Abstract
Musashi1 (Msi1) is an RNA-binding protein that is highly expressed in neural stem/progenitor cells (NS/PCs) as well as in other tissue stem cells. Msi1 binds to the 3′-UTR of its target mRNAs in NS/PCs, prevents their translation, and interferes with NS/PC differentiation. We previously showed that Msi1 competes with eIF4G to bind poly(A)-binding protein and inhibits assembly of the 80 S ribosome. Here we show that Msi1 works in concert with Lin28 to regulate post-transcriptional microRNA (miRNA) biogenesis in the cropping step, which occurs in the nucleus. Lin28 and its binding partner terminal uridylyltransferase 4 (TUT4) are known to maintain embryonic stem cell pluripotency by blocking let-7 miRNA biogenesis at the dicing step. Interestingly, we found that during early neural differentiation of embryonic stem cells, Msi1 enhanced the localization of Lin28 to the nucleus and also inhibited the nuclear cropping step of another let-7 family miRNA, miR98. These results suggest that Msi1 can influence stem cell maintenance and differentiation by controlling the subcellular localization of proteins involved in miRNA biogenesis, as well as by regulating the translation of its target mRNA.
Keywords: Embryonic Stem Cell, MicroRNA, Neural Stem Cell, Ribonuclear Protein (RNP), RNA, Importin, miRNA Biogenesis, Musashi1, Post-transcriptional Regulation
Introduction
MicroRNAs (miRNAs),2 which are small non-coding RNAs, control gene expression through sequence-specific interactions with their target mRNAs. miRNAs are involved in various biological processes (1, 2). miRNAs are first transcribed by RNA polymerase II as primary miRNA transcripts (pri-miRNAs), whose secondary structures form stem and terminal loop (3). pri-miRNAs are cleaved in the “cropping step” by Microprocessor, which contains the RNase III enzyme Drosha and the double-stranded RNA-binding protein DGCR8. The released precursor miRNAs (pre-miRNAs) are exported by exportin-5 to the cytoplasm and are subsequently cleaved in the “dicing step” by a second RNase III enzyme, Dicer, and Tar RNA-binding protein. After being loaded into the RNA-induced silencing complex, the diced, mature miRNAs guide the RNA-induced silencing complex to target mRNAs by complementary base-pairing and participate in translational repression, mRNA degradation, or both (1, 3, 4).
The biogenesis of miRNAs is often regulated post-transcriptionally (5, 6); some pri-miRNAs are not cropped and remain at high levels, and pre-miRNAs are not necessarily processed to mature miRNAs in the dicing step (7). Recent studies suggest that miRNA biogenesis is regulated at the cropping step by RNA-binding proteins, such as DDX5, DDX17, K-homology splicing regulator protein (KSRP), heterogeneous nuclear RNP (hnRNP) A1, NF90, NF45, and Lin28 (5, 6, 8, 9).
Lin28, a protein first characterized in Caenorhabditis elegans, can control stage-specific fates regulated by lin-4 (10). Mammalian Lin28 is expressed in embryonic stem cells (ESCs), in various embryonic-stage stem/progenitor cells, and in some adult-stage cell types, including heart, kidney, and muscle (11). Normally, endogenous Lin28 is predominantly in the cytoplasm, and some is also present in the nucleus, particularly the nucleolus, where DGCR8 is localized (12), in NT2 and yeast cells (10, 13). Lin28 influences cell fate by post-transcriptional mechanisms (14, 15). Furthermore, Lin28 is a key regulator in miRNA biogenesis, particularly for the let-7 family, and is involved in major cellular functions in ES and cancer cells (16, 17).
The 10 mammalian miRNAs of the let-7 family are registered in the miRBase Database: let-7a to let-7g, let-7i, miR98, and miR202. Although the expression of let-7 miRNAs is abundant in differentiated cells, it is tightly repressed in undifferentiated cells, particularly ESCs (7). In ESCs, Lin28 binds to the terminal loops of let-7 family miRNA precursors in ESCs and inhibits their processing by cropping (18, 19) or by dicing together with Lin28-binding protein, terminal uridylyltransferase 4 (TUT4), as a cofactor (20–22); however, a cofactor of Lin28 in the cropping step has not been identified. Negative regulatory feedback loops between Lin28 and let-7 family miRNAs are likely to be critical in triggering the differentiation of ES/induced pluripotent cells (23, 24).
The Musashi family is a group of neural RNA-binding proteins containing two RNA recognition motifs (RRMs). It is evolutionally conserved from invertebrates to vertebrates (25, 26). We identified mammalian Musashi1 (Msi1) and Musashi2 (Msi2) and found that Msi1 is highly expressed in NS/PCs and other somatic stem cells (27, 28). Msi1 represses the translation of its target mRNAs by binding to their 3′-UTR and helps maintain NS/PCs in the undifferentiated state (29, 30). Msi1 competes with eIF4G for poly(A)-binding protein, thereby inhibiting translation initiation (31). However, Msi1 is present not only in cytoplasm but also in the nucleus, where the details of its molecular function remain to be clarified. Msi1 is highly expressed in various stem/progenitor cells, and its role within ESCs and in their differentiation also needs clarification. In the present study, we examined the function of Msi1 in the nucleus during the early neural differentiation of mouse ESCs and found that it acts synergistically with Lin28 as a novel cofactor for the blockade of let-7 family miRNA biogenesis.
EXPERIMENTAL PROCEDURES
Vectors, Buffers, and Antibodies
Details of the plasmid constructs expressing recombinant Msi1, Lin28, and miRNAs, along with the buffers and antibodies used, are described in the supplemental tables.
Cell Culture and Transfection
The ESC culture and EB cell induction in low (10−8 m) or high (10−6 m) retinoic acid (RA) conditions were described previously (32). Low RA was used to prepare cultures for Western blotting (see Figs. 1, A and B, and 2B), immunocytochemistry (see Fig. 1C), and Northern blotting (see Fig. 2, A and C); high RA was used to generate the PCR data and the quantification shown in Fig. 4, B–E. siRNAs (5 or 30 nm each) were used to transfect ESCs with the RNAiMax reagent following standard procedures (Invitrogen) before inducing EB cells. EB cells were thereafter induced in high RA conditions (32). siControl (Ambion AM4636), siMsi1 (Ambion siRNA ID 63169), and siLin28 were described previously (Lin-28/1, 5′-GGGUUGUGAUGACAGGCAATT-3′ (15)). The 293T cell culture and transfection with GeneJuice (Novagen) were described previously (31).
FIGURE 1.
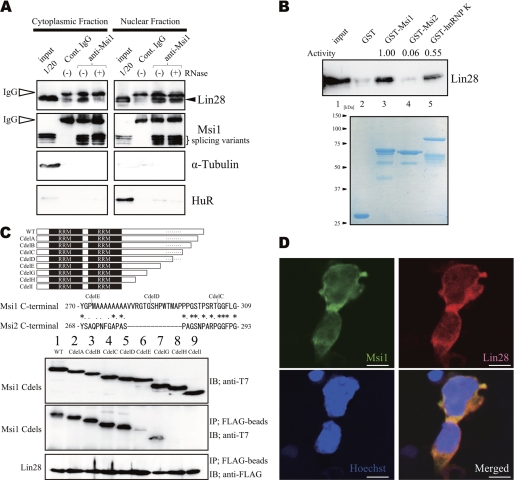
Msi1 was found in the Lin28 complex in EB cells. A, EB cell extracts of cytoplasmic and nuclear fractions, immunoprecipitated and immunoblotted with an anti-Msi1 antibody. Cont. IgG, control IgG. B, EB cell extracts were mixed with purified GST proteins, which were visualized by Coomassie Brilliant Blue staining (lanes 2–5, bottom). hnRNP-K, heterogeneous nuclear RNP-K. C, top, schema of proteins containing T7-Msi1 variants. Lanes 2–8, a series of Msi1C-terminal deletions. A dashed line indicates the gap region between Msi1 and Msi2. Middle, the alignment of Msi C-terminal proteins (see details in supplemental Fig. S2). Bottom, various T7-Msi1 mutants, bound to FLAG-Lin28. IB, immunoblot; IP, immunoprecipitates. D, Msi1 was co-localized with Lin28 in the cytoplasm and nucleus in EB cells on day 5. Green, anti-Msi1; red, anti-Lin28; blue, nuclei (Hoechst). Bars, 5 μm.
FIGURE 2.
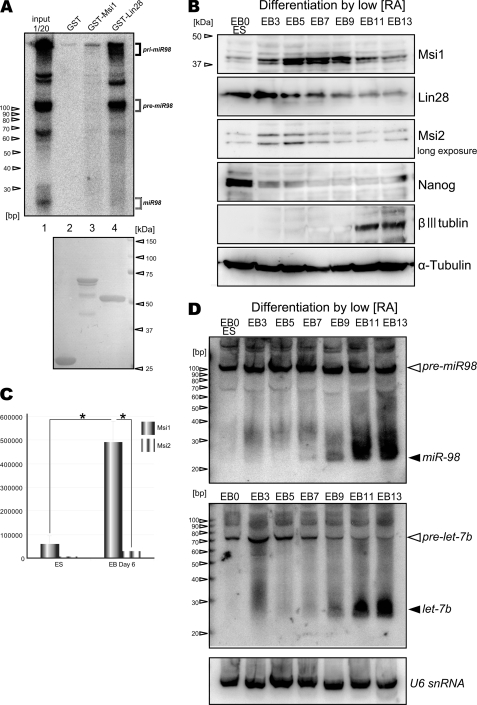
Expression profiles of Msi1, Lin28, and let-7 miRNA family during ES differentiation. A, miR98 precursor and mature species expressed in EB day 11, pulled down by various GST-tagged proteins, were detected by miR98 LNA-based Northern blotting. GST-tagged proteins used were visualized by Coomassie Brilliant Blue (lanes 2–4, bottom). B, immunoblotting during EB formation in a mouse ESC line (EB3). βIII tubulin is a neuronal marker; α-tubulin was the loading control. The numbers indicate days after the beginning of RA treatment. C, Msi1 and Msi2 mRNAs in EBs, analyzed by absolute qRT-PCR (n = 6, mean ± S.E.; *, p < 0.01). D, LNA Northern blotting showing the post-transcriptional induction of mature miR98 and let-7b during EB formation; U6 small nuclear ribonucleic acid (U6 snRNA) was used as the loading control.
FIGURE 4.
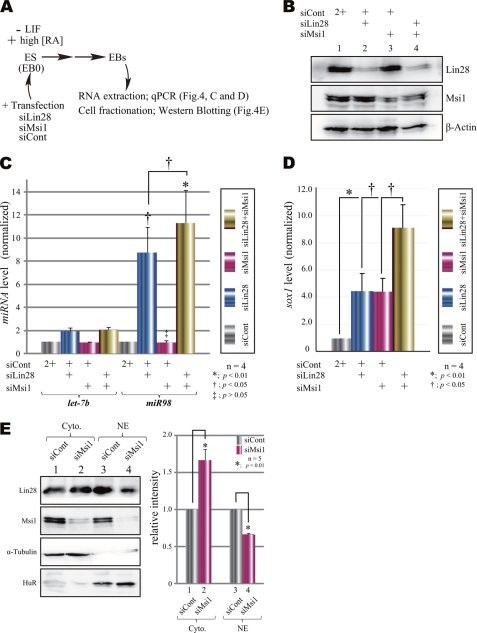
Msi1 contributes to the function of Lin28 in vivo under knockdown conditions. A, protocol for EB formation from mouse ESCs in the siRNA knockdown assay. ESCs (i.e. EB day 0) were transfected with one or more siRNA, dissociated, and cultured in the presence of high RA (10−6 m) for 3 days to induce differentiation. siCONT, small interfering control; LIF, leukemia inhibitory factor. B, endogenous Lin28, Msi1, and β-actin in siRNA-transfected EB cells. C, changes in miRNA levels upon knockdown of Msi1 and/or Lin28, analyzed by qRT-PCR. Statistical analyses were performed by Dunnett's method for multiple comparisons for the siCont group versus siLin28, siMsi1, or siLin28-siMsi1 (n = 4, mean ± S.E., *, p < 0.01, †, p < 0.05, ‡, p > 0.05), and Student's t test was used to calculate the p value for comparisons for the siLin28 group versus siLin28-siMsi1 (n = 4, mean ± S.E., †, p < 0.05). D, expression of Sox1 mRNA as an early neural maker in EBs was analyzed by qRT-PCR (n = 6, mean ± S.E.; *, p < 0.01; †, p < 0.05; Student's t test). E, immunoblotting of subcellular fractions of EB cells upon knockdown of Msi1 (lanes 2 and 4; siCont, lanes 1 and 3). Immunoblotted cytoplasmic fractions (Cyto, lanes 1 and 2) and nuclear fractions (NE, lanes 3 and 4) are shown. Hu antigen R (HuR) and α-tubulin were the loading control. The values were quantified by normalizing them to each siCont lane. Student's t test was used to calculate the p value (n = 5, mean ± S.E., *, p < 0.01).
Protein Purification, Immunoblotting, Immunoprecipitation, Co-immunoprecipitation, and GST Pulldown Assays
GST-tagged proteins were expressed in the Escherichia coli strain BL21 and purified by glutathione-Sepharose 4B resin (GE Healthcare and Invitrogen), as described previously (31). Immunoblotting was performed as described previously (31). ECL reagents (GE Healthcare) were used to visualize the signal, which was quantified using an LAS 3000 mini (Fujifilm) and its software. Immunoprecipitation of 293T cells or EB cells on day 5 and immunoblotting to detect precipitated proteins were performed as described previously (31). GST pulldown assays (see Fig. 2A) were performed in the absence of RNase A and with RNasin (400 units/ml; Promega). Lysates of EB cells on day 11 were incubated for 1 h at 4 °C and thoroughly washed, and then the total RNA was extracted for Northern blotting (see below). GST pulldown assays (see Figs. 1B and 5B) were performed in the presence of RNase A incubated with EB (see Fig. 1B) or 293T (see Fig. 5B) cell lysates. FLAG-tagged pulldown assays and co-immunoprecipitation assays were performed as described previously (31).
FIGURE 5.
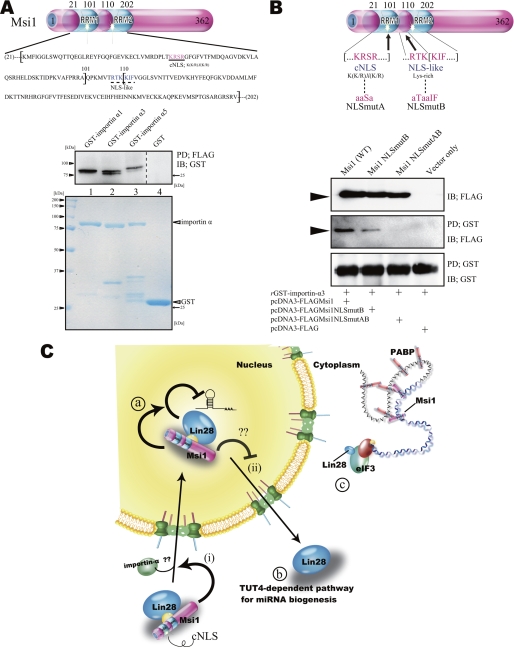
The cNLS of Msi1 as a regulated importin-α-binding domain. A, FLAG pulldown (PD) assay of bacterially purified proteins, using GST-FLAG-Msi1 (Fig. 1B, lane 3) and GST-importin-α fusion proteins (lanes 1–3, bottom panel.) IB, .immunoblot B, top, illustration of FLAG-Msi1 protein variants. Shown are Msi1NLSmutAB (NLSmutA; mutations of cNLS and NLS-like sequences in RRM1 and RRM2, respectively) and Msi1NLSmutB (NLSmutB; mutation of a NLS-like sequence in RRM2). Bottom, immunoprecipitation using Msi1 NLS mutants. Msi1NLSmutAB variants did not bind to purified GST-importin-α3 (see also panel A.) rGST, recombinant GST. aaSa, ala-ala-Ser-ala, aTaaIF, ala-Thr-ala-ala-Ile-Phe, RTKKIF, Arg-Thr-Lys-Lys-Ile-Phe. C, Msi1 could be involved in the following scenarios: (i) Lin28 may be imported into the nucleus via the Msi1 cNLS, after which Msi1 enhances the inhibitory action of Lin28 on pri-miRNA processing (a); (ii) Lin28 is exported to and/or retained in the cytoplasm. Note that the effect of Lin28 on pre-miRNA biogenesis in the cytoplasm is dependent on TUT4 (b). Msi1 regulates translation initiation (c). PABP, poly(A)-binding protein.
Immunocytochemistry
EB cells on day 5 were stained with anti-Msi1 (mAb 14H1) and anti-Lin28 as described previously (15, 31, 33). Cells were dissociated before fixing using trypsin followed by trypsin inhibitor. Alexa Fluor 488- or 555-conjugated secondary antibodies (Molecular Probes) were used to visualize the staining. Digital images were captured by a laser confocal microscope (Zeiss LSM510) using a 63×/1.2 NA water immersion objective lens.
Northern Blotting Analysis and Real-time PCR
Total RNA was extracted with TRIzol (Invitrogen) and phenol and then treated with DNase I (Invitrogen). To detect miRNAs, we used specific 32P-labeled locked nucleic acid (LNA) probes (Exiqon). Hybridization signals were detected using a BAS5000 or 2500 scanner (Fujifilm). qPCR of miRNA was performed following the manufacturer's instructions (Applied Biosystems) for the TaqMan MicroRNA RT kit and TaqMan MicroRNA assays. Each miRNA was normalized to U6 small nuclear ribonucleic acid. We performed mRNA qPCR with SYBR Premix Ex Taq II (Takara) following the manufacturer's protocol. Each mRNA was normalized to β-actin. All real-time PCR signals were detected using a 7900HT system (Applied Biosystems) and its software using the ΔΔCt or the absolute qPCR method (see Fig. 2C).
Microprocessing Assay and Cell Fractionation
The microprocessor complex was derived from pcDNA-mDrosha-FLAG and/or pcDNA-FLAG Dgcr8; the microprocessing assay was performed as described previously (19, 34). Purified GST-Msi2 (3.5 pmol/μl) or GST-Msi1 (0.25, 0.5, 3.5 pmol/μl) or proteins precipitated with FLAG, FLAG-Msi1, FLAG-Msi2, or FLAG-GST, were added to pri-miRNA along with 32P-labeled UTP (104-105 cpm), RNasin (final 1 unit/μl; Promega), and MgCl2 (final 6.4 mm) and incubated at 37 °C for 60 or 90 min as described previously (19, 34). RNAs were resolved on 11–13% Tris-borate-EDTA-urea gels and visualized by autoradiography. Mature miRNAs were not produced in this assay system because Dicer was not present. Cell fractionations were performed as described previously (35) using EB cells and the appropriate buffers (see supplemental Table S2 for buffers).
RESULTS
Msi1 Is a Component of the Lin28 Complex
We previously identified direct targets of Msi1 in high molecular weight fractions of 293T cells (31). However, we have not identified its direct target in NS/PCs or other tissue-specific stem cells or in low molecular weight fractions. Although ESCs can differentiate into several types of somatic stem cells, the function of Msi1 in this process has not been fully described. A previous report shows that Musashi is a likely component of the Lin28 protein complex in myoblasts, but how it interacts with the complex and functions in myoblasts remains elusive (15).
To address whether Msi1 is involved in Lin28-mediated regulation of ES differentiation, we investigated interactions between Msi1 and Lin28 in EB cells, which are derived from ESCs. ESCs were treated with RA to induce NS/PCs in the EB cells (32). Immunoprecipitation of the nuclear fraction from EB cell lysates with an antibody to Msi1 showed that Msi1 bound to Lin28 in RNA-independent manner in nucleus but not in cytoplasm (Fig. 1A). To examine the relative binding activity of Msi1 and Lin28, we performed a GST pulldown assay with fusion proteins between GST and Msi1, Msi2, and heterogeneous nuclear RNP-K (hnRNP-K) as a positive control (18, 21). Msi1 bound to Lin28 more strongly than did GST or Msi2 in the presence of RNase A (Fig. 1B).
To investigate the difference in Lin28 binding ability between Msi1 and Msi2, we performed co-immunoprecipitation experiments using Msi1 C-terminal deletion mutants in the presence of RNase (Fig. 1C). A search for the C-terminal regions of Msi1 and Msi2 identified an interesting difference in their amino acid sequences; there is a large gap in Msi2 corresponding to the section from Val-282 to Phe-296 in Msi1 (Fig. 1C and supplemental Fig. S2). Co-immunoprecipitation assays showed that the Lin28 binding ability of CdelA to CdelD was equal to that of wild-type Msi1, but the deletion mutants of the Msi1-specific amino acid sequence (CdelE to CdelI) lacked binding activity to Lin28 (Fig. 1C). These results suggest that the distinct amino acid sequences that are present in Msi1 but absent in Msi2 in this area may be important for the ability of Msi1 to bind Lin28. Next, we confirmed that Msi1 co-localized with Lin28 in EB cells in the cytoplasm, and to some extent, in the nucleus by immunocytochemical and cell fractionation analyses with anti-Msi1 and anti-Lin28 antibodies using EB cells on day 5 (Fig. 1D and supplemental Fig. S1A).
Msi1 Is Up-regulated and Lin28 Is Down-regulated during ESC Neural Differentiation
Lin28 post-transcriptionally represses miRNAs expression in NS/PCs and in differentiating ESCs (14, 18, 20, 21, 24, 35). Msi1 could contribute to miRNA activity based on its localization in the processing bodies and stress granules, although the precise mechanisms were unclear (31). To examine the relationship among Msi1, miRNA, and Lin28 in differentiating ESCs, we first performed a Northern blot analysis of RNAs expressed in EB cells. Precursors and mature miR98 were pulled down by individual GST-tagged proteins (GST alone, GST-Msi1, and GST-Lin28) using highly miR98-specific LNA probes to assess their binding to Msi1 or Lin28. Expression of miR98, a member of the let-7 family, is strikingly repressed in ESCs and is significantly affected by the depletion of Lin28 as compared with other members of the let-7 family (7, 18). However, its underlying regulatory mechanism has been not fully addressed. LNA probes used in this study can detect all forms of miRNA, including pri-, pre-, and mature miRNA. Consequently, although Msi1 did not bind to any forms of miR98, Lin28 bound to pre- and pri-miR98 under this condition (Fig. 2A).
Lin28 is also known as a reprogramming factor used to establish induced Pluripotent Stem cells (iPS cells) (36). Thus, we next compared the temporal expression profile of Msi1 and Lin28 during neural differentiation of ESCs. Three days after beginning differentiation with RA, Lin28 was expressed at its maximum level; it gradually decreased after day 5 and reached its baseline by day 13 (Fig. 2B). Considering the state of neural differentiation driven by RA treatment during EB formation (32), this expression pattern of Lin28 is consistent with that in the developing neural tube (11, 14) (Fig. 2B). At this stage, Msi1 expression gradually increased as cell neural differentiation increased in response to RA and reached its maximum level on day 7 of EB formation (Fig. 2B). Msi2 expression was weak as compared with that of Msi1 and Lin28 (Fig. 2B). Absolute qRT-PCR (Fig. 2C) showed that the Msi1 mRNA expression level was 16.8 times greater than that of Msi2 in the EBs on day 6.
Certain pri-miRNAs are highly expressed in mouse and human ESCs, but their processing is repressed (7, 18, 19, 23). Because Lin28 bound to pre- and pri-miR98 precursors (Fig. 2A), the expression profile of Lin28 was compared with that of maturating let-7 family miRNAs during ES differentiation (Fig. 2D), using LNA-based Northern blotting at each EB stage. The maturation of miR98 and let-7b was repressed in undifferentiated ES and in EB cells through day 7 of RA treatment (Fig. 2D). Although Lin28 expression, which strongly inhibits let-7 family miRNA biogenesis in both the cropping and the dicing steps (18–22, 24, 35), was down-regulated on days 7–9 (Fig. 2B), the expression of mature miR98 and let-7b was still repressed at this stage. Notably, Msi1 was expressed at its maximum level at this stage of differentiation (Fig. 2B). These results raised the possibility that Msi1 might compensate for the Lin28 function in let-7 family miRNA biogenesis.
Msi1 Contributes to Lin28-mediated miRNA Biogenesis
Msi1 is present not only in the cytoplasm, where it functions as a translational repressor (31), but also in the nucleus (Fig. 1, A and D, and supplemental Fig. S1A), where its role has been unclear. Recent studies demonstrate that Lin28 blocks the processing of let-7 family pri-miRNAs in the nucleus at the cropping step (18, 19). In the present study, Msi1 did not bind to TUT4 (supplemental Fig. S1B), a cofactor of Lin28, in the dicing step. Msi1 also bound to Lin28 in the nucleus independently of RNA (Fig. 1, A and B). Thus, we next analyzed whether Msi1 interacts with Lin28 to regulate miRNA processing at the cropping step.
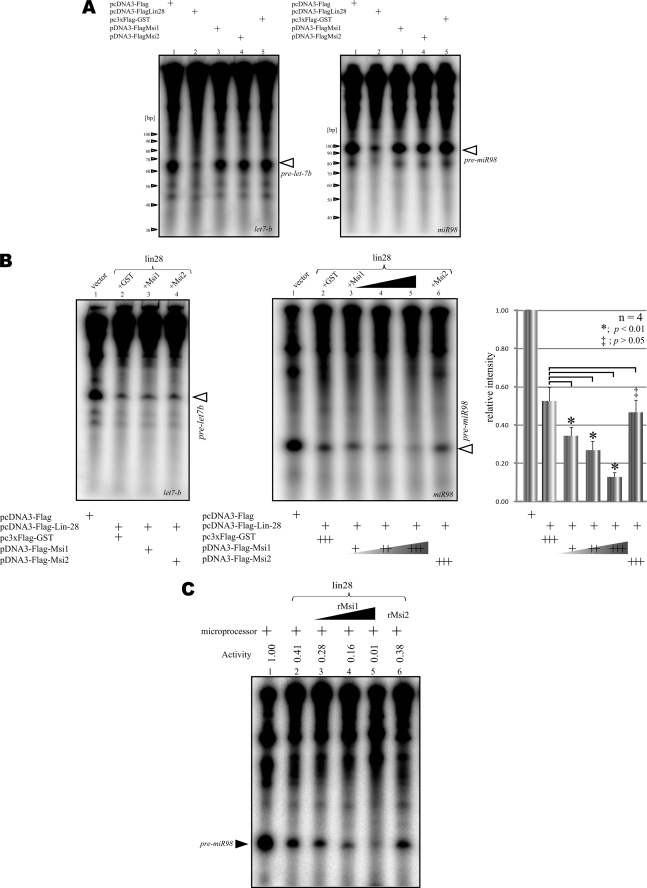
We performed an in vitro microprocessing assay in which FLAG-Lin28 was precipitated from FLAG-Lin28-expressing 293T cells and then incubated with 32P-labeled pri-miRNA and Microprocessor. Lin28 dramatically repressed the miR98 and let-7b pre-miRNAs (Fig. 3A, lanes 1 and 2) even when expressed alone, but Msi1, Msi2, and control FLAG-GST did not (Fig. 3A, lanes 3, 4, and 5), in good agreement with the previous studies using pri-let-7g (18, 19, 35). On the other hand, the presence of Msi1 strengthened Lin28-mediated inhibition of miR98 processing at the cropping step (Fig. 3B). The presence of Lin28 and GST repressed the production of pre-miR98 to 47.4% ± 7.3 of the control value (n = 4), and the presence of Lin28 and Msi1 further repressed it to 39.8% as compared with that of Lin28 and GST and to 33.9% as compared with that of Lin28 and Msi2 (Fig. 3B, right). In contrast, the presence of Msi1 and Lin28 showed little change of the production of pre-let-7b as compared with that of GST and Lin28 (Fig. 3B, left). Thus, we focused on the Lin28-mediated blockade of miR98 biogenesis to ascertain the direct effect of Msi1 in the repression of Lin28 at the cropping step; an in vitro microprocessing assay using bacterially purified GST-Msi1 showed that Msi1 directly enhanced the inhibitory action of Lin28 on the pre-miR98 biogenesis according to dosage (Fig. 3C). The results in Fig. 3, B and C, agree with previous reports showing that Lin28 selectively binds to the terminal loop region of let-7 precursors (19, 37) and participates in the cropping step of miRNA biogenesis (18). They also indicate that Msi1 enhances Lin28-mediated inhibitory effects on miR98 biogenesis at the cropping step.
FIGURE 3.
Msi1 enhances the inhibitory action of Lin28 on miRNA processing. A, the processing of pri-miR98 (left panel) and pri-let-7b (right panel), in the presence of the indicated immunoprecipitated FLAG-tagged protein. The expression of Lin28 alone reduced the amount of pre-miRNA. B, the processing of pri-let-7 family in the presence of immunoprecipitated empty pcDNA3-FLAG or FLAG-Lin28 and the indicated FLAG-tagged protein. The pri-miR98 signal was normalized to the vector immunoprecipitation lane (right panel). Statistical analyses were performed by Dunnett's method for multiple comparisons for the GST-Lin28 group versus Msi1-Lin28 or Msi2-Lin28 (n = 4, mean ± S.E., *, p < 0.01, ‡, p > 0.05). Msi1 significantly enhanced the blockade of the miR98 processing step by Lin28. C, pri-miR98 processing in the presence of FLAG-Lin28 and bacterially expressed, purified Msi1 or Msi2. As the dosage of Msi1 increased, pri-miR98 processing was more strongly inhibited by the presence of Lin28. rMsi1, recombinant Msi1.
Contribution of Msi1 to Lin28-mediated miRNA Biogenesis during EB Formation
To further investigate the in vivo function of Msi1 in miRNA biogenesis, ESCs were transfected with siRNAs for Msi1 and Lin28 and allowed to form EBs, which contain NS/PCs (38), in the presence of RA for 3 days (Fig. 4A). These siRNAs efficiently reduced the Msi1 and Lin28 expression levels as compared with cells expressing a control siRNA (Fig. 4B).
When Msi1 alone was knocked down, the levels of mature miR98 and let-7b were not significantly altered; however, when Lin28 alone was knocked down, the level of mature miR98 was increased 8.7-fold (n = 4) (Fig. 4C). The effect is consistent with a previous report showing that Lin28 knockdown in P19 cells results in increased expressions of mature let-7b and miR98 by ∼2- and 8-fold, respectively (18). Interestingly, the double knockdown of Msi1 and Lin28 increased the level of miR98 11.3-fold (Fig. 4C). Knocking down both Lin28 and Msi1 with siRNA induced a 9-fold increase in the early neural marker Sox1 over the control. Individually knocking down either Lin28 or Msi1 increased Sox1 mRNA level ∼4-fold as compared with the control (Fig. 4D). These data suggested that the combination of Msi1 and Lin28 could affect early neural EB cell differentiation.
Msi1 May Regulate the Nucleocytoplasmic Distribution of Lin28
Next, we examined the effect of Msi1 on Lin28 intracellular localization during ESC neural differentiation and EB formation in the presence of RA. In the nucleus, Lin28 expression increased from days 3–7 and then gradually decreased but was still present on day 11 (supplemental Fig. S1A). In the cytoplasm, however, the amount of Lin28 significantly decreased from day 9 and was drastically diminished by day 11 (supplemental Fig. S1A). This pattern of Lin28 levels in the nucleus is similar to the expression pattern of Msi1 (Fig. 2B) and is inversely correlated with the presence of mature miR98 (Fig. 2D) during the neural differentiation of mouse ESCs. When Msi1 was knocked down with siRNA, the quantity of Lin28 significantly increased in the cytoplasm and decreased in the nucleus as compared with cells transfected with control siRNA (Fig. 4E). These results suggest that Msi1 can enhance the formation of Lin28-containing complexes in the nucleus; it may affect an intracellular localization of Lin28 by retaining it in the nucleus or by promoting its nuclear import (Fig. 5C).
Importin-α May Regulate the Nuclear Localization of Msi1
A search for Msi1 structural motifs identified a potential classical nuclear localization signal (cNLS) with the consensus sequence K(K/R)X(K/R) (39) within the RNA recognition motif 1 (RRM1). A peptide NLS-like sequence containing a Lys-rich region was also identified within RRM2 (Fig. 5A). Although these sequences could import Msi1 from the cytoplasm into the nucleus, Msi1 is predominantly found in the cytoplasm, where it is involved in sequence-specific translational repression (29, 31). To identify the molecular mechanisms governing the nucleocytoplasmic shuttling of Msi1, we first verified whether Msi1 could bind to importin-α directly. We performed FLAG-tagged pulldown assays in the presence of RNase using purified GST-FLAG-Msi1 versus GST-importin-α isoform proteins. As shown in Fig. 5A, Msi1 bound to all of the importin-α proteins tested.
Next, we performed GST pulldown assays in the presence of RNase using purified GST-importin-α, Msi1NLSmutAB (in which three consensus NLS amino acids and a peptide NLS-like sequence are replaced by alanine residues), and Msi1NLSmutB (in which a peptide NLS-like sequence is replaced by alanine residues). Replacing the cNLS sequence of Msi1 drastically decreased its binding activity to importin-α3 as compared with wild-type Msi1 (Fig. 5B), suggesting that the interaction between Msi1 and importin-α could play an important role in the nuclear localization of Msi1.
DISCUSSION
In Fig. 1A, we show that Msi1 bound to Lin28 independently of RNA in the nucleus but not in the cytoplasm. Msi1 may need to recognize an accessible situation by Lin28. As compared with other RNA-binding proteins, Msi1 binds strongly to its target mRNA (29); Msi1 also binds directly to poly(A)-binding protein in the cytoplasm (31). Thus, the binding molecules of Msi1, including RNAs and proteins, may interfere with the binding between Lin28 and Msi1. Lin28 also binds to target mRNA and eIF3 (15). Msi1 interacts with Lin28 in the cytoplasm via a common mRNA, and moreover, may be involved in initiating translation. Both Lin28 and Msi1 co-localize into processing body and stress granule, which are tightly packed mRNPs (13, 31). Direct binding of Msi1 and TUT4, which participates in the dicing step in the cytoplasm, was not detected by a co-immunoprecipitation assay (supplemental Fig. S1B). Therefore, Msi1 may participate both in the cropping step of miRNA biogenesis in the nucleus and in translational initiation in the cytoplasm (Fig. 5C).
Recent reports indicate that Msi1 can be involved in tumorigenesis (26, 40). Notably, Lin28 is also strongly expressed in various types of cancer cells and induces tumorigenesis by repressing let-7 family miRNAs biogenesis, and let-7 miRNAs down-regulate their oncogenic targets, K-Ras, c-Myc, and HMGA2 (17, 41). Considering the facts that Msi1 is expressed in human glioma and glioblastoma (42, 43) and present results showing that Msi1 is synergistically involved in Lin28-mediated miRNA biogenesis, there might be strong correlation between Msi1 and Lin28 in the aspects of tumorigenesis via the repression of let-7 family miRNA biogenesis in cancer stem cells.
Lin28 and TUT4 contribute to the maintenance of ESCs by regulating the miRNA biogenesis (21). A feedback mechanism between the let-7 family and Lin28 is involved in triggering cell fate determination in both undifferentiated and differentiated ESCs (23), and NS/PCs (24). Additionally, our present results in Figs. 3 and 4 suggest the mutually complementing actions of Lin28 and Msi1 in the regulation of miRNA biogenesis during neural differentiation of mouse ESCs. In undifferentiated ESCs, where Lin28, but not Msi1, is strongly expressed, Lin28 by itself plays indispensable roles in suppressing miRNA biogenesis.
Considering the roles of Lin28 and Msi1 in repressing miR98 at the cropping step, their temporal expression (Fig. 2B) may finely regulate the timing of ES cell neural differentiation. In the initial induction period, around days 3–9 of EB formation, abundantly expressed Lin28 is gradually down-regulated in concert with ES cell differentiation (Fig. 2B). During this period, the repressive effect of Lin28 on miR98 may attenuate. In contrast, Msi1 expression gradually increased (Fig. 2B), compensating for the decreasing activity of Lin28 in repressing miR98 processing. Finally, Msi1 expression decreased around day 11 or after, allowing miR98 to be processed into its mature form followed by the induction of neural marker expression.
Consistent with this temporal correlation of neural differentiation with the sequential expression of Lin28 and Msi1, the knockdown of both Lin28 and Msi1 significantly induced the expression of the early neural marker Sox1 as compared with the knockdown of Lin28 or Msi1 individually (Fig. 4D). We speculate that Lin28 and Msi1 sequentially and synergistically repress miR98 processing at the cropping step and may be involved in determining the timing of neural differentiation.
With regard to the mechanisms of nucleocytoplasmic Msi1 shuttling, we found that Msi1 can bind to importin-α (Fig. 5, A and B). More importantly, the cytoplasmic localization of Msi1 can also be explained if the cNLS sequences, due to their position, are masked when Msi1 is tightly bound to a target mRNA or tightly packed as an mRNP. Indeed, we previously reported that Msi1 also localizes to the processing body and stress granule, which form tightly packed mRNPs in the cytoplasm (31). Furthermore, we reported that the RNA binding-deficient mutant Msi1-ABmut, in which six phenylalanine residues are replaced by leucine residues in the RNP-1 sequence of RRM1 and RRM2, accumulates predominantly in the nucleus (31).
A previous report (44) showed that regulation of the nuclear import and export of transcription factors in stem cells is important for cell fate determination. For example, importin-α subtype switching is involved in triggering mouse ES cells to differentiate into neurons via the selective nuclear import of the transcription factors Oct3/4, Brn2, and Sox2 (44). Moreover, it is reported that Lin28 expression overlaps with that of Msi1 in mouse neuroepithelial cells, including NS/PCs (11), and that Lin28 is a key factor for cell fate determination in both undifferentiated and differentiated mouse ES cells (23). Msi1 expression was observed after the decrease in pluripotent marker expression, such as Nanog, and before the increase in neural marker expression, such as βIII tubulin (44) (Fig. 2B), between day 3 and day 9 in our experiment (Fig. 2B). This previous finding and our present results from EB cell fractionations (Fig. 4E) suggest that Msi1 may exert control over the timing of neural differentiation via importin-α subtype switching. Thus, this model provides a functional explanation of the nucleocytoplasmic shuttling of RNA-binding proteins and its novel role in the regulation of miRNA biogenesis during stem cell maintenance and differentiation. However, it is not clear what induces nuclear import of Msi1 and Lin28. Further study is needed to investigate this nucleocytoplasmic shuttling in the future.
Supplementary Material
Acknowledgments
We are grateful to Prof. Haruhiko Siomi for critical reading of the manuscript, to Dr. Shinsuke Shibata for help and for the Msi2 protein, to Dr. Takehiko Sunabori for instruction on site-directed mutagenesis, and to Prof. Akihiko Yoshimura for the FLAG-hTUT4 vector.
This work was supported by the Funding Program for World-Leading Innovative R&D on Science and Technology from Japan Society for the Promotion of Science and by grants from the Japanese Ministry of Education, Sports and Culture of Japan (MEXT) (to H. O.) (the Leading Project for Realization of Regenerative Medicine) and (to H. K.) (Grant-in-aid for Young Scientists (B)) and a Keio University grant-in-aid for the encouragement of young medical scientists.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1 and S2.
- miRNA
- microRNA
- pre-miRNA
- precursor miRNA
- pri-miRNA
- primary miRNA
- Msi
- Musashi
- Lin28
- abnormal cell Lineage family member-28
- ESC
- embryonic stem cell
- EB
- embryoid body
- NS/PC
- neural stem/precursor cell
- RRM
- RNA recognition motif
- RA
- retinoic acid
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR
- NLS
- nuclear localization signal
- cNLS
- classical NLS
- LNA
- locked nucleic acid
- RNP
- ribonuclear protein
- mRNP
- messenger RNP
- TUT4
- terminal uridylyltransferase 4.
REFERENCES
- 1. Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yekta S., Tabin C. J., Bartel D. P. (2008) Nat. Rev. Genet 9, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 4. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 5. Newman M. A., Hammond S. M. (2010) Genes Dev. 24, 1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siomi H., Siomi M. C. (2010) Mol. Cell 38, 323–332 [DOI] [PubMed] [Google Scholar]
- 7. Thomson J. M., Newman M., Parker J. S., Morin-Kensicki E. M., Wright T., Hammond S. M. (2006) Genes Dev. 20, 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamagata K., Fujiyama S., Ito S., Ueda T., Murata T., Naitou M., Takeyama K., Minami Y., O'Malley B. W., Kato S. (2009) Mol. Cell 36, 340–347 [DOI] [PubMed] [Google Scholar]
- 9. Michlewski G., Cáceres J. F. (2010) Nat. Struct. Mol. Biol. 17, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moss E. G., Lee R. C., Ambros V. (1997) Cell 88, 637–646 [DOI] [PubMed] [Google Scholar]
- 11. Yang D. H., Moss E. G. (2003) Gene Expr. Patterns 3, 719–726 [DOI] [PubMed] [Google Scholar]
- 12. Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. (2007) Exp. Cell Res. 313, 4196–4207 [DOI] [PubMed] [Google Scholar]
- 13. Balzer E., Moss E. G. (2007) RNA Biol. 4, 16–25 [DOI] [PubMed] [Google Scholar]
- 14. Balzer E., Heine C., Jiang Q., Lee V. M., Moss E. G. (2010) Development 137, 891–900 [DOI] [PubMed] [Google Scholar]
- 15. Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E. G., Harel-Bellan A. (2007) Genes Dev. 21, 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heo I., Kim V. N. (2009) Cell 139, 28–31 [DOI] [PubMed] [Google Scholar]
- 17. Viswanathan S. R., Daley G. Q. (2010) Cell 140, 445–449 [DOI] [PubMed] [Google Scholar]
- 18. Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman M. A., Thomson J. M., Hammond S. M. (2008) RNA 14, 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagan J. P., Piskounova E., Gregory R. I. (2009) Nat. Struct. Mol. Biol. 16, 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heo I., Joo C., Kim Y. K., Ha M., Yoon M. J., Cho J., Yeom K. H., Han J., Kim V. N. (2009) Cell 138, 696–708 [DOI] [PubMed] [Google Scholar]
- 22. Lehrbach N. J., Armisen J., Lightfoot H. L., Murfitt K. J., Bugaut A., Balasubramanian S., Miska E. A. (2009) Nat. Struct. Mol. Biol. 16, 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melton C., Judson R. L., Blelloch R. (2010) Nature 463, 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R., Wulczyn F. G. (2008) Nat. Cell Biol. 10, 987–993 [DOI] [PubMed] [Google Scholar]
- 25. Okano H., Imai T., Okabe M. (2002) J. Cell Sci. 115, 1355–1359 [DOI] [PubMed] [Google Scholar]
- 26. Okano H., Kawahara H., Toriya M., Nakao K., Shibata S., Imai T. (2005) Exp. Cell Res. 306, 349–356 [DOI] [PubMed] [Google Scholar]
- 27. Sakakibara S., Imai T., Hamaguchi K., Okabe M., Aruga J., Nakajima K., Yasutomi D., Nagata T., Kurihara Y., Uesugi S., Miyata T., Ogawa M., Mikoshiba K., Okano H. (1996) Dev. Biol. 176, 230–242 [DOI] [PubMed] [Google Scholar]
- 28. Sakakibara S., Nakamura Y., Satoh H., Okano H. (2001) J. Neurosci. 21, 8091–8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Imai T., Tokunaga A., Yoshida T., Hashimoto M., Mikoshiba K., Weinmaster G., Nakafuku M., Okano H. (2001) Mol. Cell Biol. 21, 3888–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakakibara S., Nakamura Y., Yoshida T., Shibata S., Koike M., Takano H., Ueda S., Uchiyama Y., Noda T., Okano H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15194–15199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawahara H., Imai T., Imataka H., Tsujimoto M., Matsumoto K., Okano H. (2008) J. Cell Biol. 181, 639–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okada Y., Shimazaki T., Sobue G., Okano H. (2004) Dev. Biol. 275, 124–142 [DOI] [PubMed] [Google Scholar]
- 33. Kaneko Y., Sakakibara S., Imai T., Suzuki A., Nakamura Y., Sawamoto K., Ogawa Y., Toyama Y., Miyata T., Okano H. (2000) Dev. Neurosci. 22, 139–153 [DOI] [PubMed] [Google Scholar]
- 34. Lee Y., Kim V. N. (2007) Methods Enzymol. 427, 89–106 [DOI] [PubMed] [Google Scholar]
- 35. Heo I., Joo C., Cho J., Ha M., Han J., Kim V. N. (2008) Mol. Cell 32, 276–284 [DOI] [PubMed] [Google Scholar]
- 36. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin II, Thomson J. A. (2007) Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 37. Piskounova E., Viswanathan S. R., Janas M., LaPierre R. J., Daley G. Q., Sliz P., Gregory R. I. (2008) J. Biol. Chem. 283, 21310–21314 [DOI] [PubMed] [Google Scholar]
- 38. Okada Y., Matsumoto A., Shimazaki T., Enoki R., Koizumi A., Ishii S., Itoyama Y., Sobue G., Okano H. (2008) Stem Cells 26, 3086–3098 [DOI] [PubMed] [Google Scholar]
- 39. Chelsky D., Ralph R., Jonak G. (1989) Mol. Cell Biol. 9, 2487–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X. Y., Penalva L. O., Yuan H., Linnoila R. I., Lu J., Okano H., Glazer R. I. (2010) Mol. Cancer 9, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O'Sullivan M., Lu J., Phillips L. A., Lockhart V. L., Shah S. P., Tanwar P. S., Mermel C. H., Beroukhim R., Azam M., Teixeira J., Meyerson M., Hughes T. P., Llovet J. M., Radich J., Mullighan C. G., Golub T. R., Sorensen P. H., Daley G. Q. (2009) Nat. Genet. 41, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kanemura Y., Mori K., Sakakibara S., Fujikawa H., Hayashi H., Nakano A., Matsumoto T., Tamura K., Imai T., Ohnishi T., Fushiki S., Nakamura Y., Yamasaki M., Okano H., Arita N. (2001) Differentiation 68, 141–152 [DOI] [PubMed] [Google Scholar]
- 43. Toda M., Iizuka Y., Yu W., Imai T., Ikeda E., Yoshida K., Kawase T., Kawakami Y., Okano H., Uyemura K. (2001) Glia 34, 1–7 [DOI] [PubMed] [Google Scholar]
- 44. Yasuhara N., Shibazaki N., Tanaka S., Nagai M., Kamikawa Y., Oe S., Asally M., Kamachi Y., Kondoh H., Yoneda Y. (2007) Nat. Cell Biol. 9, 72–79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.