Abstract
Wilson disease (WD) is a disorder of copper metabolism caused by mutations in the Cu-transporting ATPase ATP7B. WD is characterized by significant phenotypic variability, the molecular basis of which is poorly understood. The E1064A mutation in the N-domain of ATP7B was previously shown to disrupt ATP binding. We have now determined, by NMR, the structure of the N-domain containing this mutation and compared properties of E1064A and H1069Q, another mutant with impaired ATP binding. The E1064A mutation does not change the overall fold of the N-domain. However, the position of the α1,α2-helical hairpin (α-HH) that houses Glu1064 and His1069 is altered. The α-HH movement produces a more open structure compared with the wild-type ATP-bound form and misaligns ATP coordinating residues, thus explaining complete loss of ATP binding. In the cell, neither the stability nor targeting of ATP7B-E1064A to the trans-Golgi network differs significantly from the wild type. This is in a contrast to the H1069Q mutation within the same α-HH, which greatly destabilizes protein both in vitro and in cells. The difference between two mutants can be linked to a lower stability of the α-HH in the H1069Q variant at the physiological temperature. We conclude that the structural stability of the N-domain rather than the loss of ATP binding plays a defining role in the ability of ATP7B to reach the trans-Golgi network, thus contributing to phenotypic variability in WD.
Keywords: ATPases, Copper, Membrane Proteins, NMR, Protein Stability, Protein Structure, Protein Targeting, ATP7B, Wilson Disease
Introduction
The human copper-transporting ATPase ATP7B regulates copper levels in the cytosol and within the secretory pathway (1). Genetic mutations that disrupt the transport function of ATP7B lead to Wilson disease (WD),4 a severe metabolic disorder associated with copper accumulation in the liver, brain, and other tissues (2). The WD phenotypes are rather diverse. The disease is characterized by significant variability in the age of onset, severity, and manifestations, which include hepatic, neurological, and psychiatric symptoms. More than 350 WD-causing mutations have been described. The majority of these mutations are single base pair substitutions that do not eliminate synthesis of the full-length ATP7B. Despite considerable effort, no strong correlations have been observed between specific mutations and the disease phenotype, necessitating better understanding of the effects that mutations have on ATP7B structure, function, and intracellular behavior.
ATP7B transports copper from the cytosol across cellular membranes using the energy of ATP hydrolysis. ATP binds to the cytosolic ATP-hydrolyzing domain, which consists of the N- and P-domains. The bulk of the ATP molecule is bound to the N-domain (3, 4), whereas the γ-phosphate extends to the P-domain to transiently phosphorylate the invariant aspartate residue Asp1027. The structure of the N-domain has been solved for human ATP7B (4) and several orthologs (5–8). These studies have identified invariant residues contributing to the ATP binding pocket. In ATP7B, these residues are Glu1064, Glu1068, His1069, Gly1101, Asn1150, and Gly1099 (3). The recent high resolution structure of ATP-hydrolyzing domain of CopA, a Cu-ATPase from a hyperthermophile, Archaeoglobus fulgidus, illuminated specific roles of these residues in coordinating ATP (8). By analogy with CopA (8), one can predict that in ATP7B the side chain carboxyl of Glu1064 forms hydrogen bonds with N1 and N6 of adenine, and the imidazole ring of His1069 hydrogen bonds to the α- and β-phosphates of ATP. The crystal structures of ATP-hydrolyzing domain from CopA did not show significant changes upon binding of ATP (8). In contrast, NMR analysis of the N-domains from human ATP7B and ATP7A suggested protein flexibility (4, 5). In solution, ATP binding to the N-domain appears to produce a more compact and stable structure (4, 5), an observation further supported by recent thermal denaturation and molecular dynamics simulation experiments (9).
The N-domain is the site of >30 missense mutations associated with WD. Consistent with their critical role in coordinating ATP, mutations of either Glu1064 (to Ala or Lys) or His1069 (to Gln) result in WD (10–14). The H1069Q mutation has attracted particularly close attention because it is one of the most common mutations in several major world populations. In vitro, the H1069Q substitution decreases the affinity for ATP and prevents the formation of a catalytic phosphointermediate (3, 4, 9, 15). In the cell, this mutation is associated with mistargeting of ATP7B. Normally, ATP7B is localized to the trans-Golgi network (TGN) where it delivers copper into the lumen of this compartment for biosynthetic incorporation into copper-dependent enzymes. The H1069Q mutant was found in the endoplasmic reticulum (ER) at the physiological temperature of 37 °C (16–18), and its targeting to the TGN was only observed when the protein was expressed at 28 °C or 30 °C (17, 18). Moreover, the H1069Q mutant has a decreased half-life compared with the wild-type ATP7B (17). The reasons for the retention of H1069Q mutant in the ER and its shorter half-life are not clear because the mutation does not cause noticeable structural defects in either the N-domain or the full-length ATP7B (4, 15). Similarly, in archaean CopA, an equivalent mutation was shown to change the coordination mode of ATP without affecting protein structure (8).
These observations led to a hypothesis that diminished ATP binding to the H1069Q mutant precludes ATP-dependent conformational changes that may be required for ATP7B exit from the ER. To test this hypothesis we characterized in detail a new WD mutation, E1064A. Glu1064 is located in the vicinity of His1069 (see Fig. 1A), and the substitution of alanine for Glu1064 abolishes ATP binding to the N-domain (3). We solved the structure of the E1064A N-domain variant and determined the molecular basis for the loss of ATP binding by this mutant. Further comparison of the E1064A and H1069Q mutants in vitro and in cells revealed that the N-domain thermal stability, rather than its affinity for ATP, plays a defining role in the ability of ATP7B to exit the ER and reach the TGN.
FIGURE 1.
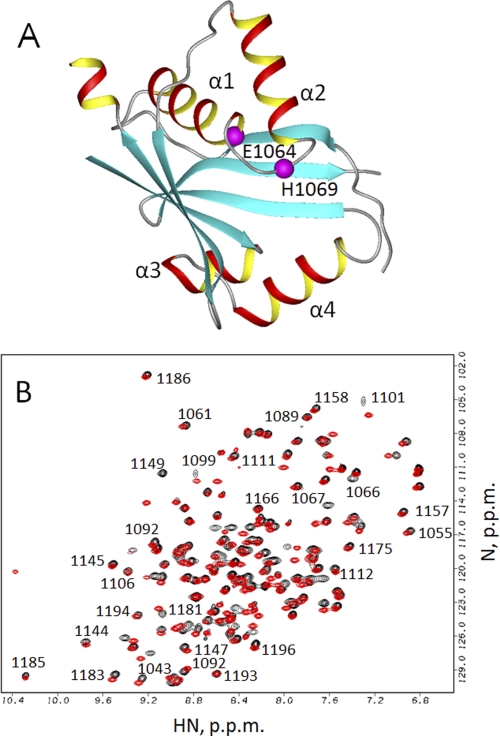
A, structure of the wild-type ATP7B N-domain (Protein Data Bank ID code 2ARF). Residues His1069 and Glu1064 are shown in magenta. The unstructured loop (residues 1115–1138) is not shown. B, overlay of the 1H,15N HSQC spectra of the Glu1064-WNDΔ1115–1138 (red) and Ala1064-WNDΔ1115–1138 (black). Some of the sequential assignments for Ala1064-WNDΔ1115–1138 are shown in the uncrowded regions of the spectrum.
EXPERIMENTAL PROCEDURES
Protein Expression for Structural Analysis
The N-domain variants carrying the E1064A mutation were expressed and purified as described previously (3). For NMR structure determination, the E1064A N-domain sequence was modified to delete the disordered loop comprising residues 1115–1138 as described for the wild-type protein (20). For isotope labeling, [13C]glucose and 15NH4Cl (Cambridge Isotope Laboratories) were used. Protein purity was checked by SDS-PAGE (supplemental Fig. 1). NMR samples contained 0.7–1.0 mm protein in 50 mm sodium phosphate buffer, pH 6.0, 5% (v/v) D2O, 5 mm DTT, 0.05 mm NaN3, and 0.5 mm 2,2-dimethyl-2-silapentane-5-sulfonic acid for chemical shift referencing.
NMR Spectroscopy and Structure Calculation
Most NMR experiments were performed on a 600 MHz Bruker spectrometer equipped with a cryoprobe with z axis pulse field gradients. NMR data collection and processing were performed essentially as described previously for the wild-type N-domain (4). Briefly, the backbone chemical shift assignments were made from HNCO, HNCA, HNCACB, HN(CO)CACB, and HN(CA)CO data. Side chain chemical shift assignments were made from three-dimensional H(CCO)NH, CC(O)NH, and 1H,15N total correlation spectroscopy experiments. Distance restraints were derived from the three-dimensional 1H,15N NOESY and three-dimensional 1H,13C NOESY experiments. The structure was validated using Protein Structure Validation Software Suite (PSVS) and deposited in the Protein Data Bank as entry 2KOY.
Temperature Stability of Recombinant N-domains
Thermal denaturation curves were recorded by measuring peak volumes of the signals of the selected backbone amide groups in the folded protein in an 1H,15N HSQC experiment as a function of temperature. Sample conditions were the same as for structure determination.
Generation of GFP-tagged E1064A and H1069Q Mutants of ATP7B
The previously described plasmid pYG7 encoding the wild-type ATP7B with a GFP tag at the N terminus (19) was used as a template. Mutations were introduced using the QuikChange XL Site-directed Mutagenesis kit (Stratagene) and the following primers: 5′-GGCTGTGGTGGGGACTGCGGCGGCCAGCAGTGAACACCCC-3′ and 5′-GGGGTGTTCACTGCTGGCCGCCGCAGTCCCCACCACAGCC-3′ for E1064A and 5′-GCGGAGGCCAGCAGTGAACAGCCCTTGGGCGTGGCAGTCAC-3′ and 5′-GTGACTGCCACGCCCAAGGGCTGTTCACTGCTGGCCTCCGC-3′ for H1069Q according to the manufacturer's protocol. The presence of desired mutations and the lack of unwanted mutations were confirmed by sequencing of the entire cDNA region.
Immunocytochemistry
Human embryonic kidney cell line HEK293TRex was maintained in minimum Eagle's medium supplemented with 10% fetal bovine serum, nonessential amino acids, penicillin, and streptomycin. For immunostaining, cells were cultured on coverslips in a 6-well plate in a CO2 incubator. Transfection was carried out at 37 °C with 2.5 μg of plasmid DNA using Turbofect (Fermentas). Six hours after transfection, coverslips with cells were transferred into a fresh medium and incubated at either 37 °C or 28 °C for 7–12 h. Cells were rinsed with PBS and fixed with a chilled acetone:methanol (1:1) fixing solution for 30 s, washed with PBS, and blocked in PBS with 1% gelatin and 1% BSA at 4 °C. Cells were then stained with the anti-TGN46 primary antibody (sheep polyclonal, 1:150; Gentex), Alexa Fluor 555 donkey anti-sheep antibody (1:250; Invitrogen) or anti-calnexin antibody (rabbit polyclonal, 1:150; Stressgen), Alexa Fluor 555 goat anti-rabbit antibody (1:500; Invitrogen) in a moist chamber. Images were analyzed using a 100× oil lens and a Pascal Confocal Scanning microscope (Carl Zeiss, Germany).
Western Blot Analysis
HEK293TRex cells were seeded on 6-cm Petri dishes and transfected with 5 μg of DNA using Turbofect (Fermentas). Cells were harvested, washed with PBS, and homogenized in the lysis buffer containing 25 mm imidazole, pH 7.5, 0.05 mm DTT, 2 mm AEBSF, 250 mm sucrose, protease inhibitors (inhibitor tablet; Roche Applied Science) in a Dounce homogenizer. The homogenate was centrifuged at 600 × g for 10 min at 4 °C. The supernatant was collected and centrifuged at 3000 × g for 10 min at 4 °C to remove mitochondria. The resulting supernatant was centrifuged at 20,000 × g for 30 min, and the pellet was resuspended in PBS containing 0.2% Triton X-100 and protease inhibitor (Roche Applied Science). Protein concentration was determined by Lowry assay, and equal amounts of membrane protein (total 3.6 μg for each wild-type or mutant sample) were analyzed on a 8.5% Laemmli gel. Following transfer to PVDF membrane, ATP7B was detected using rabbit anti-GFP antibody (Invitrogen). Equal protein loading was verified using staining with the mouse monoclonal anti-Na+/K+-ATPase α1 antibody (Millipore) and the HRP-conjugated goat anti-rabbit IgG. Intensity of protein bands was quantified by densitometry in three independent experiments and normalized to the Na+/K+-ATPase levels.
RNA Isolation for Real-time PCR
HEK293TRex cells were plated on 6-cm Petri dishes and transfected with 5 μg of DNA using Turbofect (Fermentas). Cells were harvested, washed with PBS, and RNA was extracted using an RNeasy mini kit (Qiagen). The cDNA was prepared by reverse transcription using 1 μg of RNA in a final volume of 20 μl using combination of both anchored oligo(dT)18 and random hexamer primer provided with a Transcriptor First Strand cDNA synthesis kit (Roche Applied Science) using the manufacturer's instructions.
Real-time PCR was performed using 1 μl of the prepared cDNA in a 20-μl reaction volume using Power SYBR Green PCR Master Mix (Applied Biosystems). The amounts of ATP7B transcript were measured using primers flanking the 5′ GFP of the construct to avoid a signal from endogenous ATP7B mRNA. The forward primer was 5′-ACGTAAACGGCCACAAGTTC-3′ and reverse primer 5′-AAGTCGTGCTGCTTCATGTG-3′. The reaction conditions included initial denaturation at 95 °C for 10 min followed by 40 cycles of PCR, which included 15-s melting at 95 °C and 1-min annealing/extension at 60 °C. At the end of the extension step, fluorescence of each sample was measured to allow quantification. The abundance of target mRNA in each case (wild type, E1064A, and H1069Q) was calculated in relation to the β-actin mRNA in the same sample. The amount of target mRNA relative to wild type was quantified using 2−ΔΔCT method.
RESULTS
WND-E1064AΔ1115–1138 Construct as a Model for Structure Determination
We have previously shown by isothermal titration calorimetry that the E1064A mutant of the N-domain (WND-E1064A) does not bind ATP (3). To estimate the extent of structural changes due to E1064A substitution, we first compared the 1H,15N HSQC spectra for the WND-E1064A and the wild-type N-domain in the absence of added nucleotide.
Overall similarity of these spectra (supplemental Fig. 2) indicates that the E1064A mutation does not alter the gross architecture of the N-domain (Fig. 1A), although a large number of subtle spectral changes suggested a possibility of local structural changes. Unexpectedly, in contrast to the wild-type N-domain, which is unstable in the apo state and aggregates in a matter of days, the WND-E1064A protein was stable in solution for weeks, as monitored by 1H,15N HSQC spectroscopy (data not shown). This observation suggested that some structural changes did occur as a result of mutation, which stabilized the protein and enabled us to proceed with structure determination.
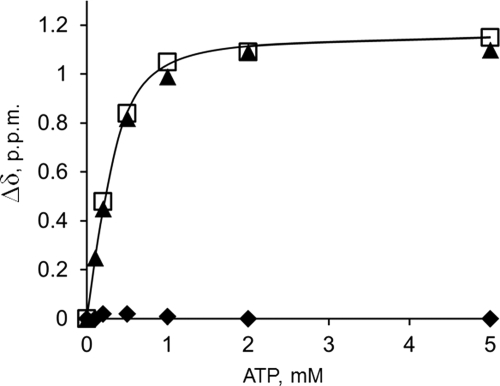
To facilitate structural analysis of WND-E1064A, we deleted residues 1115–1138, generating the construct WND-E1064A-Δ1115–1138. We have previously shown that the 1115–1138 region forms an unstructured loop (4). We reasoned that the deletion would eliminate strong and poorly dispersed signals from the residues within the loop, which complicate chemical shift and NOE assignments and produce spectra of better quality (Fig. 1B). The nucleotide titration experiments confirmed that the ATP binding affinity of the N-domain with the deletion (WND-Δ1115–1138)was indistinguishable from the wild type (Fig. 2). Also, similar to WND-E1064A, the E1064A mutation in the deletion variant led to a complete loss of ATP binding. Therefore we used the WND-E1064AΔ1115–1138 (further WND-E1064AΔ) to solve its structure by multidimensional NMR.
FIGURE 2.
Deletion of the unstructured loop 1115–1138 does not alter nucleotide binding characteristics of the N-domain. ATP binding to the wild-type N-domain (open squares), Glu1064-WNDΔ1115–1138 (triangles), and Ala1064-WNDΔ1115–1138 (diamonds) was monitored by measuring compound chemical shift change of the backbone amide group of Gly1101.
Structure of WND-E1064AΔ
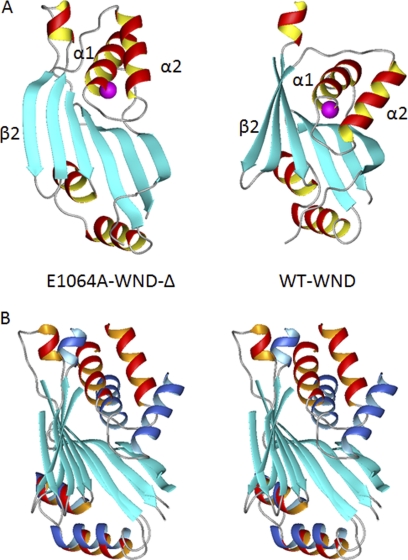
The solution structure of the Glu1064 mutant is shown in Fig. 3A. The final ensemble of 20 conformers of WND-E1064AΔ (supplemental Fig. 3) has an average backbone r.m.s.d. value of 1.35 ± 0.23Å over the residues corresponding to the 1038–1114 and 1139–1195 segments of the full-length ATP7B (the statistics data are given in supplemental Table 1). The WND-E1064AΔ structure consists of a six-stranded antiparallel β-sheet with two adjacent α-helical hairpins. The overall fold of the mutant N-domain is similar to that of the wild-type protein, and in the immediate vicinity of the E1064A mutation structural changes are minor. However, other regions of the WND-E1064AΔ domain show distinct structural differences when compared with the wild-type N-domain.
FIGURE 3.
Position of the α1–α2 hairpin differs between the wild-type WND and E1064A mutant. A, ribbon diagram of the E1064A-WNDΔ (left) and wild-type N-domain (right), with Glu/Ala1064 shown in magenta. B, stereo view of the wild-type N-domain (α-helices shown in blue) and the E1064A-WNDΔ (α-helices shown in red) aligned to minimize r.m.s.d. between the core β-sheets (cyan). The disordered loop in the wild-type N-domain is not shown (A and B).
The folded cores of the two N-domains have backbone ensemble r.m.s.d. of 4.7 Å. This difference is largely due to changes in the twist of the β-sheet and the orientation of both α-helical hairpins (Fig. 3A). The mean structures were aligned to minimize r.m.s.d. for the β-sheet cores (Fig. 3B). The greatest difference was then found in the orientation of the α1–α2 hairpin (residues 1053–1084). Although the β-sheets of the two proteins show the backbone r.m.s.d. of 2.2 Å and the α3–α4 hairpins (residues 1151–1173) have r.m.s.d. of 3.3 Å, the α1–α2 hairpins have the most dissimilar orientation resulting in r.m.s.d. of 8.9 Å (Fig. 3B).
Structure of WND-E1064AΔ Explains the Large Effect of the E1064A Mutation on ATP Binding
The less pronounced twist of the β-sheet together with the repositioning of the α1–α2 hairpin produce a more open conformation of the N-domain. Repositioning of the α1–α2 hairpin in the E1064A mutant also displaces the Cα atoms of His1069 and Ala1064 from their positions in the wild-type N-domain by ∼5.5 Å and 4 Å, respectively. As a result, in addition to the loss of hydrogen bonding to adenine due to E1064A mutation, further loss in affinity is caused by the movement of His1069 farther away from the β-sheet, which would preclude hydrogen bond formation with the phosphate groups of ATP (supplemental Fig. 4). The displacement of two invariant residues normally involved in ATP coordination explains large effect of E1064A mutation on ATP binding.
α1–α2 Hairpin Is the Most Variable Structural Element of the N-domain
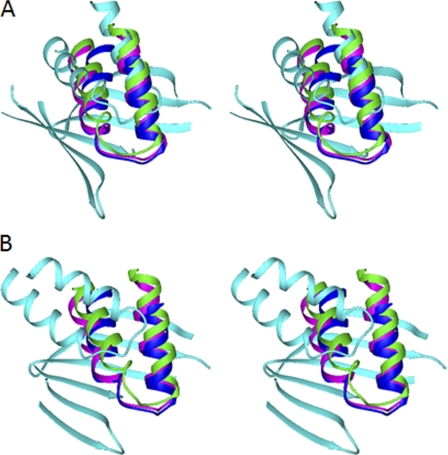
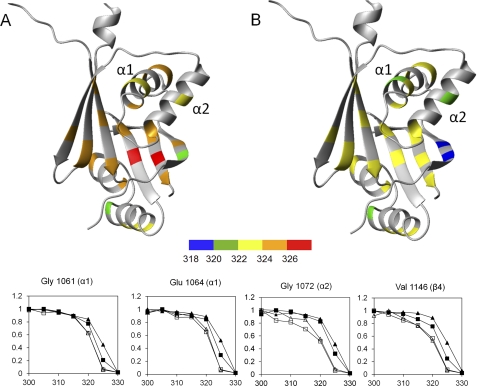
The structure of the ATP7B apo-N-domain is not yet available due to protein instability. Consequently, it was not clear whether the structure of the E1064A mutant simply represents the apo form of the N-domain or whether additional structural changes had been introduced by the mutation. To address this issue, we compared the structure of WND-E1064AΔ with those of the apo N-domains of ATP7A, CopA, and CopB. ATP7B and ATP7A share 62% sequence identity in their folded cores and are the closest structural and functional homologs, whereas sequence identity with the CopA and CopB N-domains is ∼30%. The apo structures of the N-domain orthologs, although overall very similar, all differ in the position of the α1–α2 hairpin with respect to the core (Fig. 4). This observation strongly suggests the conformational mobility of the hairpin. A conformational change was also detected in the ATP7A N-domain in response to ATP binding (5). In the E1064A mutant, the orientation of the α1–α2 hairpin is further altered and is clearly outside the range of the variations between the wild-type apo domains.
FIGURE 4.
Conformation of the α1–α2 hairpin in the N-domains of copper-transporting ATPases. Stereo view is shown. The structures of the N-domains of ATP7A (green, 2KMX), CopA from A. fulgidus (blue, 2B8E), and CopB from Sulfolobus solfataricus (magenta, 2IYE) were aligned with the structure of the wild-type WND (A; cyan, 2ARF) or E1064A-WND (B; cyan, 2KOY) to minimize r.m.s.d. between the core β-sheets. Only the α1–α2 hairpins in the four structures and the core β-sheet in WND are shown.
Intracellular Targeting of the E1064A Mutant Is Similar to That of the Wild-type ATP7B
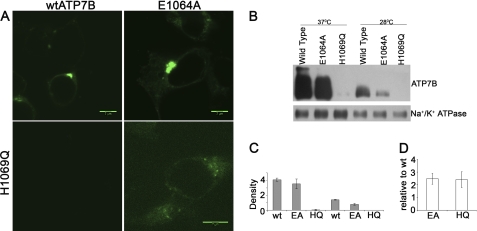
Previously, the loss of function due to the H1069Q mutation was linked to the entrapment of ATP7B in the ER and decreased half-life. Because Glu1064 is in the vicinity of His1069 and the E1064A replacement also results in the loss of ATP binding, it was interesting to examine whether in cells the ATP7B-E1064A mutant is retained in the ER. Consequently, we introduced the corresponding mutation into the full-length ATP7B containing a GFP tag at the N terminus. The wild-type GFP-tagged ATP7B was previously shown to have normal TGN targeting and function (19).
The GFP-ATP7B and GFP-ATP7B-E1064A mutants were expressed in the HEK293Trex cells, and the protein localization was examined by confocal microscopy. The fluorescence patterns were very similar for these two proteins (Fig. 5A). Tight perinuclear localization characteristic of TGN was apparent in both cases. The localization in TGN was verified by co-staining with the organelle marker TGN-46 (supplemental Fig. 5). Thus, the E1064A mutation has little, if any, effect on protein targeting. The number of cells expressing the GFP-ATP7B-E1064A mutant and the intensity of fluorescence signal in these cells were also comparable with those of the wild-type GFP-ATP7B, suggesting that the protein expression was not greatly affected by the E1064A substitution. This conclusion was confirmed by the Western blot analysis of membrane preparations (Fig. 5, B and C).
FIGURE 5.
Comparison of intracellular localization, protein, and transcript abundance for the ATP7B mutants. A, HEK293TRex cells were transfected with plasmids expressing GFP-tagged wild-type ATP7B (upper left), the E1064A mutant (upper right), or the ATP7B H1069Q mutant (lower panels). Images for the WT, E1064A, and H1069Q (lower left) were obtained using the same detector gain. Higher detector gain (lower right) was used to visualize the H1069Q mutant. B, microsomal membrane preparations were isolated from cells first transfected with the wild-type ATP7B or mutants at 37 °C and then grown at either 37 °C or 28 °C. Tagged ATP7B was detected using anti-GFP antibody; equal protein loading was verified using anti-Na+,K+-ATPase antibody. A representative Western blot is shown. C, densitometry data are shown for three independent Western blotting experiments. D, total mRNA was isolated from cells transfected with the wild-type (wt) ATP7B or ATP7B mutants, as in A, and the amount of transcripts relative to wild-type mRNA was determined by real-time PCR. The average of three independent experiments is shown.
E1069A and H1069Q Mutants Show Marked Difference in Cellular Protein Levels
The intracellular properties of ATP7B-E1064A were markedly different from the previously reported properties of the H1069Q mutant. This result was unexpected because the in vitro effect of the E1064A mutation on ATP binding to the N-domain was more severe than that of H1069Q. Because the cell lines and the detection tags used in previous studies were similar but not identical to ours, we compared the two mutants side by side. Under the same experimental conditions, the H1069Q mutant showed much lower level of expression (Fig. 5A, lower left), and a higher gain during data collection was necessary to detect the protein (Fig. 5A, lower right). We attempted to improve the H1069Q protein stability by growing the cells at 28 °C. In agreement with previous reports, partial localization in the TGN was observed for H1069Q during prolonged expression at 28 °C. However, at either 28 °C or 37 °C significant ER staining was apparent (supplemental Fig. 6A) in addition to the TGN localization (supplemental Fig. 6B) and with a shorter post-transfection period, the H1069Q mutant still showed significantly less protein compared with the wild type. Low protein levels of the H1069Q mutant at either temperature were confirmed by Western blot analysis (Fig. 5, B and C). To verify further that this difference in protein amounts was not due to varied transfection efficiency and/or mRNA levels, we quantified the amounts of E1064A and H1069Q encoding transcripts using real-time PCR. These experiments demonstrated very similar levels of mRNA for both mutants (Fig. 5D).
Differences in the N-domain Stability Correlate with the Intracellular Properties of E1064A and H1069Q Mutants
The strikingly different behavior of the two mutants indicated that the loss of ATP binding alone was not critical either for the TGN targeting or for the intracellular levels of ATP7B and that other factors played an important role in defining the mutants behavior. We considered that low stability of the H1069Q mutant could be due to disruption of important contacts between the N-domain and the other domains in ATP7B, whereas the E1064A mutation did not affect these contacts. However, this hypothesis was not well supported by the experimental data because both the structure and the nucleotide binding properties of E1064A mutant differ more from the wild type than those of H1069Q. Alternatively, we hypothesized that the E1064A and H1069Q mutations could have different effect on thermal motions and stability of the N-domain, which in turn governed the behavior of the full-length ATP7B in the cell.
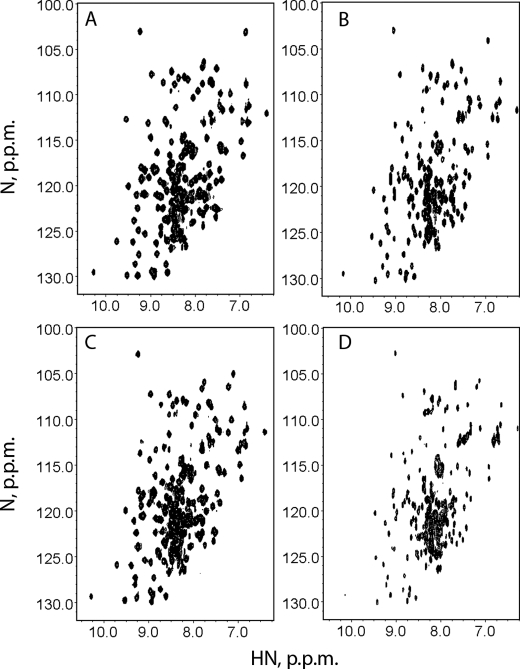
To explore this possibility we first compared the HSQC spectra of mutant N-domains with the wild-type protein at 300 K (27 °C) and then investigated stability of the individual structural elements in the N-domain by monitoring intensity of backbone amide signals in the folded form of the protein as a function of temperature (Fig. 6). As reported previously (9), both the wild type and H1069Q variant were stabilized by ATP binding. At 300 K in the presence of ATP, both N-domains have excellent backbone amide chemical shift dispersion (Fig. 6, A and C). As the temperature increased, the H1069Q mutant was found to be significantly less stable than the wild type. The fingerprint spectrum of the H1069Q mutant showed protein unfolding at 325 K (Fig. 6D), whereas the spectrum of the wild type was still dominated by the folded protein (Fig. 6B). In contrast, the stability of the E1064A variant did not differ significantly from the wild-type N-domain (data not shown). Detailed analysis of the temperature-dependent chemical shifts for the wild type and the H1069Q variant revealed that the most pronounced change in the stability of individual structural elements occurs in the α1-helix adjacent to Gln1069 and in the proximal region of the β-sheet. The midpoints of thermal denaturation curves in these regions lie about 5 K lower for the H1069Q than for the wild type (Fig. 7). These data suggest that the temperature-dependent unfolding of the α1–α2 hairpin is the initial step of the H1069Q N-domain denaturation, which then facilitates unraveling of the core β-sheet.
FIGURE 6.
H1069Q-WND is less stable than the wild type. The 1H,15N HSQC spectra of the wild type N-domain (A and B) and the H1069Q variant (C and D) were recorded at 300 K (A and C) and 325 K (B and D) in the presence of 1 mm ATP.
FIGURE 7.
Stability of the structural elements in the wild type and H1069Q-WND. A and B, midpoints of thermal denaturation curves were recorded by measuring volumes of the HSQC peaks of individual backbone amide groups in the folded forms of the wild type (A) and the H1069Q variant (B). Residues with large folding-dependent secondary chemical shifts that produced well separated peaks in the HSQC spectra (4) were chosen as reporter groups. C, relative peak volumes of several selected backbone amide groups in the wild-type WND (triangles) and the H1069Q-WND (squares) in the apo form (open symbols) and in the presence of 1 mm ATP (filled symbols) were recorded as a function of sample temperature (K).
DISCUSSION
In this study, we provide a detailed characterization of the new WD-causing mutant E1064A. Our results identify structural basis for the marked decrease of ATP binding to the N-domain caused by the E1064A substitution. In the cell, the E1064A mutant of ATP7B is stable and is properly targeted to the TGN, unlike the previously characterized H1069Q mutant. By comparing these two mutants in vitro and in the cells, we have found that their different intracellular behavior is not due to the loss of ATP binding, but rather to different effects of mutations on the stability of the N-domain. Altogether, the results provide better understanding of factors that influence ATP7B function in the cell and cause variable phenotypic manifestations in WD.
NMR structures of the wild-type N-domains from human ATP7A and ATP7B both show ATP-dependent conformational changes, whereas the x-ray structure of CopA does not. A likely explanation is that in solution, there is an equilibrium between an open and closed conformations of the N-domain. ATP binding locks the protein in the closed conformation and so does crystallization under the conditions used for the CopA ATP binding domain. In the N-domain of ATP7A, which is highly homologous to ATP7B, ATP binding primarily affects the loop connecting the α1–α2 helices and, to a lesser extent, the orientation of these helices (5). In contrast, in the structure of WND-E1064AΔ, the α1–α2 hairpin is markedly displaced compared with its position in the wild-type N-domain.
How does the E1064A substitution cause additional conformational changes of the α1–α2 hairpin? Analysis of the CopA N-domain structures with and without AMP-PCP provides some clues. In these structures, the positions of Glu457 (Glu1064 in ATP7B) are almost identical with r.m.s.d. of the side chain atoms about 0.3 Å, despite the formation of two new hydrogen bonds between the Glu457 carboxyl and the adenine ring of AMP-PCP. This unusual rigidity is apparently due to steric constraints imposed on Glu457 by neighboring bulky side chains. Substitution of β-methyl group of alanine for the much bulkier glutamate side chain may disrupt this tight packing and result in conformational change in the α1–α2 hairpin.
Our comparison of the E1064A and H1069Q mutants has also yielded insights into factors that influence the ER-to-TGN trafficking step of ATP7B. The data strongly suggest that the binding of ATP is not required for protein exit from the ER, whereas the dynamics and folding state of the N-domain play very important role in the ability of ATP7B to reach the TGN. Considering that the N-domain is cytosolic and that the ATP7B loops exposed into the ER lumen are very short, one has to conclude that the quality control machinery that regulates the ATP7B levels in cells resides in the cytosol or inside the ER membrane rather than in the ER lumen.
The H1069Q mutation is the most frequent WD causing mutation in the Caucasian population. The exact cause of impaired enzyme activity in this variant has been a matter of some controversy. The structure of the N-domain is unaffected by this mutation (3, 4, 8), but the ATP binding affinity is reduced by about an order of magnitude (Kd of approximately 1 mm). Because the ATP concentration in the cell is in the millimolar range and the full-length ATP7B has a higher affinity than the isolated N-domain, the drop in ATP binding affinity alone is unlikely to eliminate the ATP7B activity fully. The x-ray structures and molecular modeling experiments suggested that ATP may bind to the H1069Q N-domain in an incorrect orientation (8, 9), a suggestion that better explains the inability of the ATP7B H1069Q mutant to hydrolyze ATP (15). Our data lend support to an earlier observation (17) that in addition to these functional changes, the H1069Q variant of ATP7B is unstable due to temperature-dependent unfolding. We have investigated stability of individual structural elements in the N-domain by monitoring backbone amide chemical shift changes as a function of temperature. We found no significant difference in stability between the wild type and E1064A mutant. In contrast, in the H1069Q N-domain, the α1–α2 hairpin was markedly less stable than in the wild type. Unfolding of the α1–α2 hairpin in H1069Q-ATP7B may destabilize the N-domain and eventually lead to misfolding and degradation of the mutant protein in the cells.
Supplementary Material
Acknowledgments
We thank Chris O'Grady for technical assistance and Dr. Hubbard and Dr. Braiterman for the GFP-tagged ATP7B. NMR experiments were performed at the Saskatchewan Structural Sciences Center and the Center for Structural Biology at the University of Illinois, Chicago.
This work was supported by grants from Canadian Institutes of Health Research and Saskatchewan Health Research Foundation (to O. Y. D.). This work was also supported in part by National Institutes of Health Grants R01 DK071865 and P01GM067166 (to S. L.). The study also made use of the National Magnetic Resonance Facility at Madison, which is supported by funding from National Institutes of Health, NSF, United States Department of Agriculture, and the University of Wisconsin.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
The atomic coordinates and structure factors (code 2KOY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- WD
- Wilson disease
- AMP-PCP
- adenosine 5′-(β,γ-methylenetriphosphate)
- ER
- endoplasmic reticulum
- HSQC
- heteronuclear single quantum coherence
- r.m.s.d.
- root mean square deviation
- WND
- Wilson disease protein N-domain
- TGN
- trans-Golgi network.
REFERENCES
- 1. Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 2. Das S. K., Ray K. (2006) Nat. Clin. Pract. Neurol. 2, 482–493 [DOI] [PubMed] [Google Scholar]
- 3. Morgan C. T., Tsivkovskii R., Kosinsky Y. A., Efremov R. G., Lutsenko S. (2004) J. Biol. Chem. 279, 36363–36371 [DOI] [PubMed] [Google Scholar]
- 4. Dmitriev O., Tsivkovskii R., Abildgaard F., Morgan C. T., Markley J. L., Lutsenko S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banci L., Bertini I., Cantini F., Inagaki S., Migliardi M., Rosato A. (2010) J. Biol. Chem. 285, 2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lübben M., Güldenhaupt J., Zoltner M., Deigweiher K., Haebel P., Urbanke C., Scheidig A. J. (2007) J. Mol. Biol. 369, 368–385 [DOI] [PubMed] [Google Scholar]
- 7. Sazinsky M. H., Mandal A. K., Argüello J. M., Rosenzweig A. C. (2006) J. Biol. Chem. 281, 11161–11166 [DOI] [PubMed] [Google Scholar]
- 8. Tsuda T., Toyoshima C. (2009) EMBO J. 28, 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodriguez-Granillo A., Sedlak E., Wittung-Stafshede P. (2008) J. Mol. Biol. 383, 1097–1111 [DOI] [PubMed] [Google Scholar]
- 10. Caca K., Ferenci P., Kühn H. J., Polli C., Willgerodt H., Kunath B., Hermann W., Mössner J., Berr F. (2001) J. Hepatol. 35, 575–581 [DOI] [PubMed] [Google Scholar]
- 11. Cauza E., Ulrich-Pur H., Polli C., Gangl A., Ferenci P. (2000) Wien Klin. Wochenschr. 112, 576–579 [PubMed] [Google Scholar]
- 12. Stapelbroek J. M., Bollen C. W., van Amstel J. K., van Erpecum K. J., van Hattum J., van den Berg L. H., Klomp L. W., Houwen R. H. (2004) J. Hepatol. 41, 758–763 [DOI] [PubMed] [Google Scholar]
- 13. Kalinsky H., Funes A., Zeldin A., Pel-Or Y., Korostishevsky M., Gershoni-Baruch R., Farrer L. A., Bonne-Tamir B. (1998) Hum. Mutat. 11, 145–151 [DOI] [PubMed] [Google Scholar]
- 14. Folhoffer A., Horváth A., Hegedüs D., Firneisz G., Dunkel K., Willheim C., Ferenci P., Szönyi L., Abonyi M., Lakatos P. L., Szalay F. (2003) Orv. Hetil. 144, 2509–2515 [PubMed] [Google Scholar]
- 15. Tsivkovskii R., Efremov R. G., Lutsenko S. (2003) J. Biol. Chem. 278, 13302–13308 [DOI] [PubMed] [Google Scholar]
- 16. Huster D., Hoppert M., Lutsenko S., Zinke J., Lehmann C., Mössner J., Berr F., Caca K. (2003) Gastroenterology 124, 335–345 [DOI] [PubMed] [Google Scholar]
- 17. Payne A. S., Kelly E. J., Gitlin J. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van den Berghe P. V., Stapelbroek J. M., Krieger E., de Bie P., van de Graaf S. F., de Groot R. E., van Beurden E., Spijker E., Houwen R. H., Berger R., Klomp L. W. (2009) Hepatology 50, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 19. Guo Y., Nyasae L., Braiterman L. T., Hubbard A. L. (2005) Am. J. Physiol. Gastrointest. Liver Physiol. 289, G904–916 [DOI] [PubMed] [Google Scholar]
- 20. Liu L., O'Grady C., Dalrymple S. A., Prasad L., Dmitriev O. Y., Delbaere L. T. (2009) Acta Crystallogr. Sect. F. 65, 621–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.