Abstract
Progranulin (GRN) haploinsufficiency is a frequent cause of familial frontotemporal dementia, a currently untreatable progressive neurodegenerative disease. By chemical library screening, we identified suberoylanilide hydroxamic acid (SAHA), a Food and Drug Administration-approved histone deacetylase inhibitor, as an enhancer of GRN expression. SAHA dose-dependently increased GRN mRNA and protein levels in cultured cells and restored near-normal GRN expression in haploinsufficient cells from human subjects. Although elevation of secreted progranulin levels through a post-transcriptional mechanism has recently been reported, this is, to the best of our knowledge, the first report of a small molecule enhancer of progranulin transcription. SAHA has demonstrated therapeutic potential in other neurodegenerative diseases and thus holds promise as a first generation drug for the prevention and treatment of frontotemporal dementia.
Keywords: Drug Action, High-throughput Screening (HTS), Histone Deacetylase, Neurodegeneration, Resveratrol, SAHA, Frontotemporal Dementia, Progranulin
Introduction
Frontotemporal dementia (FTD)3 is a clinical syndrome characterized by progressive deterioration of decision-making abilities, control of behavior, and language, with relative early sparing of memory. It is the second most frequent presenile dementia disorder, and ∼25% of the cases are hereditary (1). The most common pathological manifestation of FTD is frontotemporal lobar degeneration with TDP-43 inclusions, familial cases of which are most frequently caused by loss-of-function mutations of the GRN gene (2–4). The protein encoded by this gene, progranulin (also called GRN protein, human granulin precursor, proepithelin, acrogranin, and PC cell-derived growth factor), is a secreted glycoprotein with growth factor-like and immunomodulatory activities (5). It was recently identified as a TNF receptor antagonist (6). Progranulin contains one half-length and seven full-length granulin domains, which are released following proteolytic cleavage. Biological effects, including promotion of neuronal survival, neurite outgrowth, and regulation of microglial inflammatory responses, have been attributed to both the full-length protein and the granulin peptides (7). To date, >60 pathogenic GRN mutations have been reported in patients with FTD, and all are expected to result in haploinsufficiency. Progranulin-deficient mice display dysregulated immune responses in the brain and recapitulate phosphorylated cytoplasmic TDP-43 aggregates seen in FTD brains (8). Furthermore, the concentration of progranulin in the serum is reported to be lower in patients and mutation carriers compared with healthy controls (9, 10), suggesting that reduced progranulin expression causes FTD. Therefore, increasing progranulin expression from the wild-type allele may prevent or slow down disease progression. Following this rationale, Capell et al. (11) recently reported that alkalizing drugs and vacuolar ATPase inhibitors increase progranulin expression through a post-transcriptional mechanism.
Many drugs in clinical use induce complex changes in gene expression (12). One of the earliest and most successful examples of altering gene expression for therapeutic benefit is the case of hydroxymethylglutaryl-CoA reductase inhibitors, commonly known as statins, which induce expression of the LDL receptor in the liver, thus clearing cholesterol from the blood (13). In addition to changing gene expression through signaling pathways, therapeutics may also act through chromatin remodeling. Thus, the role of epigenetics in the pathogenesis and therapy of neuropsychiatric disorders is an expanding area of research (14). Our objective in this study was to find small molecule enhancers of progranulin transcription by high-throughput screening (HTS) of chemical libraries.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Cell culture reagents and TRIzol® were from Invitrogen. Suberoylanilide hydroxamic acid (SAHA; vorinostat), MS-275, and CAY10591 were obtained from Cayman Chemical. Resveratrol, M344 (4-(dimethylamino)-N-(7-(hydroxyamino)-7-oxoheptyl)benzamide), PTACH, dimethyl sulfoxide (DMSO), sodium butyrate, droxinostat, trichostatin A, leflunomide, and sodium valproate were from Sigma. SRT1720 and MC1568 were from Selleck Chemicals. Tubastatin A was obtained from BioVision Research Products (Mountain View, CA). Tubacin was a gift from Stuart L. Schreiber (funded by the Initiative for Chemical Genetics, NCI). Rabbit antibodies were generated against linker-3 anti-mouse progranulin peptide ((C)VPWMKKVIAPLRLPDPQIL, amino acid residues 353–371) conjugated to keyhole limpet hemocyanin.
Plasmids and Cell Lines
The firefly luciferase coding sequence was fused by bacterial recombination to the authentic human GRN start codon in exon 2 (NM_002087.2) on a bacterial artificial chromosome (BACPAC RP11-812N09), and stably transfected Neuro-2a cells were derived.
Cell Culture and Drug Treatments
Neuro-2a and HEK293 cells were grown in DMEM and 10% FBS. Sodium valproate was dissolved in PBS. All other drugs were dissolved in DMSO (10–50 mm stock solutions kept at −80 °C) and diluted in cell culture medium to a final DMSO concentration of 0.2–0.5%.
Human Cell Lines
All experiments pertaining to collection of human samples were approved by the University of California San Francisco Committee on Human Research. The human subjects and family members were recruited at the University of California San Francisco Memory and Aging Center, and written informed consent was obtained. Genotypes were confirmed by direct sequencing. To obtain human dermal fibroblasts, skin biopsy samples were cut into small pieces, placed under a coverslip, and grown in DMEM containing glutamine, sodium pyruvate, nonessential amino acids, 10% FBS, penicillin, streptomycin, and amphotericin B for ∼3 weeks. Amphotericin B was omitted for further passages. The cells were used at passage 3 or 4.
Immortalized human lymphoblastoid cells were prepared as described (15). Briefly, white blood cells were obtained by Ficoll gradient centrifugation of the Buffy coat from donor blood and transformed in growth medium containing 25% FCS, 1% phytohemagglutinin, and 10% Epstein-Barr virus supernatant. Rapidly growing cultures were maintained in RPMI 1640 medium and 10% FBS.
Library Screening and Luciferase Reporter Assays
Neuro-2a cells were assayed in 384-well plates (3000 cells/well). 6 h after cell plating, 1200 Prestwick Chemical Library® compounds in DMSO, including internal controls, were dispensed using a BioMek FX system to final concentrations of 2.5 μm compound and 1% DMSO (unless indicated otherwise). Sodium butyrate (9 mm) was used as a positive control on each plate for initial screening. Luciferase activity was measured 24 h after compound addition using Bright-GloTM reagent (20 μl/well; Promega). Each well was normalized to the average luminescence from DMSO-treated wells on the same plate.
Determination of Cell Viability
Neuro-2a cells were seeded in 384-well plates (3000 cells/well). After 24 h of drug treatment, the ATP content of each well was measured with the CellTiter-Glo® luminescent cell viability assay (Promega) according to the manufacturer's instructions.
RNA Extraction and Quantitative PCR
Cells in 6-well plates were lysed in 500 μl of TRIzol® reagent/well. cDNA was reverse-transcribed with MultiScribeTM (Applied Biosystems). For some experiments, the Quick-RNA MiniPrep system (Zymo Research, Irvine, CA) was used to isolate total RNA. Primer sequences were as follows: human U36B-F, 5′-CGAGGGCACCTGGAAAAC-3′; human U36B-R, 5′-CACATTCCCCCGGATATGA-3′; human GRN-S, 5′-CAGGGACTTCCAGTTGCTGC-3′; human GRN-A, 5′-GCAGCAGTGATGGCCATCC-3′; mouse cyclophilin QF1S, 5′-GGAGATGGCACAGGAGGAA-3′; mouse cyclophilin QR1A, 5′-GCCCGTAGTGCTTCAGCTT-3′; mouse GRNS, 5′-AGTTCGAATGTCCTGACTCCGCCA-3′; mouse GRNA, 5′-AAGCCACTGCCCTGTTGGTCCTTT-3′; intronic GRN_F1, 5′-CCGGCTACTGTCCAGAGGTCC-3′; and intronic GRN_R1, 5′-CTAGGGGAGTTTCAAGAGGCAGGT-3′.
Quantitative PCRs (qPCRs; 10 μl) contained 20 ng of cDNA, 150 nm primer, and 5 μl of Fast SYBR Green PCR Master Mix (Applied Biosystems) and were performed in triplicate on an Applied Biosystems PRISM 7500 Fast sequence detection system. Relative mRNA levels were calculated using U36B or cyclophilin Q primers as internal controls.
Immunoblotting and Quantification
Cells were lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and Roche Complete protease inhibitor mixture) and cleared by centrifugation at 20,000 × g for 10 min. 0.5-ml cell culture supernatants containing 1% FBS were concentrated by centrifugation at 14,000 × g for 50 min in Millipore Amicon Ultra devices (3-kDa cutoff). 15–20 μg of total protein was separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% milk for 1 h and probed overnight with primary antibodies at 4 °C. Mouse progranulin was detected with linker-3 anti-progranulin antibody at 1:5000 dilution, β-actin with anti-β-actin antibody (Sigma A2228) at 1:5000 dilution, human progranulin with anti-PC cell-derived growth factor antibody (Invitrogen) at 1:1000 dilution, and GAPDH with anti-GAPDH antibody (Sigma) at 1:10,000 dilution. Bound IgG was detected by ECL. For quantitative immunoblotting, secondary antibodies labeled with IRDye® infrared dyes and an Odyssey® infrared imager (LI-COR Biosciences) were used following the manufacturer's instructions.
Statistical Analysis
Statistical analysis was done with SigmaPlot 11 software. For comparison between multiple treatment groups, analysis of variance was followed by Dunnett's or Tukey's test. All data are presented as means ± S.E.
RESULTS
Identification of Potential Enhancers of Progranulin Expression by Chemical Library Screening
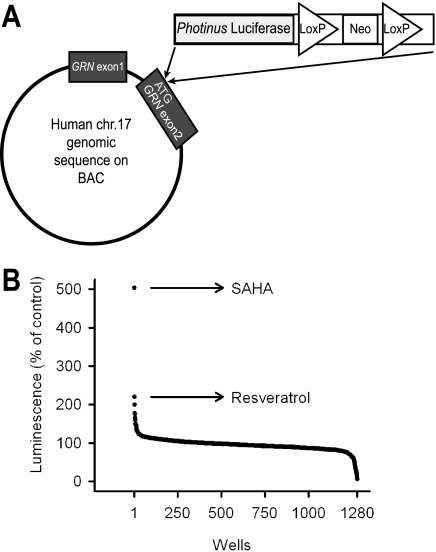
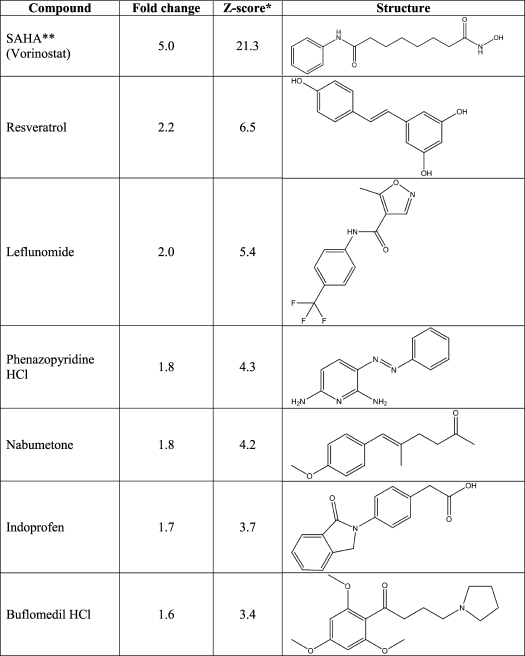
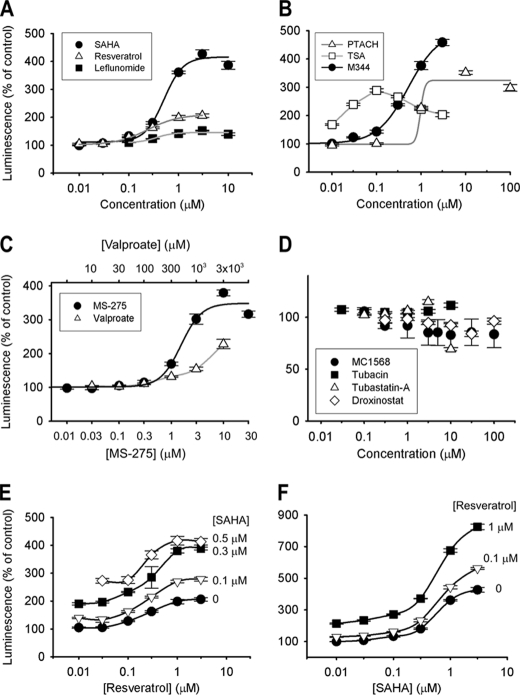
First, we sought to identify small molecule enhancers of GRN expression by HTS. To generate a reporter of GRN promoter activity, a luciferase reporter was inserted by homologous recombination into the human GRN gene on a bacterial artificial chromosome (Fig. 1A). Utilizing cotransfection with a plasmid encoding Zeocin resistance, we stably integrated this construct into the neuronally derived mouse cell line Neuro-2a. Several independent clones robustly and stably expressed luciferase from the GRN promoter/enhancer under base-line conditions (data not shown). Next, we sought to validate our automated 384-well HTS strategy using sodium butyrate, a histone deacetylase (HDAC) inhibitor, as a positive control. The coefficients of variation were 5.3% for treatment wells and 8% for DMSO control wells, whereas the Z′ value was 0.68, indicating a favorable signal/noise ratio. Utilizing this HTS assay, we screened the Prestwick Chemical Library, comprising 1200 marketed drugs, and identified multiple compounds that increased GRN promoter activity by >3 standard deviations above the mean, as summarized in Fig. 1B and Table 1. >98% of the compounds in this chemical library had no significant effect on luciferase activity, indicating high specificity of the screening assay. The highest levels of activation resulted from addition of SAHA (a class I and II HDAC inhibitor), resveratrol (a putative class III HDAC activator), and leflunomide (a pyrimidine synthesis inhibitor). We established the dose-response curves for these three compounds, as shown in Fig. 2A. SAHA and resveratrol dose-dependently increased luciferase activity in the reporter assay with submicromolar EC50 values of 0.51 μm for SAHA and 0.24 μm for resveratrol. Leflunomide had minimal activity and was not pursued further.
FIGURE 1.
Luciferase reporter-based HTS for small molecule enhancers of progranulin expression. A, schematic representation of a bacterial artificial chromosome (BAC)-based reporter construct. Luciferase coding sequence was inserted into exon 2 of the human GRN gene, replacing the original start codon with the first codon of luciferase cDNA. chr.17, chromosome 17. B, results from screening 1200 compounds composing the Prestwick Chemical Library®. Neuro-2a cells stably expressing the reporter were treated with the compounds at 2.5 μm for 24 h, and luciferase activity was measured afterward. Arrows indicate the compounds that resulted in the top two highest activities.
TABLE 1.
Activating compounds identified by screening the Prestwick Chemical Library
*, Z score = (compound normalized activity − average normalized activity)/population S.D.; **, SAHA, suberoylanilide hydroxamic acid.
FIGURE 2.
HDAC inhibitors and resveratrol dose-dependently increase luciferase reporter activity. The Neuro-2a luciferase reporter cell line used for screening was also utilized in follow-up experiments. A, dose-response relationships for HTS hits SAHA (●), resveratrol (△), and leflunomide (■). EC50 values were 0.51 μm for SAHA and 0.24 μm for resveratrol. B, dose-response relationships for HDAC inhibitors M344 (●), PTACH (△), and trichostatin A (TSA; □). C, dose-response relationships for class I specific HDAC inhibitors MS-275 (●) and sodium valproate (△). D, selective HDAC inhibitors MC1568 (class II selective; ●), tubacin (HDAC6-specific; ■), tubastatin A (HDAC6-specific; △), and droxinostat (HDAC3-, HDAC6-, and HDAC8-selective; ♢) did not increase GRN promoter reporter activity. E, SAHA and resveratrol have additive effects. Resveratrol concentrations tested are shown on the x axis. SAHA concentrations tested are noted next to the dose-response curves. ●, resveratrol alone; △, resveratrol + 0.1 μm SAHA; ■, resveratrol + 0.3 μm SAHA; ♢, resveratrol + 0.5 μm SAHA. F, SAHA and resveratrol have additive effects. SAHA concentrations tested are shown on the x axis. Resveratrol concentrations tested are noted next to the dose-response curves. ●, SAHA alone; △, SAHA + 0.1 μm resveratrol; ■, SAHA + 1 μm resveratrol.
HDAC Inhibitors Trichostatin A, M344, and PTACH Are Active in the Luciferase-based Progranulin Promoter Activity Assay
Because SAHA is a known HDAC inhibitor, we tested whether other HDAC inhibitors would have the same effect in our reporter assay. The pan-HDAC inhibitors trichostatin A, PTACH, and M344 significantly increased luciferase activity (Fig. 2B), although the effect of trichostatin A was limited by toxicity at concentrations ≥0.1 μm.
Selective HDAC Inhibition Is Not Sufficient to Enhance Progranulin Promoter Activity
We tested whether selective inhibitors of class I HDACs would be effective in the luciferase reporter assay. Valproate and MS-275 enhanced luciferase activity only at the highest end of the pharmacological concentrations (Fig. 2C). This effect further supports our hypothesis that HDAC inhibition is the mechanism of action responsible for enhanced progranulin expression. Because these compounds could inhibit HDACs less selectively at these concentrations, we cannot conclude that class I HDAC inhibition is sufficient for enhanced expression.
We also tested M344, a putatively selective inhibitor of HDAC6, and observed dose-dependent activation of the luciferase reporter at submicromolar concentrations. The EC50 value (0.52 μm) and maximal effect size (∼4-fold) of M344 were similar to the values observed for SAHA (Fig. 2B). However, two other HDAC6-specific inhibitors, tubacin (16) and Tubastatin A (17), did not change luciferase activity (Fig. 2D). M344, although 3-fold selective for HDAC6 over HDAC1, nevertheless inhibits HDAC1 with an IC50 of 250 nm (18). Therefore, we conclude that HDAC6 inhibition is not sufficient to enhance progranulin reporter activity. We then tested droxinostat (a selective inhibitor of HDAC3, HDAC6, and HDAC8) and MC1568 (a class II selective HDAC inhibitor), but we did not observe any effects on luciferase reporter activity (Fig. 2D).
Because activation of sirtuins is one of the supposed biological effects of resveratrol, we also tested SRT1720 (0.01–1 μm) and CAY10591 (0.5 or 5 μm), other putative activators of sirtuins. These compounds had no effect on luciferase activity in the progranulin reporter assay (data not shown), suggesting that resveratrol may be acting through a different pathway.
SAHA and Resveratrol Have Additive Effects in the Luciferase-based Progranulin Promoter Activity Assay
We next tested the interaction between these two compounds utilizing the reporter assay. As shown in Fig. 2 (E and F), we observed an upward shift in the dose-response curves when a fixed submaximal concentration of either drug was tested in combination with a range of different concentrations of the other. The maximal effect size was ∼4-fold with SAHA alone, ∼2-fold with resveratrol alone, and ∼8-fold with a combination of the maximal effective concentrations of both (Fig. 2F).
SAHA Moderately Inhibits Cell Proliferation
To assess the toxicity of SAHA, resveratrol, and trichostatin A, we treated Neuro-2a cells with these compounds in 384-well plates and measured cell titers 24 h later with a luciferase-based assay that measures ATP content of the wells. SAHA at concentrations ≥0.3 μm reduced cell viability by ∼25% (Fig. 3), consistent with previously reported effects (19).
FIGURE 3.
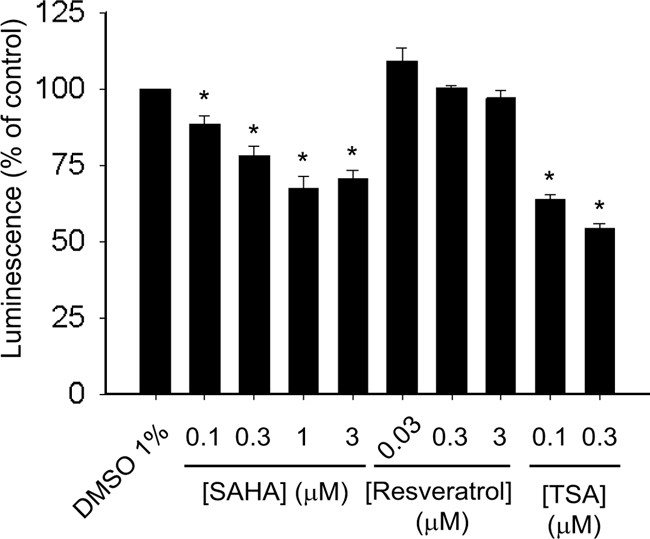
HDAC inhibitors moderately decrease cell viability. Neuro-2a cells were treated with the indicated drugs for 24 h, and cell titers were measured with a luciferase-based assay. Lower luminescence values correspond to lower ATP concentrations in the well. Error bars indicate S.E. *, p < 0.05 versus the vehicle control. TSA, trichostatin A.
SAHA Increases Progranulin mRNA and Protein Levels in Neuro-2a Cells
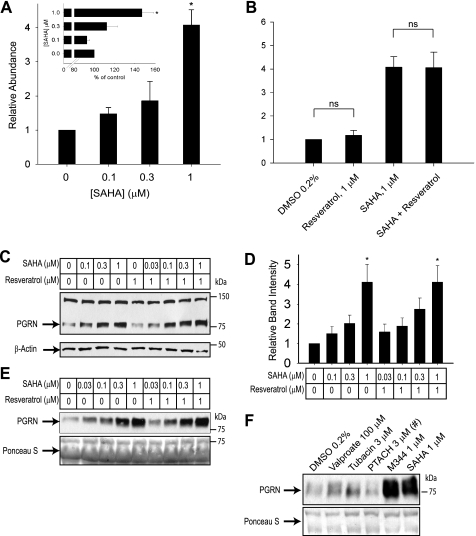
To test whether SAHA and resveratrol augment transcription from the endogenous GRN promoter, we treated Neuro-2a cells with these drugs for 24 h and performed qPCR to measure the relative levels of progranulin mRNA. 1 μm SAHA significantly increased the relative abundance of GRN mRNA (Fig. 4A). We repeated this experiment with primers designed to amplify GRN pre-mRNA but not mature mRNA and saw the same trend we observed using regular qPCR primers (Fig. 4A, inset). This suggests that the increase in GRN mRNA is at least partially due to increased transcription.
FIGURE 4.
SAHA enhances progranulin expression in cultured cells. A and B, qPCR analysis measured the relative abundance of GRN mRNA in Neuro-2a cells treated with SAHA or resveratrol for 24 h. 1 μm SAHA significantly increased GRN expression, whereas 1 μm resveratrol did not have any effect either alone or in combination with 1 μm SAHA. In the inset in A, the same samples were analyzed using qPCR primers designed to detect pre-mRNA. *, p < 0.05; ns, p > 0.05 versus the vehicle control or for the indicated comparisons. C–F, Neuro-2a whole cell lysates (C; quantified in D) and cell culture supernatants (E and F) were analyzed for progranulin (PGRN) protein expression by immunoblotting after a 24-h application of indicated drugs. #, significant toxicity was observed with the indicated treatment.
In contrast to the luciferase reporter assay, we did not observe increased GRN mRNA expression with resveratrol treatment either alone or in combination with SAHA (Fig. 4B). It is possible that resveratrol may have direct effects on luciferase enzyme stability or activity in the reporter assay. Alternatively, the reporter may have been artificially inserted near a regulatory element responsive to resveratrol. These results emphasize the general importance of secondary validation of HTS hits.
We next tested whether increased GRN mRNA expression translates into increased protein levels. We analyzed total cell lysates from Neuro-2a cells treated with SAHA, resveratrol, or a combination of both by Western blotting. After 24 h of treatment with SAHA alone or with resveratrol, the intensity of the progranulin immunoreactive band was dose-dependently increased compared with the loading control (Fig. 4, C and D). In line with our qPCR results, the effect of resveratrol was muted. Progranulin is a secreted protein; therefore, we also tested whether increased GRN expression translates into increased secretion. For this experiment, we treated Neuro-2a cells as indicated in Fig. 4E, collected and concentrated the conditioned medium, and analyzed it by Western blotting. Treatment with increasing concentrations of SAHA resulted in more intense progranulin reactive bands, replicating our results from our qPCR and total cell lysate Western blot analyses.
We tested whether the effects we observed in the luciferase reporter assay with different HDAC inhibitors would correlate with increased progranulin levels. As shown in Fig. 4F, M344, but not PTACH, robustly increased secreted progranulin levels. At the concentrations required to increase transcription from the GRN promoter, PTACH reduced cell viability by ∼20% and exhibited overt toxicity on the remaining cells (data not shown). This pronounced toxicity readily accounts for the reduced accumulation of progranulin in the medium.
SAHA Treatment Can Normalize Progranulin mRNA Levels in GRN+/− Human Cells
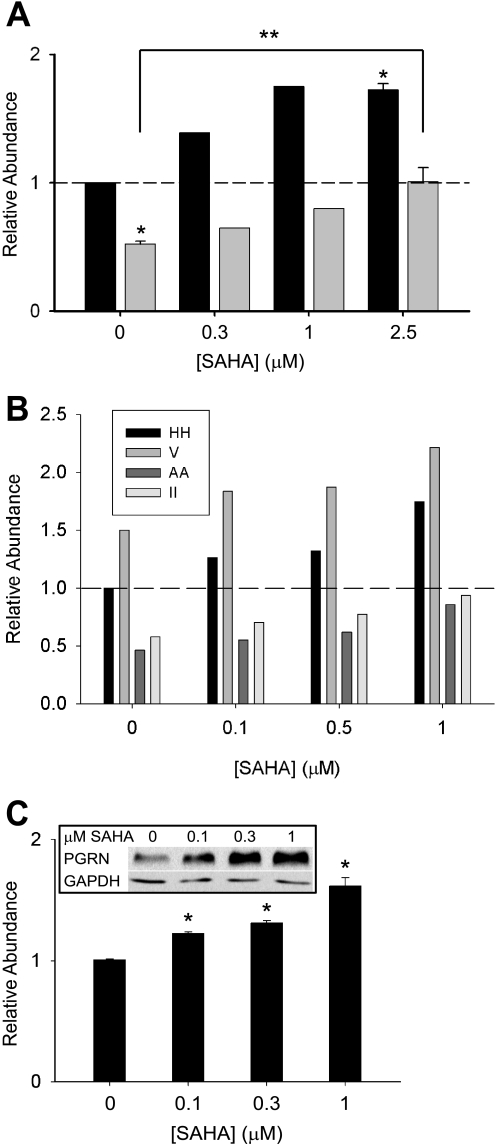
Because GRN null mutations lead to familial FTD, probably through GRN haploinsufficiency, we next tested whether SAHA could correct this progranulin deficit in human cells that contain only one wild-type allele of the GRN gene. For the experiment summarized in Fig. 5A, we used Epstein-Barr virus-immortalized human lymphoblastoid cells derived from a subject with a GRN null mutation (R493X) and similarly prepared cells from a wild-type relative. GRN mRNA expression was reduced by ∼50% in heterozygous lymphoblasts compared with control cells. Treatment with SAHA dose-dependently increased GRN mRNA levels in both haploinsufficient and control lymphoblasts (Fig. 5A). After 24 h of treatment with 2.5 μm SAHA, the relative abundance of GRN mRNA in haploinsufficient cells was normalized to wild-type levels.
FIGURE 5.
SAHA enhances progranulin expression in haploinsufficient human cells. A, Epstein-Barr virus-immortalized human lymphoblastoid cells from a subject with a nonsense GRN R493X mutation (gray bars) and from a relative carrying wild-type alleles (black bars) were treated with SAHA for 24 h, and the relative abundance of GRN mRNA was measured by qPCR (n = 2 for 0.3 and 1 μm SAHA; n = 3 for control and 2.5 μm SAHA). *, p < 0.05 versus the GRN+/+ control; **, p < 0.05 for the indicated comparison. The dashed line indicates the relative abundance of 1. B, human dermal fibroblasts from two subjects heterozygous for GRN nonsense mutations (AA and II) and from two control subjects (HH and V) were treated as indicated, and the relative abundance of GRN mRNA was measured by qPCR. All samples were processed in parallel, and ΔΔCt values were normalized to one of the control samples (HH). The dashed line indicates the relative abundance of 1. C, cumulative data from the four cell lines shown in B plus an independent human dermal fibroblast line with a heterozygous R493X mutation is shown (n = 6 for 0 and 1 μm SAHA; n = 4 for 0.1 and 0.3 μm SAHA). *, p < 0.05 versus the vehicle control. The inset shows Western blot analysis of total cell lysates from HEK293 cells treated with the indicated concentrations of SAHA for 24 h. PGRN, progranulin.
We also obtained human dermal fibroblasts from two subjects with nonsense or frameshift GRN mutations (Q300X (II) and S226WfsX28 (AA)) and their unaffected siblings. We treated these fibroblasts with SAHA or vehicle and measured the relative abundance of GRN mRNA 24 h later. Four cell lines were treated in parallel, and as shown in Fig. 5B, the heterozygous cell lines had a relative insufficiency of GRN. SAHA dose-dependently increased GRN mRNA in all cell lines, nearly normalizing GRN levels in the heterozygous cells at a concentration of 1 μm. Cumulative data from all four cell lines plus another human dermal fibroblast line from a subject with an R493X mutation treated with only vehicle or 1 μm SAHA are shown in Fig. 5C. Statistically significant increases in GRN mRNA were observed at all tested concentration (0.1–1 μm). The qPCR results from primary human cells were replicated at the protein level. Because basal expression of progranulin in the primary cells was too low to obtain consistent results with immunoblotting, we treated human HEK293 with SAHA. As shown in Fig. 5C (inset), progranulin protein expression was robustly increased by the drug.
DISCUSSION
FTD is a devastating and often fatal disease. Median survival after diagnosis is <10 years (20). Current treatment options are limited to management of emotional and behavioral aspects with antidepressants and social interventions (21). Identification of drug targets that can be exploited to slow down or reverse the cognitive decline will probably depend on a better understanding of the molecular pathogenesis. Familial forms of the disease with known pathogenic mutations provide an opportunity for fast-track development of new therapeutics for FTD.
Because loss-of-function mutations in one GRN allele cause familial and sporadic FTD, we undertook an HTS approach to identify small molecule enhancers of progranulin expression. We chose the Prestwick library, comprising 1200 marketed drugs, for our initial screening, assuming that the identification of a Food and Drug Administration-approved compound would accelerate the search for a cure for this disease. Indeed, we discovered that SAHA, an HDAC inhibitor currently in clinical use for cutaneous T-cell lymphoma (22), enhances progranulin expression in a variety of relevant cell types in culture.
Physiological regulation of progranulin expression is largely unknown. Two recent studies have identified microRNAs miR-107 (23) and miR-29b (24) as negative regulators of GRN expression. Also, a common variant in a putative miR-659-binding site of GRN was identified as a risk factor for FTD (25). Previously, Bhandari et al. (26) analyzed the promoter region of GRN and identified various possible binding sites for transcription factors. Interestingly, two of the most prominent of these, GATA1 and SP1, have been reported to interact with HDACs (27, 28), with SP1 also being involved in the same regulatory network with miR-29b.
Recently, Capell et al. (11) reported that an inhibitor of vacuolar ATPase, bafilomycin A1, and the alkalizing drugs chloroquine, bepridil, and amiodarone increased secreted progranulin levels from haploinsufficient primary human lymphoblasts, although only bafilomycin A1 increased the protein to near-normal levels. Bafilomycin exerts its effect through a post-transcriptional mechanism, suggesting a potential synergistic application with SAHA for FTD prevention depending on toxicity and tolerated dose.
HDACs regulate the acetylation status of cellular proteins, which is emerging as an important post-translational modification in cell regulation (29). Small molecule inhibitors of HDACs are being developed as drugs for cancer and neurological disorders, such as Rubinstein-Taybi syndrome, Friedreich's ataxia, and fragile X syndrome (30). Several compounds are now in clinical trials for cancer therapy, and SAHA has been approved by the Food and Drug Administration for treatment of cutaneous T-cell lymphoma. SAHA has been investigated in a very similar paradigm for the treatment of motor neuron disease, and it was shown to enhance the expression of SMN2 in brain slices, potentially substituting for the loss of SMN1 expression, the gene mutated in spinal muscular atrophy (31). SAHA crosses the blood-brain barrier, and treatment with SAHA was shown to improve rotarod performance in a mouse model of Huntington disease (32). Furthermore, inhibition of HDAC2 by SAHA was reported to enhance memory formation in mice (33). HDAC inhibitors also have anti-inflammatory properties (34–36). Interestingly, changes in inflammatory markers induced by SAHA treatment in a rodent septic shock model (37) were similar to the effects of recombinant progranulin and a progranulin-mimetic peptide in a rodent inflammatory arthritis model (6). These findings suggest two possibilities: first, that increasing the expression of progranulin, a protein with anti-inflammatory growth-promoting activities, may underlie some of the therapeutic effects observed in other conditions, perhaps by antagonizing TNF signaling; and second or alternatively, SAHA may have secondary beneficial effects in FTD besides normalizing progranulin deficiency.
We were unable to elucidate in the last detail the mechanism and specific contribution of individual HDACs to the increased GRN expression resulting from SAHA treatment. Chemically similar (trichostatin A) and structurally unrelated (M344) HDAC inhibitors, as well as sodium butyrate, also increased GRN expression, supporting our conclusion that HDAC inhibition is critical to the underlying mechanism, although we cannot completely rule out off-target effects. HDAC inhibitors have global effects on gene expression. Although SAHA is Food and Drug Administration-approved, the targets responsible for its anticancer effects are also incompletely understood (38).
Which HDAC isoform might be responsible for the effectiveness of SAHA? HDACs are classified into five classes (I, IIa, IIb, III, and IV) (30). SAHA inhibits class I (HDAC1–3 and HDAC8), class IIa (HDAC4, HDAC5, HDAC7, and HDAC9), and class IIb (HDAC6 and HDAC10) enzymes. In the luciferase reporter assay, HDAC6 or class II specific inhibitors were not sufficient to enhance expression, whereas class I inhibitors MS-275 and valproate increased expression only at excessively high concentrations at which cross-inhibition of other HDACs may have occurred. Although we cannot rule out the possibility that class I HDAC inhibition might be sufficient, we interpret our data as supporting a complex mechanism requiring inhibition of several HDACs. Further experiments utilizing RNA interference could potentially answer this question. However, our pharmacological data suggest that knockdown of a single HDAC isoform would be unlikely to account for increased progranulin expression.
The second most efficacious hit in our screen was resveratrol, a plant polyphenol currently marketed as a dietary supplement. Resveratrol is implicated in neuroprotection against diverse types of injury (39–41). Although resveratrol has been suggested to be an activator of sirtuins, this has been called into question recently (42), and the mechanism of action remains poorly understood (43, 44). We confirmed that resveratrol was effective in the luciferase-based assay; however, we did not observe a significant increase in progranulin mRNA or protein levels by qPCR or Western blotting after treatment with resveratrol in any cell type. There can be several explanations for this discrepancy. First, resveratrol may have direct effects on the luciferase enzyme stability or activity, thus representing a false positive hit. Given the small number of hits from our library screen, we chose not to do a counterscreen for luciferase activation from a different promoter, opting instead to validate hand-picked hits directly as we show here for SAHA. Second, resveratrol is reported to have diverse effects on gene expression and chromatin remodeling (45, 46); therefore, the actual integration site of the reporter in the genomic DNA of the stable cell line may have resulted in an artifactual result. Last, albeit unlikely, the sensitivity of qPCR and Western blotting may not be high enough to detect <2-fold changes in progranulin levels under our experimental conditions.
In summary, we have demonstrated that one of the hits in our chemical library screen, SAHA, robustly enhances GRN expression at both the mRNA and protein levels in cells from relevant mouse or human lineages. Because SAHA is already in clinical use for an unrelated indication, conducting human trials with SAHA as a therapy or prevention strategy for FTD is facilitated. Our results also validate the present HTS strategy for performing screens of larger and more diverse chemical libraries to identify additional drug leads amenable to optimization and clinical development.
Acknowledgments
We thank Haydn Ball (Protein Chemistry Technology Center, University of Texas Southwestern Medical Center at Dallas), Lauren Herl, Richard Gibson (deceased), Michael Geschwind, Anna Karydas, and Laura Mitic for help with peptide synthesis, fibroblast isolation, cell culture, antibody production, and testing. We are indebted to Stuart L. Schreiber and the support of the NCI (to S. L. S.) for the generous gift of tubacin. We also thank the staff of the HTS facility at the Simmons Cancer Center for technical help and acknowledge the assistance of the Southwestern Small Animal Imaging Resource (supported in part by the National Institutes of Health).
This work was supported by the National Institutes of Health, the Consortium for Frontotemporal Dementia Research, the California Institute for Regenerative Medicine, the National Cell Repository for Alzheimer's Disease, the Ted Nash Long Life Foundation, the Welch Foundation, the Humboldt Foundation, and the American Health Assistance Foundation.
- FTD
- frontotemporal dementia
- HTS
- high-throughput screening
- SAHA
- suberoylanilide hydroxamic acid
- DMSO
- dimethyl sulfoxide
- qPCR
- quantitative PCR
- HDAC
- histone deacetylase.
REFERENCES
- 1. van Swieten J. C., Heutink P. (2008) Lancet Neurol. 7, 965–974 [DOI] [PubMed] [Google Scholar]
- 2. Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J. J., van Duijn C., Peeters K., Sciot R., Santens P., De Pooter T., Mattheijssens M., Van den Broeck M., Cuijt I., Vennekens K., De Deyn P. P., Kumar-Singh S., Van Broeckhoven C. (2006) Nature 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 3. Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A. D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C. A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., Hutton M. (2006) Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 4. Gass J., Cannon A., Mackenzie I. R., Boeve B., Baker M., Adamson J., Crook R., Melquist S., Kuntz K., Petersen R., Josephs K., Pickering-Brown S. M., Graff-Radford N., Uitti R., Dickson D., Wszolek Z., Gonzalez J., Beach T. G., Bigio E., Johnson N., Weintraub S., Mesulam M., White C. L., 3rd, Woodruff B., Caselli R., Hsiung G. Y., Feldman H., Knopman D., Hutton M., Rademakers R. (2006) Hum. Mol. Genet. 15, 2988–3001 [DOI] [PubMed] [Google Scholar]
- 5. Ahmed Z., Mackenzie I. R., Hutton M. L., Dickson D. W. (2007) J. Neuroinflammation 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tang W., Lu Y., Tian Q. Y., Zhang Y., Guo F. J., Liu G. Y., Syed N. M., Lai Y., Lin E. A., Kong L., Su J., Yin F., Ding A. H., Zanin-Zhorov A., Dustin M. L., Tao J., Craft J., Yin Z., Feng J. Q., Abramson S. B., Yu X. P., Liu C. J. (2011) Science, in press [Google Scholar]
- 7. Eriksen J. L., Mackenzie I. R. (2008) J. Neurochem. 104, 287–297 [DOI] [PubMed] [Google Scholar]
- 8. Yin F., Banerjee R., Thomas B., Zhou P., Qian L., Jia T., Ma X., Ma Y., Iadecola C., Beal M. F., Nathan C., Ding A. (2010) J. Exp. Med. 207, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sleegers K., Brouwers N., Van Damme P., Engelborghs S., Gijselinck I., van der Zee J., Peeters K., Mattheijssens M., Cruts M., Vandenberghe R., De Deyn P. P., Robberecht W., Van Broeckhoven C. (2009) Ann. Neurol. 65, 603–609 [DOI] [PubMed] [Google Scholar]
- 10. Finch N., Baker M., Crook R., Swanson K., Kuntz K., Surtees R., Bisceglio G., Rovelet-Lecrux A., Boeve B., Petersen R. C., Dickson D. W., Younkin S. G., Deramecourt V., Crook J., Graff-Radford N. R., Rademakers R. (2009) Brain 132, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Capell A., Liebscher S., Fellerer K., Brouwers N., Willem M., Lammich S., Gijselinck I., Bittner T., Carlson A. M., Sasse F., Kunze B., Steinmetz H., Jansen R., Dormann D., Sleegers K., Cruts M., Herms J., Van Broeckhoven C., Haass C. (2011) J. Neurosci. 31, 1885–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iorio F., Bosotti R., Scacheri E., Belcastro V., Mithbaokar P., Ferriero R., Murino L., Tagliaferri R., Brunetti-Pierri N., Isacchi A., di Bernardo D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14621–14626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma P. T., Gil G., Südhof T. C., Bilheimer D. W., Goldstein J. L., Brown M. S. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8370–8374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Narayan P., Dragunow M. (2010) Br. J. Pharmacol. 159, 285–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neitzel H. (1986) Hum. Genet. 73, 320–326 [DOI] [PubMed] [Google Scholar]
- 16. Haggarty S. J., Koeller K. M., Wong J. C., Grozinger C. M., Schreiber S. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butler K. V., Kalin J., Brochier C., Vistoli G., Langley B., Kozikowski A. P. (2010) J. Am. Chem. Soc. 132, 10842–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heltweg B., Dequiedt F., Marshall B. L., Brauch C., Yoshida M., Nishino N., Verdin E., Jung M. (2004) J. Med. Chem. 47, 5235–5243 [DOI] [PubMed] [Google Scholar]
- 19. Yin D., Ong J. M., Hu J., Desmond J. C., Kawamata N., Konda B. M., Black K. L., Koeffler H. P. (2007) Clin. Cancer Res. 13, 1045–1052 [DOI] [PubMed] [Google Scholar]
- 20. Hodges J. R., Davies R., Xuereb J., Kril J., Halliday G. (2003) Neurology 61, 349–354 [DOI] [PubMed] [Google Scholar]
- 21. Kirshner H. S. (2010) Curr. Neurol. Neurosci Rep. 10, 504–511 [DOI] [PubMed] [Google Scholar]
- 22. Mann B. S., Johnson J. R., Cohen M. H., Justice R., Pazdur R. (2007) Oncologist 12, 1247–1252 [DOI] [PubMed] [Google Scholar]
- 23. Wang W. X., Wilfred B. R., Madathil S. K., Tang G., Hu Y., Dimayuga J., Stromberg A. J., Huang Q., Saatman K. E., Nelson P. T. (2010) Am. J. Pathol. 177, 334–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiao J., Herl L. D., Farese R. V., Gao F. B. (2010) PLoS ONE 5, e10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rademakers R., Eriksen J. L., Baker M., Robinson T., Ahmed Z., Lincoln S. J., Finch N., Rutherford N. J., Crook R. J., Josephs K. A., Boeve B. F., Knopman D. S., Petersen R. C., Parisi J. E., Caselli R. J., Wszolek Z. K., Uitti R. J., Feldman H., Hutton M. L., Mackenzie I. R., Graff-Radford N. R., Dickson D. W. (2008) Hum. Mol. Genet. 17, 3631–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhandari V., Daniel R., Lim P. S., Bateman A. (1996) Biochem. J. 319, 441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watamoto K., Towatari M., Ozawa Y., Miyata Y., Okamoto M., Abe A., Naoe T., Saito H. (2003) Oncogene 22, 9176–9184 [DOI] [PubMed] [Google Scholar]
- 28. Liu S., Wu L. C., Pang J., Santhanam R., Schwind S., Wu Y. Z., Hickey C. J., Yu J., Becker H., Maharry K., Radmacher M. D., Li C., Whitman S. P., Mishra A., Stauffer N., Eiring A. M., Briesewitz R., Baiocchi R. A., Chan K. K., Paschka P., Caligiuri M. A., Byrd J. C., Croce C. M., Bloomfield C. D., Perrotti D., Garzon R., Marcucci G. (2010) Cancer Cell 17, 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 30. Kazantsev A. G., Thompson L. M. (2008) Nat. Rev. Drug Discov. 7, 854–868 [DOI] [PubMed] [Google Scholar]
- 31. Hahnen E., Eyüpoglu I. Y., Brichta L., Haastert K., Tränkle C., Siebzehnrübl F. A., Riessland M., Hölker I., Claus P., Romstöck J., Buslei R., Wirth B., Blümcke I. (2006) J. Neurochem. 98, 193–202 [DOI] [PubMed] [Google Scholar]
- 32. Hockly E., Richon V. M., Woodman B., Smith D. L., Zhou X., Rosa E., Sathasivam K., Ghazi-Noori S., Mahal A., Lowden P. A., Steffan J. S., Marsh J. L., Thompson L. M., Lewis C. M., Marks P. A., Bates G. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2041–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guan J. S., Haggarty S. J., Giacometti E., Dannenberg J. H., Joseph N., Gao J., Nieland T. J., Zhou Y., Wang X., Mazitschek R., Bradner J. E., DePinho R. A., Jaenisch R., Tsai L. H. (2009) Nature 459, 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin H. S., Hu C. Y., Chan H. Y., Liew Y. Y., Huang H. P., Lepescheux L., Bastianelli E., Baron R., Rawadi G., Clément-Lacroix P. (2007) Br. J. Pharmacol. 150, 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glauben R., Batra A., Fedke I., Zeitz M., Lehr H. A., Leoni F., Mascagni P., Fantuzzi G., Dinarello C. A., Siegmund B. (2006) J. Immunol. 176, 5015–5022 [DOI] [PubMed] [Google Scholar]
- 36. Halili M. A., Andrews M. R., Labzin L. I., Schroder K., Matthias G., Cao C., Lovelace E., Reid R. C., Le G. T., Hume D. A., Irvine K. M., Matthias P., Fairlie D. P., Sweet M. J. (2010) J. Leukocyte Biol. 87, 1103–1114 [DOI] [PubMed] [Google Scholar]
- 37. Finkelstein R. A., Li Y., Liu B., Shuja F., Fukudome E., Velmahos G. C., deMoya M., Alam H. B. (2010) J. Surg Res. 163, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lane A. A., Chabner B. A. (2009) J. Clin. Oncol. 27, 5459–5468 [DOI] [PubMed] [Google Scholar]
- 39. Araki T., Sasaki Y., Milbrandt J. (2004) Science 305, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 40. Vingtdeux V., Dreses-Werringloer U., Zhao H., Davies P., Marambaud P. (2008) BMC Neurosci. 9, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang F., Liu J., Shi J. S. (2010) Eur. J. Pharmacol. 636, 1–7 [DOI] [PubMed] [Google Scholar]
- 42. Pacholec M., Bleasdale J. E., Chrunyk B., Cunningham D., Flynn D., Garofalo R. S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., Withka J., Ahn K. (2010) J. Biol. Chem. 285, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Calamini B., Ratia K., Malkowski M. G., Cuendet M., Pezzuto J. M., Santarsiero B. D., Mesecar A. D. (2010) Biochem. J. 429, 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang B. L. (2010) Brain Res. Bull. 81, 359–361 [DOI] [PubMed] [Google Scholar]
- 45. Kai L., Samuel S. K., Levenson A. S. (2010) Int. J. Cancer 126, 1538–1548 [DOI] [PubMed] [Google Scholar]
- 46. Yang J., Kong X., Martins-Santos M. E., Aleman G., Chaco E., Liu G. E., Wu S. Y., Samols D., Hakimi P., Chiang C. M., Hanson R. W. (2009) J. Biol. Chem. 284, 27042–27053 [DOI] [PMC free article] [PubMed] [Google Scholar]