Abstract
C1q is the recognition subunit of the first component of the classical complement pathway. It participates in clearance of immune complexes and apoptotic cells as well as in defense against pathogens. Inappropriate activation of the complement contributes to cellular and tissue damage in different pathologies, urging the need for the development of therapeutic agents that are able to inhibit the complement system. In this study, we report heme as an inhibitor of C1q. Exposure of C1q to heme significantly reduced the activation of the classical complement pathway, mediated by C-reactive protein (CRP) and IgG. Interaction analyses revealed that heme reduces the binding of C1q to CRP and IgG. Furthermore, we demonstrated that the inhibition of C1q interactions results from a direct binding of heme to C1q. Formation of complex of heme with C1q caused changes in the mechanism of recognition of IgG and CRP. Taken together, our data suggest that heme is a natural negative regulator of the classical complement pathway at the level of C1q. Heme may play a role at sites of excessive tissue damage and hemolysis where large amounts of free heme are released.
Keywords: Complement, Heme, Immunochemistry, Protein-Protein Interactions, Thermodynamics, C-reactive Protein, IgG, C1q, Complement Inhibitors
Introduction
The complement system is among the first lines of defense against pathogens and is a sensor for altered self (1, 2). A set of recognition molecules detect foreign and altered self structures and trigger one of the three complement pathways, referred to as classical, lectin, and alternative (3). The three pathways merge at the level of the central component C3 to execute a common terminal pathway leading to inflammation, pathogen opsonization, and lysis. When the complement cascade is triggered on altered self, it is tightly regulated to proceed to clearance of apoptotic cells and debris without inducing inflammation (3, 4). The recognition molecule of the classical complement pathway C1q has a complex topology, consisting of six globular (gC1q)5 heterotrimer domains and a collagen-like region (5). C1q detects pathogen associated patterns (6). It also recognizes immunoglobulins (IgG and IgM) and pentraxins (such as C-reactive protein, CRP) (7) bound to their targets. CRP exists as a pentamer (8, 9) in solution and is considered to dissociate to monomers once bound to a membrane (10, 11). C1q was reported to recognize both forms of CRP. C1q also binds and participates in the clearance of host apoptotic cells and debris. Complement activation and regulation are balanced in physiological conditions, but complement dysregulation is responsible for severe tissue damage in a variety of pathological conditions. Therefore, specific inhibitors of complement are needed for clinical practice (12).
Pathological activation of complement has been associated with ischemic tissue after reperfusion, autoimmune hemolytic anemia, lupus nephritis, and etc. (13–15). In these conditions the alternative and/or lectin pathways are mostly involved. In these conditions, tissue or cellular damages are often accompanied by the release of large amounts of intracellular macromolecules and low molecular weight compounds including heme (16, 17). Heme (iron protoporphyrin IX) is a macrocyclic compound utilized as prosthetic group by many proteins for gas transport and oxidative metabolism. Tissue damage and hemolysis were shown to result in release of heme from hemoproteins such as hemoglobin, myoglobin, and cytochromes (16–19). Free heme is redox-active and cytotoxic (16, 20). Therefore, it must be scavenged and removed quickly from the circulation. This is achieved by heme-binding plasma proteins such as hemopexin, α-microglobulin and albumin (16). However, as a result of excessive tissue damage and hemolysis, heme-binding proteins are saturated, and high local and/or systemic concentrations of heme (>20 μm) may be achieved (18, 21). In such situations, heme accumulates in the endothelial cells membranes and plasma lipoproteins, imposing oxidative stress and inflammation (21, 22). Heme also interacts with various plasma proteins and modulates their functions (23–26). We have demonstrated previously that in vitro exposure of C1q to heme results in concentration-dependent inhibition of C1q binding to its main targets, IgG and CRP (27). The underlying mechanisms of this inhibition, however, remain poorly understood. In the present study, we investigated the functional aspect and the molecular mechanisms of inhibition of C1q by heme. We identify heme as an endogenous negative regulator of the classical complement pathway activation that acts at the level of C1q and may play a role at sites of excessive tissue damage and hemolysis.
EXPERIMENTAL PROCEDURES
Materials
The oxidized form of heme (ferriprotoporphyrin IX chloride) was obtained from Fluka (Taufkirchen, Germany), hematoporphyrin IX was obtained from Sigma-Aldrich. The Zn(II) protoporphyrin IX (Zn(II)PP), Mg(II)PP, Mn(III)PP, Ni(II)PP, Sn(IV)MP, Cr(IV)PP, and Co(III)PP were obtained from Frontier Scientific (Logan, UT). In all assays, ferriprotoporphyrin IX chloride was dissolved in dimethyl sulfoxide (designated in the text as hemin) or 0.05 n NaOH (designated as hematin) and kept in the dark before use. Human C1q isolated from plasma and human C-reactive proteins isolated from ascites were obtained by Calbiochem. Rituximab (Mabtera, Roche) was used as a source of human IgG1 with high purity. Endobulin (Baxter, Austria) was used as a source of pooled human IgG. Pneumococcal C-polysaccharide (PnC) was obtained from the Staten Serum Institute (Copenhagen, Denmark). Human serum depleted of C1q and the kit for determination of sC5b-9 (MicroVue sC5b-9 Plus EIA kit) were obtained from Quidel (San Diego, CA). All buffers were prepared with ACS reagent grade chemicals.
Assessment of Classical Complement Pathway Activation in Presence of Heme
The level of complement activation in presence of C1q, exposed to increased concentrations of hemin was evaluated by C3 deposition ELISA, after addition of C1q-depleted serum. Microtiter plates were coated with 10 μg/ml pneumococcal C-polysaccharide as a natural ligand for CRP or 5 μg/ml tetanus toxoid as an antigen for IgG-containing immune complex formation. After blocking with 1% BSA, the wells were washed with 10 mm Tris, pH 7.4, 140 mm NaCl, 5 mm CaCl2, 0.05% Tween 20 for the plate coated with PnC or PBS with 0.05% Tween 20 for the plate coated with tetanus toxoid. CRP was applied at 1 μg/ml in 1% BSA-containing washing buffer for 1 h at 37 °C. Pooled human IgG at 0.5 mg/ml was applied on the tetanus toxoid-coated plate to form immune complexes. Anti-tetanus toxoid antibodies are present in the immunoglobulin preparation due to common vaccination against tetanus. After washing of all plates with 10 mm Hepes, pH 7.4, 100 mm NaCl, 5 mm CaCl2, 1 mm MgCl2 0.05% Tween 20, intact or hemin-treated C1q was applied. For treatment of C1q with hemin, purified protein at 0.1 μm was incubated with increasing concentrations (0–15 μm) of hemin for 20 min at 4 °C in the dark. After incubation, the samples were diluted with washing buffer to a final C1q concentration of 1 nm (C1q molecular mass, 460 kDa) and applied at 50 μl/well to the microtiter plates. After incubation for 1 h at 37 °C, the plates were washed and either developed with anti-C1q-HRP (Abcam), or 50 μl of C1q-depleted serum (Quidel) diluted 1/400 with the washing buffer was added. The plates were then incubated for 1 h at 37 °C. After washing, the plates were incubated with biotinilated rabbit polyclonal anti-C3 antibody (diluted 1/500) or chicken anti-human C3 (Biodesign, diluted 1/10,000) for 1 h at 37 °C, followed by incubation with streptavidin-HRP for 30 min or goat anti-chicken HRP at 37 °C. Alternatively, anti-membrane attack complex (MAC) neoantigen-specific antibody-HRP (Quidel) was added to reveal the MAC formation. The reaction was developed by using tetramethylbenzidine (Sigma-Aldrich) substrate system and stopped by adding 1 m H2SO4. The level of C3 deposition in the presence of C1q, without hemin was considered as 100%, and the percentage of inhibition was scored for each hemin concentration. The optimal CRP, IgG, C1q concentration, and C1q-depleted serum dilution were determined prior running the experiment. To assure that the observed C3 deposition is due to activation of the classical pathway without being amplified by the alternative pathway, a serum dilution factor of 1/400 was used. To determine the level of soluble C5b-9 released in the fluid phase during the incubation with C1q-depleted serum, the supernatants from the wells were transferred to sC5b-9-specific ELISA (Quidel) and developed according to the manufacturer's instructions. Wells, where coating with IgG1 or CRP was omitted, served as a control for the baseline level of sC5b-9 present in the C1q-depleted serum.
Thermodynamics of Interaction of Native and Heme-exposed C1q to IgG1 and mCRP
The kinetic constants of the binding of C1q to IgG1 and CRP were determined as a function of temperature by using BIAcore 2000 system. Human IgG1 and CRP were immobilized on research grade CM5 sensor chip, as described in the supplemental data. Kinetic experiments were performed using PBS as sample dilution and running buffer. C1q (0.1 μm) was treated with 2.5 μm hemin or with vehicle for 5 min at 4 °C. Native and hemin-exposed C1q were injected sequentially over the sensor surface at concentrations of 4, 2, 1, 0.5, 0.25, 0.125, 0.0625, 0.0312, and 0.0156 nm. The samples were always injected with flow rate of 10 μl/min. The association and dissociation were monitored for 5 min. Regeneration of the chip surface was performed by a 30-s exposure to 1.25 m guanidine·HCl. All kinetic measurements were performed at temperatures of 10, 15, 20, 25, 30, and 35 °C. BIAevaluation 4.1 software (Biacore) was used for the evaluation of the rate constants by global analysis of the experimental data using a model of Langmuir binding with correction of the drifting baseline.
Evaluation of the thermodynamic parameters (ΔH, TΔS, and ΔG) of the binding of C1q to its targets was performed as described previously (28). Further details are given in supplemental data.
Absorbance Spectroscopy
Absorbance spectra of hemin were recorded by using UNICAM Helios b, UV-visible spectrophotometer. Human C1q was diluted to 0.2 or 0.4 μm in PBS, pH 7.4. Aliquots of hemin, resulting in final concentrations in the range of 0.01 to 20.5 μm were added to an optical cell containing C1q and to a reference optical cell containing PBS only. The absorbance spectra in the wavelength range of 350–700 nm were read after addition of each aliquot of hemin and incubation for 2 min in the dark at 20 °C. The spectra were recorded at a rate of 1500 nm/minute. The data of difference of the absorbance of protein bound hemin from free hemin at absorbance maxima (λ = 395 nm) in the Soret region were used to build titration binding curves. The data were fitted by using GraphPad Prism software (version 5.0) to the single binding site hyperbolic equation: y = ΔAmax x/(Kd + x), where x is the concentration of hemin, ΔAmax is the maximal change in the absorbance intensity, and KD is the apparent equilibrium dissociation constant. The effect of the imidazole on the spectral characteristics of hemin was studied by addition of imidazole at final concentration of 2 mm to optical cell containing 12.8 μm hemin only or 12.8 μm hemin in the presence of 0.2 μm C1q. The absorbance spectra were measured as described above.
Fluorescence Spectroscopy
Quenching of Intrinsic C1q Fluorescence by Heme
Fluorescence spectra were recorded with a Hitachi F-2500 fluorescence spectrophotometer (Hitachi Instruments, Inc., Wokingham, UK). C1q was diluted to 0.5 μm in PBS, pH 7.4. The emission spectra of tyrosine and tryptophans after excitation at 280 nm and at 295 nm, respectively, were recorded in a 1-cm quartz cell. The excitation and emission slits were 10 nm. The spectra were recorded in a wavelength range of 300–500 nm with a scan speed of 1500 nm/min. Increasing aliquots of hemin stock solutions were added directly into the optical cell containing C1q solution. After intensive homogenization and a 2-min incubation in the dark, the emission spectra were recorded. All measurements were performed at 20 °C.
Fluorescence of Zn(II) Protoporphyrin IX Bound to C1q
Spectrofluorimetric analyses of the binding of Zn(II)PP to human C1q were performed in PBS, pH 7.4, by using a Hitachi F-2500 fluorescence spectrometer. Aliquots of Zn(II)PP dissolved in dimethyl sulfoxide was added to C1q solution (0.1 μm) or to buffer only to increasing concentrations of 0.010–5.12 μm. Samples were then homogenized and allowed to stand in the dark for 2 min at 20 °C. Emission spectra of Zn(II)PP in the wavelength range 550–700 nm were recorded after excitation of at 410 nm. The excitation and emission slits were set to 10 nm. The rate of wavelength scan was 1500 nm/min. All measurements were performed in a 1-cm quartz cell at 20 °C.
Measurement of Binding of Heme to C1q by Surface Plasmon Resonance
The interaction of heme with C1q was studied by surface plasmon resonance (BIAcore 2000). C1q was immobilized on a research-grade CM5 sensor chip using an amino-coupling kit (Biacore). In brief, C1q were diluted in 5 mm maleic acid, pH 5, to a final concentration of 5 μg/ml and injected on an EDC/NHS-activated flow cell. The experiments were performed using PBS, pH 7.4. Hemin solutions were prepared by diluting the stock in PBS just before injection. Hemin was injected manually on immobilized C1q at concentrations of 0.05, 0.1, 0.25, 0.5, 0.75, and 1 μm with a flow rate of 20 μl/min. The association and dissociation phases of the interaction of hemin with C1q were monitored for 7 and 10 min, respectively. Regeneration of the chip surface was achieved by brief exposure (30 s) to a solution containing 0.05 m NaOH and 0.5 m NaCl. The binding to the surface of the control flow cell was always subtracted from the binding to the C1q-coated flow cells. BIAevaluation software (version 4.1; Biacore) was used for the evaluation of the kinetic rate constants.
Size-exclusion Chromatography
The molecular composition of native and hemin exposed C1q was analyzed by using an FPLC Akta Purifier 10 (GE Healthcare Europe) system with a Superose-12 column equilibrated with PBS, pH 7.4. C1q (2.17 μm) was incubated for 10 min on ice with 43.4 μm hemin, giving 20-fold molar excess. One hundred μl of native or heme-exposed C1q were diluted to final volume of 1 ml with PBS. The samples were loaded to 30 ml Superose 12 column. Detection was set at a dual wavelength, at 280 nm for presence of protein and 405 nm for presence of hemin.
Molecular Docking of Heme to gC1q
The atomic coordinates of gC1q (Protein Data Bank code 1PK6) containing a Ca2+ ion (29) were used for the molecular docking. An apoform was created by omitting the Ca2+ ion, as described previously (30). AutoDock (version 3.0.5) (31) was used to accommodate the heme molecule to gC1q. Options for rigid protein and flexible heme were applied. Independently, Hex 6.1 and HexServer (32, 33) were utilized considering or not the electrostatic interactions. Because both docking partners are rigid-body structures, heme molecules (both in normal and in upset planes) from Protein Data Bank codes of different hemoproteins were selected. The criteria for final gC1q-heme complex selection were based on the total energy of binding. The final representative complexes were chosen statistically.
Electrostatic Analysis of gC1q-Heme Complex
Electric moments determinations were performed using the PHEMTO sever (Institute of Organic Chemistry, Biophysical Chemistry Lab at Bulgarian Academy of Sciences) as described previously (34, 35). PHEMTO (PH-dependent Electric Moments of proteins TOols) uses three different approaches to calculate three-dimensional electrostatic potentials grid(s) of proteins and their complexes. As a main function, it predicts explicitly the scalar and the orientation of electric moments, μe (in particular the dipole moment (μd) when Z = 0, i.e. in the isoelectric point, pI) based on space integration of the electrostatic potential. A variety of molecular viewers (RasMol, JMol, and Chimera) were used for results visualization and analysis.
RESULTS
Heme Is an Inhibitor of Classical Complement Pathway
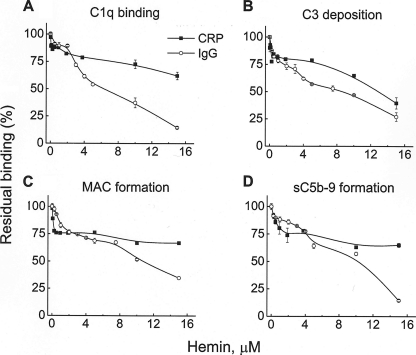
Immunoglobulins and CRP are important targets for C1q binding and subsequent classical pathway activation, leading to C3 deposition. Here, we assessed whether native and heme-exposed C1q have different abilities to activate the classical complement pathway on IgG and CRP-coated surfaces. C1q was allowed to interact with ligand-bound CRP (PnC-CRP as described in Ref. 36) or IgG-containing immune complexes. Fifteen μm hemin inhibited up to 90% of the binding of C1q to the immune complexes and ∼30% of the binding of C1q to PnC-CRP complexes (Fig. 1A). Furthermore, C1q-depleted serum was added as a source of complement proteins. Exposure of C1q to different doses of hemin resulted in decreased deposition of C3, reduced MAC deposition, and decreased formation of sC5b-9 (Fig. 1). The inhibition of the C3 deposition on IgG-containing immune complexes reached >80% at 15 μm hemin and 50% at 6 μm hemin. Deposition of C3 on PnC-CRP was inhibited ∼60% in the presence of 15 μm hemin (Fig. 1B). IgG-containing immune complexes activated the terminal complement pathway more strongly than PnC-CRP. Fifteen μm hemin resulted in a >50% inhibition of the MAC (Fig. 1C) and sC5b-9 (Fig. 1D) formation on IgG-containing immune complexes. Inhibition of the terminal complement pathway on PnC-CRP was also detected but was less efficient compared with the one on immune complexes.
FIGURE 1.
Inhibition of the classical complement pathway activation by hemin. The capacity of native and hemin-exposed C1q, bound to IgG-containing immune complexes or ligand-bound CRP to activate the classical complement pathway, was evaluated by ELISA. C1q (0.1 μm) was exposed to increasing concentrations of hemin (0–15 μm) and was allowed to interact with immune complexes (tetanus toxoid, anti-tetanus toxoid antibodies from an IVIg preparation) and PnC-bound CRP. C1q-depleted serum was added as a source of complement proteins. A, C1q binding; B, C3 deposition; C, MAC formation; D, sC5b-9 release in the fluid phase, measured by Quidel ELISA kit. The results for the native C1q were taken as 100%. Each point represents mean ± S.D. of three repetitions.
Different Metaloporphyrins Affect Differentially the Binding of C1q to IgG1 and CRP
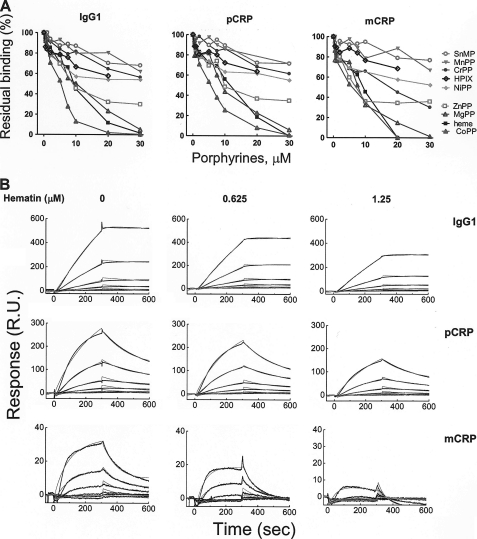
Furthermore, we investigated the interactions of C1q to its targets, IgG1 and CRP, by biosensor real time interaction analyses, as described in the supplemental “Experimental Procedures” (Fig. 2). Both pentameric and monomeric forms of CRP were used because the two forms are suggested to have a physiological relevance and are able to interact with C1q (10, 11, 37, 38). Exposure of C1q to increasing concentrations of heme, resulted in a dose-dependent decrease in the interaction with IgG1, mCRP, and pCRP (Fig. 2, A and B). The exposure of C1q to a heme analog hematoporphyrin IX, a compound with a structure similar to that of heme but devoid of central iron ion and with modified vinyl groups, resulted in considerably lower inhibitory activity of C1q interactions (Fig. 2A). Furthermore, we studied the contribution of the central metal ions in porphyrins for the inhibition of C1q interactions. To this end, different metalloporphyrins were used that differ in their coordination chemistries. Co(III)PP, Mg(II)PP, and Zn(II)PP were found to inhibit strongly the interactions of C1q with its targets. In a contrast exposure of C1q to Sn(IV)MP, Mn(III)PP and Cr(III)PP had only negligible inhibitory effect (Fig. 2A), and Ni(II)PP had an intermediate inhibitory activity on the IgG and CRP binding.
FIGURE 2.
Inhibition of the interaction of C1q with IgG1 and CRP by heme and different porphyrins. A, the inhibitory activity of different porphyrins on the binding of C1q to pCRP, mCRP, and IgG1 was assessed by real-time interaction analyses. C1q (0.1 μm) was exposed to increasing concentrations of metalloporphyrins (0–30 μm) and injected for 5 min over immobilized IgG, pCRP, and mCRP. The graph depicts the percentage of residual binding of C1q as a function of the porphyrin concentration. The binding of native C1q is considered as 100%. All stock solutions of metalloporphyrins were in dimethyl sulfoxide except Mg(II)PP, which was dissolved in water. B, real-time profiles of the interaction of native and hematin-exposed C1q to IgG1, pCRP, and mCRP. C1q at 0.1 μm was exposed to the indicated concentrations of hematin or to vehicle only. The profiles were generated by injecting increasing concentrations of C1q (5, 2.5, 1.25, 0.625, 0.312, and 0.156 nm) over sensor chips immobilized with IgG1, mCRP, and pCRP. The time-dependant binding of C1q in the indicated conditions is presented in resonance units (R.U.). The experimental binding curves (black lines) and curves generated by fitting the data to Langmuir binding with drifting baseline model (gray lines) are presented. SnMP, Sn(IV)MP; MgPP, Mg(II)PP; MnPP, Mn(III)PP; HPIX, hematoporphyrin IX; CrPP, Cr(IV)PP; NiPP, Ni(II)PP; CoPP, Co(III)PP.
To rule out the possibility that heme causes changes in the surface-bound IgG or CRP that further affect C1q binding, we pretreated surface-bound IgG and CRP with the highest concentration of hemin introduced with C1q in our analyses. The injection of native C1q afterward showed binding profiles identical with the binding of native C1q on the untreated chip surface (data not shown).
Kinetic and Thermodynamic Characterization of Heme-mediated Inhibition of C1q Interactions
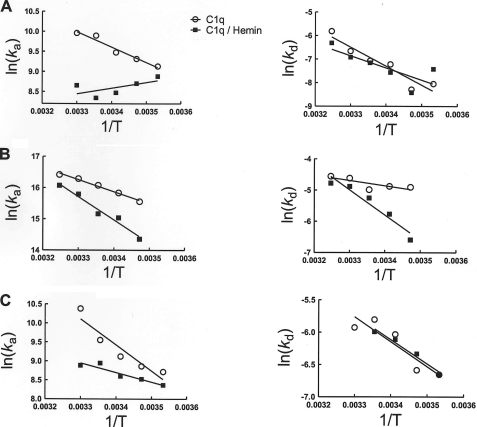
To understand the mechanism of heme-mediated inhibition of C1q interactions, we performed kinetic and thermodynamic analyses. The association and dissociation rate constants of binding of native and hemin-exposed C1q (0.1 μm C1q exposed to 2.5 μm of hemin) to IgG1, mCRP, and pCRP were evaluated as a function of the temperature. The association rate constants of the interaction of native C1q with IgG1 and both forms of CRP showed identical temperature dependence. Thus, for the binding of C1q to proteins, the increase in the temperature resulted in a concomitant increase in the association rate (i.e. negative slopes in left panels of Fig. 3). Exposure of C1q to hemin resulted in qualitative and quantitative differences in the temperature dependence of ka of the interaction with IgG1 (Fig. 3); the association rate was reduced in the whole temperature range, and the temperature dependence was opposite to that of native C1q (Fig. 3A, left panel). Thus, an increase in the temperature resulted in the decrease of the association constant of hemin-exposed C1q to IgG1. The change in the temperature had similar effect on ka for hemin-exposed C1q and native C1q when the binding to mCRP was studied (Fig. 3B, left panel). However, the slope of the Arrhenius plot was greater in the case of hemin-treated C1q, implying a higher temperature sensitivity of the association rate. In contrast, the hemin exposure decreased the temperature sensitivity of association rate constant for the binding to pCRP.
FIGURE 3.
Temperature dependence of the binding kinetics of C1q to IgG1 and CRP. Arrhenius plots depict the temperature dependence of the association and dissociation rate constants that characterize the binding of native (open circles) and hemin-exposed C1q (filled squares) to IgG1 (A), to monomeric CRP (B), and to pentameric CRP (C). C1q diluted to 0.1 μm in phosphate buffer, pH 7.4, and was treated with 2.5 μm of hemin or with the same volume of vehicle. After 5 min of incubation, C1q was diluted to 4, 2, 1, 0.5, 0.25, 0.125. 0.0625, 0.0312, and 0.0156 nm in phosphate buffer and injected over the sensor chip. The kinetic values were obtained by global analyses of at least six binding curves. To determine the slopes of the Arrhenius analyses, the kinetic data were subjected to linear regression analysis.
The effect of the temperature on the dissociation rate constant was similar for all studied interactions. The increase of the temperature resulted always in an increase in the rates of dissociation (Fig. 3, right panels).
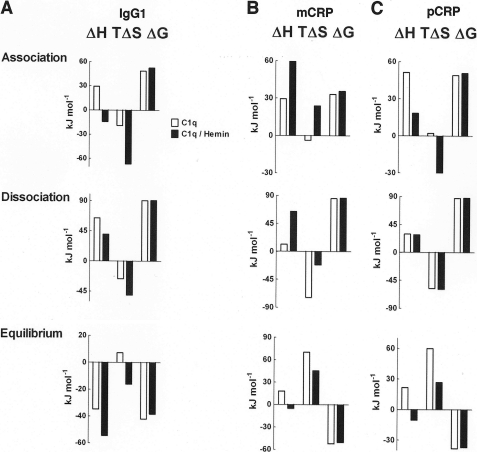
Differences in the temperature sensitivity of the rate constants of native and hemin-exposed C1q reflect differences in molecular mechanism of targets recognition. To characterize the biophysical mechanism of C1q interactions, we determined the changes in the thermodynamic parameters. The changes in enthalpy (ΔH), entropy (TΔS), and free energy (ΔG) were evaluated for the association and dissociation phases, as well as for equilibrium of the binding of native and hemin-exposed C1q to IgG1 and both forms of CRP (Fig. 4). The exposure of C1q to hemin resulted in qualitative and quantitative changes in the thermodynamic parameters of the association in case of the three studied targets (Fig. 4). In contrast, the energetic changes during the dissociation phase of C1q interactions were less affected by the presence of hemin (Fig. 4). The thermodynamic changes at equilibrium indicated that C1q binding to mCRP and pCRP occur by identical mechanism that differed qualitatively from those typical for IgG1 (Fig. 4).
FIGURE 4.
Thermodynamics of the binding of C1q to IgG1 and CRP. Changes in the thermodynamic parameters, enthalpy (ΔH), entropy (TΔS), and Gibbs free energy (ΔG), during the association, dissociation, and equilibrium phases of the binding of native (white bars) and hemin-exposed (black bars) C1q to IgG1 (A), monomeric CRP (B), and pentameric CRP (C). Reported values were obtained by applying thermodynamic calculations (see “Experimental Procedures”) to the Arrhenius plots shown in Fig. 3. The thermodynamic parameters were determined by using a reference temperature of 298 K (25 °C).
Heme Binds to C1q
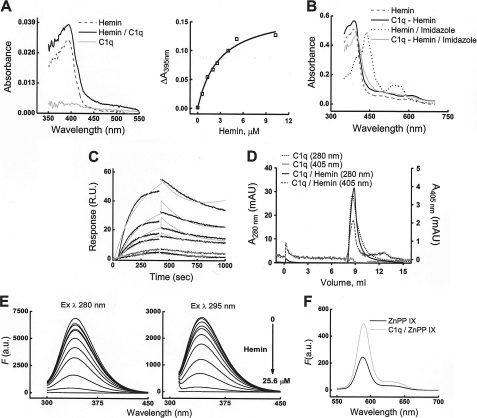
We hypothesized that the inhibition of C1q recognition of its targets may result from a direct interaction of heme with C1q. The specific electronic characteristics of heme allow investigating its binding to proteins by spectroscopic approaches. A UV-visible absorbance spectrum of oxidized heme showed an increase in the molar extinction coefficient at the Soret band (∼400 nm) in the presence of C1q (Fig. 5A). The titration of C1q with increasing concentrations of hemin resulted in saturable (at 12-fold heme excess) increase in the differential spectrum at the Soret band. These data indicate that hemin binds to C1q molecule and that each C1q molecule binds 12 hemin molecules (i.e. two hemin molecules per gC1q). Furthermore, we studied the effect of imidazole on the hemin electronic properties (Fig. 5B). Incubation of hemin with imidazole resulted in a red-shift in the Soret band and the appearance of two absorbance peaks in the α/β-region of the spectrum. These changes are consistent with a transition of the heme iron to a bis-imidazole hexacoordinated low spin state. The addition of imidazole to C1q preincubated with hemin resulted in less pronounced changes in the absorbance spectrum of hemin, indicating the engagement of hemin in an interaction with the protein that may reduce the ability of imidazole to coordinate the hemin iron.
FIGURE 5.
Binding of heme to C1q. A, left panel, absorbance spectra (350–550 nm) of 0.2 μm C1q (solid gray curve), of 0.2 μm C1q preincubated for 5 min with 0.8 μm hemin (solid black curve), and of 0.8 μm hemin alone (dashed black curve). The absorbance spectra were measured in phosphate buffer, pH 7.4, at 20 °C by using optical cells with a 1-cm path. A, right panel, the specific binding of heme to C1q was estimated by addition of increasing concentrations of hemin (0–20.5 μm) to an optical cell containing C1q (0.4 μm) and using optical cells containing phosphate buffer as a reference. The binding curve was generated by plotting the difference in absorbance at 395 nm (AC1q/hemin − Ahemin) versus the molar concentration of hemin. Data were fitted with the single binding-site hyperbola equation. B, absorbance spectra (350–750 nm) of hemin (12.8 μm) and C1q/hemin (12.8/0.2 μm) in the absence (dashed black and solid black curves, respectively) or presence of 2 mm imidazole (dotted black and solid gray curves, respectively). The samples were incubated with imidazole for 5 min at 20 °C before measurement of the absorbance spectra. C, detection of heme binding to C1q by biosensor measurements. Real-time interaction profiles generated after injections of increasing concentrations of hemin (0.05, 0.1, 0.25, 0.5, 0.75, and 1 μm) over immobilized C1q. The association and dissociation of hemin to C1q were followed for 7 and 10 min, respectively. The specific heme-binding responses to C1q-coated chip surface were generated by subtracting the response from a reference surface. The experimental data were fitted to a Langmuir model with drifting base-line (Chi2, 1.95). D, size-exclusion chromatography for determination of molecular composition of hemin-exposed C1q. Elution profiles of native and hemin-treated C1q from a Superose 12 column were recorded at 280 and 405 nm. Dotted black and solid black lines represent the profiles of native C1q and heme-treated C1q, respectively, detected at 280 nm (protein absorbance). Solid gray and dashed black lines refer to native C1q and heme-treated C1q, respectively, detected at 405 nm (heme absorbance). E, quenching of the fluorescence of C1q by hemin. Fluorescence spectra of C1q (0.5 μm in phosphate buffer, pH 7.4) were recorded after addition of increasing concentrations of hemin (0–25.6 μm). After each addition of hemin, samples were incubated for 2 min in the dark. The left panel shows the emission spectra of C1q after excitation at 280 nm. The right panel shows the emission spectra after excitation at 295 nm (tryptophan fluorescence). The gray curves on the graphs represent the background emission from the phosphate buffer. F, emission spectra of Zn(II)PP (0.320 μm) after excitation at 410 nm in the absence (black line) and presence (gray line) of 0.1 μm C1q. The spectra were recorded 2 min after addition of Zn(II)PP to solution of C1q in phosphate buffer. R.U., resonance units; a.u., arbitrary units; ZnPP IX, Zn(II) protoporphyrin IX.
To further confirm binding of hemin to C1q and to characterize the kinetics of the interaction, we applied a surface plasmon resonance-based technique. Injection of hemin over immobilized C1q resulted in a concentration-dependent increase in hemin binding to C1q (Fig. 5C). Evaluation of the kinetic parameters of the interaction revealed that hemin binds to C1q with an affinity in the 1–2 μm range. The interaction was characterized by a slow association, ka of 3.2 × 103 m−1 s−1 and a relatively fast dissociation, kd of 3.44 × 10−3 s−1. By using size-exclusion chromatography (Fig. 5D), we analyzed the molecular integrity of C1q after exposure to hemin at concentrations that are able to significantly inhibit C1q binding to its targets (Figs. 1 and 2). The elution profile of hemin-C1q did not differ from that of C1q alone, indicating that heme treatment is not inducing aggregation of C1q. Size-exclusion chromatography analyses also confirmed that heme forms stable complexes with C1q. Thus, when C1q was treated with hemin, the elution of the protein, detected by absorbance at 280 nm, coincided with elution of hemin, detected by its specific absorbance at 405 nm (Fig. 5D).
Another evidence for hemin binding to C1q was provided by using fluorescence spectroscopy. Titration of C1q with increasing concentrations of hemin resulted in quenching of the intrinsic fluorescence of aromatic residues (mainly of Tyr and Trp excited at 280 nm) and more specifically of Trp (excited at 295 nm) in C1q (Fig. 5E). These results suggested that hemin binds in close vicinity to aromatic residues.
Finally, we studied the ability of the fluorescence compound Zn(II)PP, which differs from heme only by the nature of its central metal, to bind to C1q. Spectroscopy analyses revealed that incubation of C1q in the presence of Zn(II)PP resulted in an increase in the fluorescence intensity of the porphyrin (Fig. 5F), which was saturated at the highest concentration used (data not shown). These data indicate that C1q is able to bind Zn(II)PP and highlight the importance of the porphyrin ring in the interaction.
Heme-induced Alteration of Electrostatic Properties of gC1q
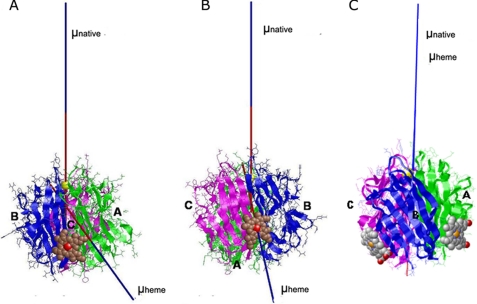
The attempt to dock heme to gC1q by using AutoDock resulted in a large number of possible structures with close values of the binding energy. The preferred binding site appeared to be located in the lower part of the heterotrimer, close to the collagen-like part. The complexes obtained using Hex and HexServer with and without electrostatic contributions gave less scattering of the possible binding sites and allowed us to select two models (Fig. 6), representing the most probable site (with lowest value of energy) with (Fig. 6A) and without (Fig. 6B) taking into account the electrostatic contributions. In both cases, heme is located at the lower part of gC1q, bound to the A-chain (Fig. 6A, A-model) or to the C-chain (Fig. 6B, C-model).
FIGURE 6.
Heme-induced alteration of the direction of the electric moment vectors of gC1q. The direction of the electric moment vectors (μ) two most probable gC1q-heme docking complexes are represented in a native form (μ native), as calculated from the Protein Data Bank code 1PK6 structure and in presence of heme (μ heme). The docking complexes where heme is bound to the A-chain (A-model, panel A) and to the C-chain (C-model, panel B), or both (panel C) are depicted. The three chains of gC1q are colored as follows: A, green; B, blue; and C, magenta. The heme is shown in sphere representation.
The residues that make a direct contact with heme in the A-model are as follows: ValA114, IleA115, AsnA117, GluA119, GluA120, AsnA124, ArgB159, LysB188, and GluB190 (supplemental Fig. 1A). The closest aromatic residue is TyrA122, which is at 3 Å from the heme moiety. The contact residues for heme in the C-model are as follows: ValB118, IleB119, ThrB120, AsnB121, MetB122, AspC156, ArgC182, LeuC183, GlnC184, GluC187, and AspC217, with TrpC190 being at less than 10 Å away from the heme (supplemental Fig. 1B). The two binding sites may be occupied simultaneously.
To get an insight to the mode of heme-induced inhibition, the value and the direction of the electric moment vectors were calculated for the free gC1q and the two selected heme-containing complexes in a large pH range. At a pH interval of 5–11, all vectors are directed toward heme moieties, mainly through the A-chain for the A-model and fixed through the iron-heme atom on the C-model. The position of the vectors does not depend on the presence or absence of the Ca2+ ion. These directions strongly differ from the ones in the native gC1q in presence of Ca2+ (directed toward the apex of gC1q) or in a Ca2+-free state (toward the lateral surface of the B-chain). In case of simultaneous occupancy of the two binding sites, the electric moment vector remains directed toward the apex of the heterotrimer as in the native (Ca2+-bound) form, but cannot be redirected when the Ca2+ ion is removed.
DISCUSSION
In this study, we demonstrated that heme is able to inhibit IgG- and CRP-mediated complement activation by direct binding to C1q. The inhibitory effect of heme resulted from alterations in the molecular mechanism that drives the interaction of C1q with its targets. Spectroscopic and biosensor analyses revealed that heme binds to C1q with micromolar equilibrium affinity. Heme binding to C1q was saturable with two binding sites for each of the six gC1q domains. The spectral data obtained by absorbance and fluorescence spectroscopy suggest that binding of heme to C1q is not accompanied by changes in its molecular environment. The absence of shifts in the absorbance maxima implies that heme most probably bind to the surface of the protein. Heme is a hydrophobic compound and has a tendency to interact with hydrophobic regions of proteins. The marked quenching of the intrinsic Trp and Tyr fluorescence of C1q by hemin indicates that heme forms contacts at regions that are rich in aromatic residues. The collagen-like region of C1q is composed primarily of glycine-hydroxyproline-hydroxylysine repeats. The majority of the aromatic residues are found in the globular domains, the surface of which consists of charged and hydrophobic patches (5, 29). In accordance with the experimental results, the data obtained by molecular docking of heme to the structure of gC1q suggested that heme binds to the surface of the protein, in close proximity to TyrA122 (for the binding site in the A-chain) and to TrpC190 (for the binding site in the C-chain).
The thermodynamic analyses of interactions of native and heme-exposed C1q indicate that reduced binding of heme-C1q to its targets is mainly due to unfavorable changes in association thermodynamics. The changes in the thermodynamic parameters that characterize intermolecular recognition reflects the formation of noncovalent contacts (changes in enthalpy) and the presence of structural alternations in the interacting proteins (changes in entropy) (39–41). Negative values of the changes in the entropy during association are usually associated with “induced fit” binding of macromolecules, where the conformational heterogeneity (high entropy) of free binding partners is reduced (decrease in entropy) upon complex formation (40–42). Interestingly, the physicochemical mechanisms underlying the inhibitory effect of heme were not identical when the interactions of C1q with IgG1, and both forms of CRP were studied. The marked increase in the negative value of TΔS observed for the association of heme-bound C1q to IgG1 and to pCRP suggests that heme increases the structural flexibility of C1q; the energetic penalty of the structural adjustments of heme-bound C1q to IgG increases the activation energy of association and thus negatively influences the overall binding affinity. In contrast, the binding of heme-C1q to mCRP was characterized by a favorable contribution from TΔS (positive sign), implying the absence of structural changes during association. The latter interaction can be categorized as obeying to the “lock-and-key” recognition model, an interaction of preformed rigid binding sites. The reduction of the binding affinity of C1q for mCRP derives mainly from a highly unfavorable change in the association enthalpy. Thus, the binding of heme most probably prevents vital noncovalent interactions between C1q and mCRP, resulting in a reduction of the binding affinity.
The thermodynamic analyses further provide important evidence about the mechanism of target recognition by the native C1q. These data indicate that native C1q associates to IgG monomeric and pentameric CRP by similar physicochemical mechanisms. However, quantitative differences in the association and the dissociation phase for the targets results in qualitative alteration of the equilibrium thermodynamics. Thus, in the case of IgG1, the interaction is driven by favorable changes in equilibrium enthalpy. In contrast, the binding to mCRP and pCRP is driven by favorable changes in equilibrium entropy. These data are supported by previous observations indicating that the binding sites for IgG and CRP may overlap but are not identical (30, 43, 44). The observed differences in the binding mechanism of native C1q also reflect the different nature of the target proteins.
Heme is not the only porphyrin capable of inhibiting the binding of C1q to IgG and CRP. We observed that Co(III)PP, Mg(II)PP, and Zn(II)PP also have potent inhibitory activity on the interactions of C1q. In contrast, porphyrins containing Cr(III), Mn(III), or Sn(IV) ions, or devoid of metal ions, such as hematoporphyrin IX, possess much lower potential to inhibit C1q. This result suggests that coordination of the central metal in the porphyrins by C1q plays an essential role for inhibitory activity. Indeed, the differences in the coordination geometries of different metals would result in different preferences and energies of coordination by particular amino acid residues in the protein.
Previous studies have demonstrated that other heterocyclic compounds structurally similar to heme are also able to inhibit C1q functions. Thus, unconjugated bilirubin, which is a product of heme catabolism, found at high concentrations in the case of liver diseases or of hemolytic anemia, binds to C1q and inhibits the classical complement pathway (45, 46). Moreover, synthetic heterocyclic compounds such as bisphenol disulfates, sulfated steroids, and triterpenoids also inhibit the classical complement pathway at the level of C1q (47, 48). Two of these compounds, betulin disulfate and 9,9-bis(4′-hydroxyphenyl) fluorene disulfate, were studied in detail (43). They bind to gC1q and inhibit its interaction with IgG1 and CRP by modulating the electrostatic properties of the protein, reducing the scalar values, and altering the orientation of the electric moment vectors of gC1q. These dipole/electric moment effectors impair the electrostatic properties of C1q and consequently inhibit the complement activation. Here, we provide evidence that similar electric moments inhibition occurs with natural heterocyclic compounds such as heme. The iron ion appears to be important for this activity, as hematoporphyrin, which has the similar macrocyclic backbone but lacks the metal ion, inhibits C1q binding to a much smaller extent. Interestingly, heme, like bisphenol disulfates, sulfated steroids, triterpenoids, and bilirubin, share common structural features: two negative charges, separated by 8–12 Å by bulky hydrophobic groups in the central part of the molecules. These characteristics were demonstrated to be of importance for inhibition of the classical complement pathway (47, 48) and may explain the inhibitory effect of heme. The binding sites for heme on C1q, predicted by molecular docking, are different from the ones predicted in the case of betulin disulfate and 9,9-bis(4′-hydroxyphenyl) fluorene disulfate, but the effect on the electric moments orientation is similar. Yet, the binding of heme also results in a reorientation of the electric moment vectors of gC1q. We therefore hypothesize that heme is an endogenous electrostatic effector, which alters the binding properties from C1q by modulation of its physicochemical characteristics.
A complement is a double-edged sword, capable of destroying invading pathogens but also of damaging self-tissues, as seen in many pathological conditions. Inappropriate complement activation has been shown to induce tissue injury in the context of severe inflammation, ischemia/reperfusion, transplantation, stroke, myocardial infarction, hemolytic anemia, immune complex diseases, malaria, and etc. (13–15, 49). Several natural inhibitors of the early steps of the classical pathway activation, like C1 inhibitor (50, 51) and C1q inhibitor (chondroitin-4 sulfate proteoglycan) (52), can counteract the deleterious effects induced by undesired complement triggering. However, in many situations, these systems are insufficient to prevent attack by the complement, and there is an urgent need for developing synthetic inhibitors of the early stages of classical pathway activation (12, 53, 54). A good understanding of the interaction between C1q and its target molecules is an indispensable step toward designing proficient inhibitors of the classical complement pathway. Based on our results, it is tempting to speculate that heme is an endogenous negative-feedback regulator of the activation of the classical complement pathway. Indeed, in cases where high levels of free heme are present, like malaria and ischemia/reperfusion, the classical complement pathway plays only a minor role (if any) despite of the presence of damaged cells (ligands for C1q) and activation of other pathways (49). The heme that is released during cellular damage may modulate the classical pathway triggered by immune complexes and CRP. This anti-inflammatory activity of free heme may be a natural last chance for rescue in such severe, life-threatening conditions. Novel, optimized molecules based on the heme scaffold could be candidates for future therapeutic agents.
Supplementary Material
This work was supported by the Institut National de la Santé et de la Recherche Médicale (France), by Centre National de la Recherche Scientifique (France), and by Université Pierre et Marie Curie (Paris 6).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Fig. 1.
- gC1q
- globular C1q
- MAC
- membrane attack complex
- PP
- protoporphyrin IX
- PnC
- pneumococcal C-polysaccharide.
REFERENCES
- 1. Walport M. J. (2001) N. Engl. J. Med. 344, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 2. Walport M. J. (2001) N. Engl. J. Med. 344, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 3. Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010) Nat. Immunol. 11, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sjöberg A. P., Trouw L. A., Blom A. M. (2009) Trends Immunol. 30, 83–90 [DOI] [PubMed] [Google Scholar]
- 5. Gaboriaud C., Thielens N. M., Gregory L. A., Rossi V., Fontecilla-Camps J. C., Arlaud G. J. (2004) Trends Immunol. 25, 368–373 [DOI] [PubMed] [Google Scholar]
- 6. Kang Y. H., Tan L. A., Carroll M. V., Gentle M. E., Sim R. B. (2009) Adv. Exp. Med. Biol. 653, 117–128 [DOI] [PubMed] [Google Scholar]
- 7. Agrawal A., Singh P. P., Bottazzi B., Garlanda C., Mantovani A. (2009) Adv Exp Med Biol 653, 98–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thompson D., Pepys M. B., Wood S. P. (1999) Structure 7, 169–177 [DOI] [PubMed] [Google Scholar]
- 9. Volanakis J. E. (2001) Mol. Immunol. 38, 189–197 [DOI] [PubMed] [Google Scholar]
- 10. Eisenhardt S. U., Habersberger J., Murphy A., Chen Y. C., Woollard K. J., Bassler N., Qian H., von Zur Muhlen C., Hagemeyer C. E., Ahrens I., Chin-Dusting J., Bobik A., Peter K. (2009) Circ. Res. 105, 128–137 [DOI] [PubMed] [Google Scholar]
- 11. Ji S. R., Wu Y., Zhu L., Potempa L. A., Sheng F. L., Lu W., Zhao J. (2007) Faseb J. 21, 284–294 [DOI] [PubMed] [Google Scholar]
- 12. Roos A., Ramwadhdoebé T. H., Nauta A. J., Hack C. E., Daha M. R. (2002) Immunobiology 205, 595–609 [DOI] [PubMed] [Google Scholar]
- 13. Weiser M. R., Williams J. P., Moore F. D., Jr., Kobzik L., Ma M., Hechtman H. B., Carroll M. C. (1996) J. Exp. Med. 183, 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Griselli M., Herbert J., Hutchinson W. L., Taylor K. M., Sohail M., Krausz T., Pepys M. B. (1999) J. Exp. Med. 190, 1733–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. del Zoppo G. J. (1999) Nat. Med. 5, 995–996 [DOI] [PubMed] [Google Scholar]
- 16. Kumar S., Bandyopadhyay U. (2005) Toxicol. Lett. 157, 175–188 [DOI] [PubMed] [Google Scholar]
- 17. Wagener F. A., Volk H. D., Willis D., Abraham N. G., Soares M. P., Adema G. J., Figdor C. G. (2003) Pharmacol. Rev. 55, 551–571 [DOI] [PubMed] [Google Scholar]
- 18. Muller-Eberhard U., Javid J., Liem H. H., Hanstein A., Hanna M. (1968) Blood 32, 811–815 [PubMed] [Google Scholar]
- 19. Sears D. A. (1970) J. Clin. Invest. 49, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeney V., Balla J., Yachie A., Varga Z., Vercellotti G. M., Eaton J. W., Balla G. (2002) Blood 100, 879–887 [DOI] [PubMed] [Google Scholar]
- 21. Balla J., Jacob H. S., Balla G., Nath K., Eaton J. W., Vercellotti G. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9285–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wagener F. A., Eggert A., Boerman O. C., Oyen W. J., Verhofstad A., Abraham N. G., Adema G., van Kooyk Y., de Witte T., Figdor C. G. (2001) Blood 98, 1802–1811 [DOI] [PubMed] [Google Scholar]
- 23. Morgan W. T. (1985) Biochemistry 24, 1496–1501 [DOI] [PubMed] [Google Scholar]
- 24. Green D., Furby F. H., Berndt M. C. (1986) Thromb. Haemost. 56, 277–282 [PubMed] [Google Scholar]
- 25. Dimitrov J. D., Roumenina L. T., Doltchinkova V. R., Mihaylova N. M., Lacroix-Desmazes S., Kaveri S. V., Vassilev T. L. (2007) J. Biol. Chem. 282, 26696–26706 [DOI] [PubMed] [Google Scholar]
- 26. Dimitrov J. D., Vassilev T. L. (2009) Nat. Biotechnol. 27, 892. [DOI] [PubMed] [Google Scholar]
- 27. Dimitrov J. D., Roumenina L. T., Doltchinkova V. R., Vassilev T. L. (2007) Scand. J. Immunol. 65, 230–239 [DOI] [PubMed] [Google Scholar]
- 28. Dimitrov J. D., Lacroix-Desmazes S., Kaveri S. V., Vassilev T. L. (2007) Mol. Immunol. 44, 1854–1863 [DOI] [PubMed] [Google Scholar]
- 29. Gaboriaud C., Juanhuix J., Gruez A., Lacroix M., Darnault C., Pignol D., Verger D., Fontecilla-Camps J. C., Arlaud G. J. (2003) J. Biol. Chem. 278, 46974–46982 [DOI] [PubMed] [Google Scholar]
- 30. Roumenina L. T., Kantardjiev A. A., Atanasov B. P., Waters P., Gadjeva M., Reid K. B., Mantovani A., Kishore U., Kojouharova M. S. (2005) Biochemistry 44, 14097–14109 [DOI] [PubMed] [Google Scholar]
- 31. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 32. Macindoe G., Mavridis L., Venkatraman V., Devignes M. D., Ritchie D. W. (2010) Nucleic Acids Res. 38, W445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ritchie D. W., Venkatraman V. (2010) Bioinformatics 26, 2398–2405 [DOI] [PubMed] [Google Scholar]
- 34. Kantardjiev A. A., Atanasov B. P. (2006) Nucleic Acids Res. 34, W43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kantardjiev A. A., Atanasov B. P. (2009) Nucleic Acids Res. 37, W422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agrawal A., Shrive A. K., Greenhough T. J., Volanakis J. E. (2001) J. Immunol. 166, 3998–4004 [DOI] [PubMed] [Google Scholar]
- 37. Ji S. R., Wu Y., Potempa L. A., Liang Y. H., Zhao J. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 935–941 [DOI] [PubMed] [Google Scholar]
- 38. Bíró A., Rovó Z., Papp D., Cervenak L., Varga L., Füst G., Thielens N. M., Arlaud G. J., Prohászka Z. (2007) Immunology 121, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Janin J. (1995) Biochimie 77, 497–505 [DOI] [PubMed] [Google Scholar]
- 40. Stites W. E. (1997) Chem. Rev. 97, 1233–1250 [DOI] [PubMed] [Google Scholar]
- 41. Amzel L. M. (2000) Methods Enzymol. 323, 167–177 [DOI] [PubMed] [Google Scholar]
- 42. Manivel V., Sahoo N. C., Salunke D. M., Rao K. V. (2000) Immunity 13, 611–620 [DOI] [PubMed] [Google Scholar]
- 43. Roumenina L., Bureeva S., Kantardjiev A., Karlinsky D., Andia-Pravdivy J. E., Sim R., Kaplun A., Popov M., Kishore U., Atanasov B. (2007) J. Mol. Recognit. 20, 405–415 [DOI] [PubMed] [Google Scholar]
- 44. Roumenina L. T., Ruseva M. M., Zlatarova A., Ghai R., Kolev M., Olova N., Gadjeva M., Agrawal A., Bottazzi B., Mantovani A., Reid K. B., Kishore U., Kojouharova M. S. (2006) Biochemistry 45, 4093–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arriaga S. M., Mottino A. D., Almará A. M. (1999) Biochim. Biophys. Acta 1473, 329–336 [DOI] [PubMed] [Google Scholar]
- 46. Basiglio C. L., Arriaga S. M., Pelusa H. F., Almará A. M., Roma M. G., Mottino A. D. (2007) Biochim. Biophys. Acta 1770, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 47. Bureeva S., Andia-Pravdivy J., Petrov G., Igumnov M., Romanov S., Kolesnikova E., Kaplun A., Kozlov L. (2005) Bioorg. Med. Chem. 13, 1045–1052 [DOI] [PubMed] [Google Scholar]
- 48. Bureeva S., Andia-Pravdivy J., Symon A., Bichucher A., Moskaleva V., Popenko V., Shpak A., Shvets V., Kozlov L., Kaplun A. (2007) Bioorg. Med. Chem. 15, 3489–3498 [DOI] [PubMed] [Google Scholar]
- 49. Pawluczkowycz A. W., Lindorfer M. A., Waitumbi J. N., Taylor R. P. (2007) J. Immunol. 179, 5543–5552 [DOI] [PubMed] [Google Scholar]
- 50. Castellano G., Melchiorre R., Loverre A., Ditonno P., Montinaro V., Rossini M., Divella C., Battaglia M., Lucarelli G., Annunziata G., Palazzo S., Selvaggi F. P., Staffieri F., Crovace A., Daha M. R., Mannesse M., van Wetering S., Paolo Schena F., Grandaliano G. (2010) Am. J. Pathol. 176, 1648–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kirschfink M., Mollnes T. E. (2001) Expert Opin. Pharmacother. 2, 1073–1083 [DOI] [PubMed] [Google Scholar]
- 52. Galanakis D. K., Ghebrehiwet B. (1994) J. Clin. Invest. 93, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roos A., Nauta A. J., Broers D., Faber-Krol M. C., Trouw L. A., Drijfhout J. W., Daha M. R. (2001) J. Immunol. 167, 7052–7059 [DOI] [PubMed] [Google Scholar]
- 54. Pepys M. B., Hirschfield G. M., Tennent G. A., Gallimore J. R., Kahan M. C., Bellotti V., Hawkins P. N., Myers R. M., Smith M. D., Polara A., Cobb A. J., Ley S. V., Aquilina J. A., Robinson C. V., Sharif I., Gray G. A., Sabin C. A., Jenvey M. C., Kolstoe S. E., Thompson D., Wood S. P. (2006) Nature 440, 1217–1221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.