogated (Fig. 4D).
OA-NO2 Decreases MMP-9 Expression in Atherosclerotic Plaque of ApoE-deficient (ApoE−/−) Mice
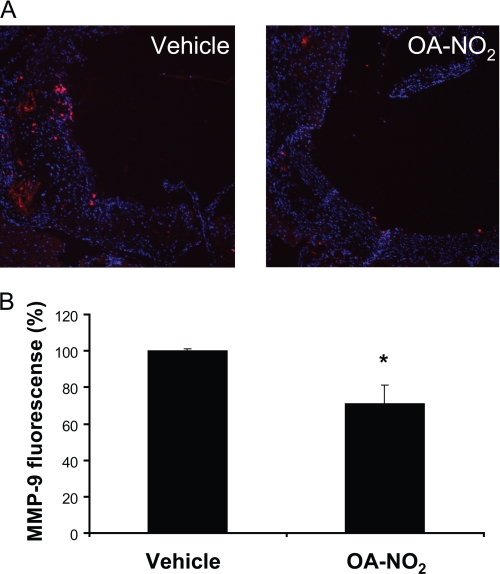
The effect of OA-NO2 administration in vivo on MMP-9 expression in developing atherosclerotic lesions of ApoE−/− mice was examined by immunofluorescence analysis of protein expression. There was a significant decrease in MMP-9 expression when compared with vehicle-treated mice (Fig. 5, A and B) supporting in vivo regulation of MMP-9 expression by OA-NO2.
FIGURE 5.
OA-NO2 decreased MMP-9 expression in atherosclerotic plaques. A, representative aortic sections stained by MMP-9 (magnification ×20, scale bar indicate 100 nm, blue = nuclei, red = MMP-9). B, quantitative analysis of MMP-9 expression in the atherosclerotic lesion area. OA-NO2 significantly reduces MMP-9-positive areas (*, p < 0.05 versus vehicle control). Data are presented as mean ± S.E. of three independent experiments.
DISCUSSION
The electrophilic fatty acid OA-NO2 modulates the proteolytic activity of MMP-9 and MMP-7 by reaction with and alkylation of the cysteine switch domain thiol, converting the pro-enzyme to the active form. In addition, OA-NO2 suppresses PMA-induced human THP-1 macrophage MMP-9 mRNA gene expression to an extent similar to the PPARγ agonist Rosiglitazone. This effect was inhibited by the PPARγ receptor antagonist GW9662. These actions are of potential clinical significance, because MMPs strongly influence turnover and remodeling of the vascular extracellular matrix. Thus, electrophilic fatty acids can transduce the vessel remodeling promoted by physiologic modulators of MMP expression and activity that include inflammatory cytokines, reactive oxygen species, and hemodynamic stresses (32).
Inflammatory stimuli induce elevated rates of generation of reactive oxygen and NO-derived species, which in turn contribute to the nitration of unsaturated fatty acids. For example, oleic and linoleic acid nitration are induced in cardiac tissue and mitochondria following ischemia-reperfusion events and in inflammatory mediator-activated macrophages (2, 28, 29). The endogenous concentrations of these electrophilic species that can accumulate during inflammatory responses depends on multiple factors including nitration rates, hydrophobic versus aqueous compartmentalization of reactions, free versus esterified unsaturated fatty acid levels, lipase activities, acyl chain metabolism via β-oxidation, extents of electrophilic fatty acid adduction to susceptible nucleophilic residues of proteins and glutathione, and finally, the cellular export of electrophilic fatty acid-GSH adducts via multidrug resistance transport proteins (33, 34). Although the predominant mechanisms leading to the formation of NO2-FA in vivo remain incompletely defined, fatty acid nitration can also arise from acid-catalyzed reactions of nitrite (NO2−), both a dietary constituent and a product of NO metabolism. The protonation of NO2− yields nitrous acid (HNO2), which decomposes to a broad range of oxidizing, nitrating, and nitrosating species (35, 36). In addition, nitration reactions can be mediated by the nitrogen dioxide (•NO2) that arises from homolytic decomposition of peroxynitrous acid (ONOOH) and the CO2 reaction product of peroxynitrite, nitrosoperoxocarbonate (ONOOCO2). Finally, fatty acid nitration can be induced by one electron oxidation of NO2− to •NO2 by heme peroxidases such as leukocyte myeloperoxidase (37, 38).
One of the most distinctive properties of NO2-FA is an ability to undergo kinetically rapid and reversible Michael addition with nucleophilic amino acids of proteins (e.g. Cys, His), an event leading to post-translational protein modification and subsequent modulation of protein structure, function, and distribution (8). In this regard, the limited proteolytic cleavage of the MMP prodomain is viewed as the principal mechanism for MMP activation (16). Recent evidence also support that oxidants and electrophilic species can activate pro-MMP (39, 20). In this context acrolein, an electrophilic constituent of cigarette smoke (39), and the myeloperoxidase product hypochlorous acid (HOCl) both stimulate MMP activity (19, 40). Contradictory observations have been reported regarding the activation of MMP by NO and its secondary superoxide reaction product peroxynitrite (22, 41).
A central aspect of many inflammatory and degenerative diseases is the dysregulation of MMP activity, a consequence of a compromised proteolytic balance arising from increased MMP expression, zymogen activation, and the action of protease inhibitors (16). MMPs are secreted by diverse cell types, with their expression dependent on cell type and regulation by a broad spectrum of mediators. For example, in the vessel wall MMP-9 is secreted by fibroblasts, endothelial cells, and macrophages. In atherosclerotic tissues, macrophage-derived foam cells are the primary source of the proMMP-9 that dictates the stability of the atherosclerotic plaque (23, 42). The factors that stimulate MMP-9 activity in atheroma-associated macrophages are unclear; however, activation by oxidative stress and up-regulation of expression by oxidized LDL are implicated (43).
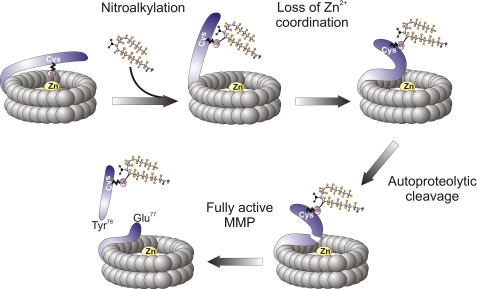
The catalytic activity of proMMP-7 and proMMP-9 was modulated by OA-NO2 in a dose-dependent manner (Fig. 1). The mechanism accounting for proMMP-7 activation by OA-NO2 was identified by LC-MS/MS, which revealed Michael addition of OA-NO2 to the cysteine switch domain thiol (C(OA-NO2)GVPDVAE77Y78SLFPNSPK). An alternative mechanism for MMP activation has also been characterized, autoproteolytic cleavage at Glu77–Tyr78 and the subsequent release of the prodomain fragment. The products of this autoproteolytic event were investigated, and revealed an increase in the formation of the 78YSLFPNSPK peptide upon OA-NO2 treatment. The appearance of this peptide supports that OA-NO2 adduction also triggered the catalytic activation of pro-MMP and thus reveals a new mechanism for the post-translational regulation of MMP catalytic activity (Fig. 6).
FIGURE 6.
OA-NO2 regulation of MMP activation.
In aggregate, this data supports that OA-NO2 and other inflammatory or dietary-derived electrophiles can exert the activation of proMMP-7 and -9 at low concentrations, while inhibiting MMP at higher concentrations (14). Patterns of MMP activation and inhibition will be influenced by steady state OA-NO2 concentrations and target molecule exposure times. The MMP cysteine switch domain is viewed to represent the primary target of electrophile reaction that leads to the zymogen activation. This was further confirmed by an affinity labeling strategy using biotinylated OA-NO2. In this context, biotin-OA-NO2 underwent rapid reaction with proMMP-9, but did not react with active MMP-9 at lower and more physiologically relevant concentrations (Fig. 3). Incubation with biotin-OA-NO2 at concentrations >10 μm promoted reaction with catalytically active forms of MMP, reflecting OA-NO2 adduction with less reactive nucleophilic residues such as histidine.
Current data indicate that NO2-FA play critical roles in modulating inflammation by inducing both tissue-protective and anti-inflammatory responses. Nitroalkene derivatives of fatty acids are robust PPARγ ligands that lead to suppression of MMP transcription. Notably, NO2-FA covalently react with the PPARγ ligand binding domain Cys285 residue, a phenomenon that will significantly impact Koff reactions (11). The extended exposure of PPARγ to low concentrations of a covalently adducting ligand can result in significant accumulation of bound ligand and receptor activation. This mode of PPARγ adduction also induces unique patterns of coregulatory protein interactions when contrasted to those induced by Rosiglitazone, thus uniquely impacting patterns of downstream PPARγ-dependent gene expression (11). PPARγ agonists such as Rosiglitazone and 15-deoxy-Δ12,14-prostaglandin J2 inhibit inflammatory cytokine (IL-1β, IL-6, and TNFα), inducible nitric-oxide synthase, and MMP-9 expression in monocytes by a transrepression mechanism, wherein PPAR-retinoic acid receptor heterodimers bind to the transcription factor AP-1, STAT-1, and NF-κB to inhibit pro-inflammatory transcriptional activity (44). The present results may be explained by these events, because OA-NO2 or Rosiglitazone treatment of PMA-induced human THP-1 monocytes suppressed both MMP-9 mRNA expression and protein levels (Fig. 4). Thus, OA-NO2 also modulates MMP-9 expression at the transcriptional level through activation of PPARγ, a signaling event not previously demonstrated for endogenous PPARγ ligands.
Recently, the analysis of atherosclerotic vascular lesions in ApoE−/− mice revealed that in vivo treatment with OA-NO2 significantly attenuated inflammatory cell accumulation within the lesion and decreased adhesion molecule expression (27). These data also suggested that OA-NO2 might improve plaque stability by its ability to increase collagen and α-smooth muscle actin content in plaque material. Because the degradation of matrix proteins and collagen by MMPs have also been associated with plaque instability and rupture (26), the present study extended these observations by showing a significant suppression of MMP-9 expression in atherosclerotic lesions of ApoE−/− mice by OA-NO2 (Fig. 5). The decreased plaque region MMP-9 expression also affirmed in vitro observations in THP-1 cells. Thus, NO2-FA regulation of expression and activation of MMP-9 appears to play a critical role in modulating macrophage infiltration, plaque formation, vessel wall remodeling, and plaque stability. In aggregate, these results reveal a novel attribute of electrophilic fatty acids, decreasing plaque formation and increasing plaque stability.
In summary, conditions favoring the generation of electrophilic fatty acid derivatives can influence MMP activity either via PPARγ-dependent transcriptional regulation or by post-translational modification of pro-MMP. Specifically, the mechanism of pro-MMP activation involves Michael addition of the nitroalkene with a critical nucleophilic cysteine thiol, disruption of the thiolate-Zn2+ ion interaction and the subsequent autocatalytic release of the pro-domain. The functional consequences of these electrophilic lipid signaling actions in the context of inflammation could involve an early MMP activation necessary for remodeling and promotion of cell migration to the site of injury, followed by inhibition of MMP expression in support of the resolution of inflammatory processes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL58115 and R01 HL64937 (to B. A. F.), American Diabetes Association Grant 7-08-JF-52 (to F. J. S.), American Heart Association Grant 0665418U (to F. J. S.), and a grant from the Deutsche Herzstiftung (to V. R.). B. A. F. acknowledges financial interest in Complexa, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- LNO2
- nitro-linoleic acid
- OA-NO2
- nitro-oleic acid
- PPAR
- peroxisome proliferator-activated receptor.
REFERENCES
- 1. Baker P. R., Schopfer F. J., O'Donnell V. B., Freeman B. A. (2009) Free Radic. Biol. Med. 46, 989–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rudolph V., Freeman B. A. (2009) Circ. Res. 105, 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schopfer F. J., Lin Y., Baker P. R., Cui T., Garcia-Barrio M., Zhang J., Chen K., Chen Y. E., Freeman B. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2340–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui T., Schopfer F. J., Zhang J., Chen K., Ichikawa T., Baker P. R., Batthyany C., Chacko B. K., Feng X., Patel R. P., Agarwal A., Freeman B. A., Chen Y. E. (2006) J. Biol. Chem. 281, 35686–35698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ichikawa T., Zhang J., Chen K., Liu Y., Schopfer F. J., Baker P. R., Freeman B. A., Chen Y. E., Cui T. (2008) Endocrinology 149, 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kansanen E., Jyrkkänen H. K., Volger O. L., Leinonen H., Kivelä A. M., Häkkinen S. K., Woodcock S. R., Schopfer F. J., Horrevoets A. J., Ylä-Herttuala S., Freeman B. A., Levonen A. L. (2009) J. Biol. Chem. 284, 33233–33241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Serhan C. N., Chiang N., Van Dyke T. E. (2008) Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Batthyany C., Schopfer F. J., Baker P. R., Durán R., Baker L. M., Huang Y., Cerveñansky C., Branchaud B. P., Freeman B. A. (2006) J. Biol. Chem. 281, 20450–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker L. M., Baker P. R., Golin-Bisello F., Schopfer F. J., Fink M., Woodcock S. R., Branchaud B. P., Radi R., Freeman B. A. (2007) J. Biol. Chem. 282, 31085–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rudolph V., Rudolph T. K., Schopfer F. J., Bonacci G., Woodcock S. R., Cole M. P., Baker P. R., Ramani R., Freeman B. A. (2010) Cardiovasc. Res. 85, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schopfer F. J., Cole M. P., Groeger A. L., Chen C. S., Khoo N. K., Woodcock S. R., Golin-Bisello F., Motanya U. N., Li Y., Zhang J., Garcia-Barrio M. T., Rudolph T. K., Rudolph V., Bonacci G., Baker P. R., Xu H. E., Batthyany C. I., Chen Y. E., Hallis T. M., Freeman B. A. (2010) J. Biol. Chem. 285, 12321–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kansanen E., Bonacci G., Schopfer F. J., Linna S., Tong K. I., Leinonen H., Woodcock S. R., Yamamoto M., Carlberg C., Yla-Herttuala S., Freeman B. A., Levonen A. L. (2011) J. Biol. Chem. 286, 14019–14027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Villacorta L., Zhang J., Garcia-Barrio M. T., Chen X. L., Freeman B. A., Chen Y. E., Cui T. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin D., Saleh S., Liebler D. C. (2008) Chem. Res. Toxicol. 21, 2361–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rudolph T. K., Freeman B. A. (2009) Sci. Signal. 2, re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ra H. J., Parks W. C. (2007) Matrix Biol. 26, 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Wart H. E., Birkedal-Hansen H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemaître V., D'Armiento J. (2006) Birth Defects Res. C Embryo Today 78, 1–10 [DOI] [PubMed] [Google Scholar]
- 19. Fu X., Kassim S. Y., Parks W. C., Heinecke J. W. (2001) J. Biol. Chem. 276, 41279–41287 [DOI] [PubMed] [Google Scholar]
- 20. Okamoto T., Akaike T., Sawa T., Miyamoto Y., van der Vliet A., Maeda H. (2001) J. Biol. Chem. 276, 29596–29602 [DOI] [PubMed] [Google Scholar]
- 21. Gu Z., Kaul M., Yan B., Kridel S. J., Cui J., Strongin A., Smith J. W., Liddington R. C., Lipton S. A. (2002) Science 297, 1186–1190 [DOI] [PubMed] [Google Scholar]
- 22. McCarthy S. M., Bove P. F., Matthews D. E., Akaike T., van der Vliet A. (2008) Biochemistry 47, 5832–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dollery C. M., Libby P. (2006) Cardiovasc. Res. 69, 625–635 [DOI] [PubMed] [Google Scholar]
- 24. Newby A. C. (2007) Trends Cardiovasc. Med. 17, 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson J. L., George S. J., Newby A. C., Jackson C. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15575–15580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newby A. C., George S. J., Ismail Y., Johnson J. L., Sala-Newby G. B., Thomas A. C. (2009) Thromb. Haemost. 101, 1006–1011 [PMC free article] [PubMed] [Google Scholar]
- 27. Rudolph T. K., Rudolph V., Edreira M. M., Cole M. P., Bonacci G., Schopfer F. J., Woodcock S. R., Franek A., Pekarova M., Khoo N. K., Hasty A. H., Baldus S., Freeman B. A. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 938–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferreira A. M., Ferrari M. I., Trostchansky A., Batthyany C., Souza J. M., Alvarez M. N., López G. V., Baker P. R., Schopfer F. J., O'Donnell V., Freeman B. A., Rubbo H. (2009) Biochem. J. 417, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nadtochiy S. M., Baker P. R., Freeman B. A., Brookes P. S. (2009) Cardiovasc. Res. 82, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baker P. R., Lin Y., Schopfer F. J., Woodcock S. R., Groeger A. L., Batthyany C., Sweeney S., Long M. H., Iles K. E., Baker L. M., Branchaud B. P., Chen Y. E., Freeman B. A. (2005) J. Biol. Chem. 280, 42464–42475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baker P. R., Schopfer F. J., Sweeney S., Freeman B. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11577–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manicone A. M., McGuire J. K. (2008) Semin. Cell Dev. Biol. 19, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alexander R. L., Bates D. J., Wright M. W., King S. B., Morrow C. S. (2006) Biochemistry 45, 7889–7896 [DOI] [PubMed] [Google Scholar]
- 34. Rudolph V., Schopfer F. J., Khoo N. K., Rudolph T. K., Cole M. P., Woodcock S. R., Bonacci G., Groeger A. L., Golin-Bisello F., Chen C. S., Baker P. R., Freeman B. A. (2009) J. Biol. Chem. 284, 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jain K., Siddam A., Marathi A., Roy U., Falck J. R., Balazy M. (2008) Free Radic. Biol. Med. 45, 269–283 [DOI] [PubMed] [Google Scholar]
- 36. O'Donnell V. B., Eiserich J. P., Chumley P. H., Jablonsky M. J., Krishna N. R., Kirk M., Barnes S., Darley-Usmar V. M., Freeman B. A. (1999) Chem. Res. Toxicol. 12, 83–92 [DOI] [PubMed] [Google Scholar]
- 37. Casella L., Monzani E., Roncone R., Nicolis S., Sala A., De Riso A. (2002) Environ. Health Perspect. 110, Suppl. 5, 709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gladwin M. T., Kim-Shapiro D. B. (2008) Blood 112, 2636–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deshmukh H. S., Shaver C., Case L. M., Dietsch M., Wesselkamper S. C., Hardie W. D., Korfhagen T. R., Corradi M., Nadel J. A., Borchers M. T., Leikauf G. D. (2008) Am. J. Respir. Cell Mol. Biol. 38, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu X., Kassim S. Y., Parks W. C., Heinecke J. W. (2003) J. Biol. Chem. 278, 28403–28409 [DOI] [PubMed] [Google Scholar]
- 41. Okamoto T., Akaike T., Nagano T., Miyajima S., Suga M., Ando M., Ichimori K., Maeda H. (1997) Arch. Biochem. Biophys. 342, 261–274 [DOI] [PubMed] [Google Scholar]
- 42. Newby A. C. (2006) Cardiovasc. Res. 69, 614–624 [DOI] [PubMed] [Google Scholar]
- 43. Rajagopalan S., Meng X. P., Ramasamy S., Harrison D. G., Galis Z. S. (1996) J. Clin. Invest. 98, 2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ricote M., Li A. C., Willson T. M., Kelly C. J., Glass C. K. (1998) Nature 391, 79–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.