Abstract
The serine/threonine kinase RAF is a central component of the MAPK cascade. Regulation of RAF activity is highly complex and involves recruitment to membranes and association with Ras and scaffold proteins as well as multiple phosphorylation and dephosphorylation events. Previously, we identified by molecular modeling an interaction between the N-region and the RKTR motif of the kinase domain in RAF and assigned a new function to this tetrapeptide segment. Here we found that a single substitution of each basic residue within the RKTR motif inhibited catalytic activity of all three RAF isoforms. However, the inhibition and phosphorylation pattern of C-RAF and A-RAF differed from B-RAF. Furthermore, substitution of the first arginine led to hyperphosphorylation and accumulation of A-RAF and C-RAF in plasma membrane fraction, indicating that this residue interferes with the recycling process of A-RAF and C-RAF but not B-RAF. In contrast, all RAF isoforms behave similarly with respect to the RKTR motif-dependent dimerization. The exchange of the second arginine led to exceedingly increased dimerization as long as one of the protomers was not mutated, suggesting that substitution of this residue with alanine may result in similar a structural rearrangement of the RAF kinase domain, as has been found for the C-RAF kinase domain co-crystallized with a dimerization-stabilizing RAF inhibitor. In summary, we provide evidence that each of the basic residues within the RKTR motif is indispensable for correct RAF function.
Keywords: MAP Kinases (MAPKs), Protein Kinases, Protein Phosphorylation, Raf, Signal Transduction, RAF Dimerization
Introduction
RAF proteins, originally discovered as the oncogenic product of mouse sarcoma virus (1), belong to the family of serine/threonine-specific protein kinases that control cell proliferation, transformation, differentiation, and apoptosis (2–5). They serve as signaling modules for the classical mitogenic pathway, functioning to connect the receptor tyrosine kinase via MEK and ERK with nuclear transcription factors. Unlike lower eukaryotes that express only one RAF gene, vertebrates inherit at least three members of the RAF family, A-RAF, B-RAF, and C-RAF (6–9). B-RAF is likely to be the original RAF kinase in the evolution of vertebrates, as its nucleotide sequence is more closely related to all other eukaryotic RAF homologues than either A-RAF or C-RAF. B-RAF also plays an important role in cancer progression. This is underlined by the fact that in several cancer types like melanoma and colorectal cancer, a single amino acid exchange in B-RAF (V600E, formerly termed V599E) is one of the most commonly found mutations (3, 10, 11).
All RAF isoforms share three conserved regions (CR1, CR2, and CR3) and can be divided in two functional parts, the N-terminal regulatory domain and the C-terminal kinase domain (see Fig. 1A). Although CR1 (residues 51–194 in C-RAF) and CR2 (residues 254–269 in C-RAF) are positioned within the regulatory domain, CR3 represents the kinase domain extending in C-RAF from residue 347 to 613. CR1 includes two subdomains, the Ras binding domain followed by the cysteine-rich domain. The serine/threonine-rich CR2 contains a conserved 14-3-3 binding motif (12).
FIGURE 1.
Molecular model for interaction between the N-region and the catalytic domain in B-RAF and C-RAF. A, shown is a schematic presentation of A-, B-, and C-RAF proteins and sequence alignment of the N-region and the RKTR motif. Conserved regulatory phosphorylation sites within the N-region and 14-3-3 binding segments are indicated. Light-shaded boxes, non-conserved parts of the sequence; dark-shaded boxes, conserved regions. CR1 comprises the Ras binding (RBD) and the cysteine-rich domain (CRD). CR2 is the serine-threonine rich domain, and CR3 is the kinase domain. The position of the N-region is highlighted in blue, whereas that of the RKTR motif is in red. B and C, modeling of the spatial orientation of the N-region relative to the RKTR motif of the kinase domain in B-RAF (A) and C-RAF (B). Although the structure of the kinase domain is highlighted in green, the N-region peptide is highlighted in light blue. Positions of the amino acids within the N-region and the amino acids of the kinase domain making interactions with the N-region are indicated.
The molecular mechanism of RAF activation is highly complex and, despite intensive investigations, it is not completely understood. Although experimental support is missing, it has been generally accepted that in quiescent cells C-RAF exists in an inactive conformation maintained by autoinhibitory interactions occurring between the N-terminal regulatory and the C-terminal catalytic domains. This inactive conformation is supposed to be stabilized by binding of 14-3-3 dimers to the conserved phosphoserines within the internal and the C-terminal 14-3-3 binding sites of RAF (4, 13). There is evidence that a fraction of RAF molecules exists as membrane-prebound being targeted to the plasma membrane primarily by association with cholesterol and ceramides (raft microdomains) (14, 15). Additionally, the association of RAF with the membranes may be facilitated through the binding of the RAF catalytic domain to phosphatidic acid (PA)2 (16, 17). Thus, this model implicates that RAF association with membrane lipids represents the initial step in RAF activation (14). Upon stimulation of cell surface receptors, association with activated small G protein Ras (Ras-GTP) occurs in the plane of the inner leaflet of the plasma membrane (18–20). This interaction initiates further activation events, such as multiple phosphorylation and dephosphorylation steps as well as dimer formation (3, 21–23). RAF dimerization, originally discovered by Weber et al. (21), proved to be one of the most decisive steps in the regulation of RAF activity, particularly in light of recent findings concerning the paradoxical behavior of some RAF inhibitors in cancer treatment (24–29).
Treatment of cells with growth factors induces phosphorylation at multiple sites. Among others, phosphorylation within the N-region (the name is derived from negative-charge regulatory region) is essential for activation of RAF kinases. The N-region is a short conserved sequence in front of the kinase domain (see also Fig. 1A). It contains one highly conserved serine, which is present in all three RAF isoforms (Ser-299 in A-RAF, Ser-338 in C-RAF, and Ser-446 in B-RAF) and two conserved tyrosines (Tyr-301/Tyr-302 in A-RAF and Tyr-340/Tyr-341 in C-RAF) at positions where in B-RAF aspartates are located (Asp-448/Asp-449). Stimulation-dependent phosphorylation of these sites positively regulates catalytic activity of RAF kinases (3, 30–32). Recently, we reported that non-conserved amino acids within the N-region determine low basal activity and limited inducibility of the A-RAF kinase, as substitution of the non-conserved Tyr-296 to arginine led to a constitutively active kinase, suggesting that Tyr-296 may be responsible for the low kinase activity toward MEK (33). Moreover, we suggested an interaction between the N-region and the catalytic domain in A-RAF and C-RAF based on results of molecular modeling (33). The segment of the kinase domain, which in our reconstruction model makes contact to the N-region, is part of the sequence that has been described as the phosphatidic acid binding region of RAF kinases (17).
The phosphatidic acid binding region is a highly conserved 36-amino acid sequence containing a cluster of basic residues (RKTR motif, see Fig. 1A) as well as a short segment of hydrophobic residues (405–408 in C-RAF) (17, 34). Mutation of one of these residues to alanine (R398A) was sufficient to reduce the ability of C-RAF to associate with PA using a microtiter plate binding assay as reported by Rizzo et al. (16). An opposing observation was seen by Ghosh et al. (35), who reported that single site R398A or R401A replacements did not affect PA binding when compared with wild type protein, whereas K399A replacement resulted in reduced C-RAF binding to PA. Furthermore, developmental defects were observed in zebra fish embryos microinjected with mutant C-RAF RNA containing triple substitution R398A/K399A/R401A, underlining the physiological importance of the RKTR motif (35). In addition, the crystal structure of a C-RAF homodimer composed of two kinase domains lacking the N-terminal regulatory part of the protein revealed that Arg-398, Lys-399, and Arg-401 are parts of the dimerization interface, assigning a new function to the RKTR motif (26, 27). Dimerization deficiency of the B-RAF kinase domain-containing the R509H mutation (residue Arg-509 is equivalent to Arg-401 in C-RAF; see also Fig. 1A) confirmed the involvement of the RKTR motif in dimer formation (27). On the other hand, based on data of Lu et al. (36), who reported that mutations in the PA binding site do not alter the transforming activity of v-RAF, Rizzo et al. (16) concluded that kinase activity of C-RAF was unaffected by the mutations within the RKTR motif. Moreover, these mutations did not alter the ability of the constructs to associate with endogenous C-RAF and did not disrupt the basal activity of the kinase in the study of Rizzo et al. (16). Taken together, the overview of the published data discloses inconsistence and incompleteness of available information; however, there is a high need for new insights into regulation of RAF kinases by the RKTR motif with respect to development of potent and specific RAF inhibitors. Therefore, in the present study, we analyzed systematically the impact of the single mutations within the RKTR motif of all three RAF isoforms on kinase activity, subcellular distribution, and dimerization behavior of the full-length proteins in vivo.
Data presented here reveal that each single substitution of the basic residues within the RKTR motif results in inhibited kinase activity of RAF isoforms. However, the inhibition and phosphorylation pattern of C-RAF and A-RAF differs from B-RAF. We found also that substitution of Arg-398 and Arg-359 led to hyperphosphorylation and accumulation of A-RAF and C-RAF, respectively, in plasma membrane fraction, suggesting that this residue is involved in the recycling process of A-RAF and C-RAF but not B-RAF. Concerning the RKTR motif-dependent dimerization, all RAF isoforms behave similarly. Whereas substitution of the lysine residue to alanine resulted in a moderate inhibition of dimer formation, exchange of the second arginine residue led to a highly increased dimerization. This implies that substitution of the second arginine residue for alanine may result in similar structural conformation of the RAF kinase domain, as has been found for the C-RAF kinase domain co-crystallized with a close analog of the selective RAF inhibitor GDC-0879 (26). Collectively, selective replacement of the basic residues within the RKTR motif resulted basically in different phenotypes for A-RAF and C-RAF versus B-RAF, thereby indicating a multifunctional role for this regulatory segment.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: anti-c-Myc (9E10), anti-pERK (E-4), anti-ERK2 (C-14), anti-Lck (3A5), anti-A-RAF (C-20), anti-C-RAF (RAF-1, C-12), anti-B-RAF (C-19), and anti-HA (12CA5) from Santa Cruz; anti-H-Ras (#R02120) from BD Transduction Laboratories; anti-M2PK (DF4) from Schebo Biotech; anti-phospho-C-RAF-Ser-338 (56A6, was also used for detection of phospho-Ser-446 in B-RAF), anti-phospho-A-RAF-Ser-299 (#4431) and anti-phospho-C-RAF-Ser-259 (#9421, was also used for detection of phospho-Ser-365 in B-RAF and phospho-Ser-214 in A-RAF) from Cell Signaling Technology; anti-phospho-C-RAF-Ser-621 (6B4, was also used for detection of phospho-Ser-729 in B-RAF and phospho-Ser-582 in A-RAF) has been described previously (37).
DNA Constructs and Site-directed Mutagenesis
Cloning of C-terminal Myc-tagged human RAF cDNA was performed as described previously (33). The site-specific mutations of RAF were introduced using QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The correctness of the RAF mutant cDNA was confirmed by DNA sequencing.
Immunoprecipitation and in Vitro Kinase Assay
COS7 cells were transfected with Myc-tagged RAF constructs using jetPEI transfection reagent (Polyplus Transfection). Cells were lysed 24 h after transfection with buffer containing 50 mm Tris-HCl, pH 8.0, 137 mm NaCl, 2 mm EDTA, 2 mm EGTA, 10% (v/v) glycerol, 0.1% (v/v) β-mercaptoethanol, 25 mm β-glycerophosphate, 10 mm sodium pyrophosphate, 1 mm Na3VO4, 25 mm NaF, 1% Nonidet P-40, and proteinase inhibitors for 45 min at 4 °C. The lysates were clarified by centrifugation at 27,000 × g for 15 min and incubated for 1 h at 4 °C with the anti-Myc antibody. After the addition of protein G-agarose, the incubation was continued for 2 h at 4 °C. The agarose beads were washed twice with lysis buffer containing 0.2% Nonidet P-40 and once with kinase assay buffer. The kinase assay was carried out directly with immunoprecipitated proteins in 25 mm Hepes, pH 7.6, 150 mm NaCl, 25 mm β-glycerophosphate, 10 mm MgCl2, 1 mm dithiothreitol, 1 mm Na3VO4, and 500 μm ATP buffer (50 μl final volume). Recombinant MEK and ERK-2 were used as substrates. After incubation for 30 min at 30 °C, the kinase assay mixtures were supplemented with Laemmli buffer, boiled for 5 min at 100 °C, and applied to SDS-PAGE. After Western blotting, the extent of ERK phosphorylation was determined by an anti-phospho-ERK antibody.
Isolation of RAF Dimer Complexes
cDNA encoding Myc-tagged RAF together with untagged or HA-tagged RAF was transfected into COS7 cells using jetPEI transfection reagent (Polyplus Transfection). RAF dimerization was amplified by co-transfection with H-RasV12 and Lck. Cells were lysed 24 h after transfection with buffer containing 10 mm Tris-HCl, pH 8.0, 50 mm NaCl, 30 mm sodium pyrophosphate, 100 μm Na3VO4, 1% Triton X-100, and proteinase inhibitors for 45 min at 4 °C. The lysates were clarified by centrifugation at 27,000 × g for 15 min and incubated for 1 h at 4 °C with the anti-Myc antibody. After the addition of protein G-agarose, the incubation was continued for 2 h at 4 °C. The agarose beads were washed 3 times with lysis buffer containing 0.1% Triton X-100. The immunoprecipitates were supplemented with Laemmli buffer, boiled for 5 min at 100 °C, and applied to SDS-PAGE. After Western blotting, the isolated RAF dimers were visualized by appropriate antibodies.
Subcellular Fractionation
Cell fractions were isolated using the ProteoExtract subcellular proteome extraction kit (Calbiochem). COS7 cells were grown on 10-cm Petri dishes and transfected with cDNA encoding Myc-tagged RAF variants together with H-RasV12 and Lck under starvation conditions. The cells were fractionated into four subproteomic fractions (cytoplasmic and nuclear fractions, fractions of whole membranes, and cytoskeleton) according to the manufacturer's protocol. Proteins were separated by SDS-PAGE and blotted onto nitrocellulose membrane. The recombinant RAF proteins were detected by an anti-Myc antibody. The selectivity of subcellular extraction was documented by immunoblotting against marker proteins (M2PK for cytosolic fraction, Lck for membrane fraction, PARP (poly(ADP-ribose) polymerase) for nuclear fraction, and vimentin for cytoskeletal fraction).
Molecular Modeling of the N-region Interaction with the Catalytic Part of RAF Kinases
For modeling the published crystal structure of B-RAF catalytic domain was used (38). The N-terminal extension (residues Gly-442 to Ser-447 in B-RAF) was first built with an extended β-strand conformation for the backbone atoms. Backbone and side-chain atoms of the N-terminal extension were then optimized by energy minimization using 500 steps of AdoptedRaphson Newton algorithm and the Charmm22 force field employing only geometrical energy terms and no electrostatics. For the final step, a full energy term minimization was performed for the N-terminal extension, and all other residues were kept fixed, employing a distance-dependent dielectric treatment for the electrostatics. The structure model for the C-RAF kinase domain was adopted from our previous study (33).
RESULTS
Molecular Modeling Reveals Substantial Differences with Respect to the Interaction between the N-region and the RKTR motif in B-RAF and C-RAF
Based on molecular modeling we proposed previously contact points between the catalytic domain and the N-region of C-RAF and A-RAF protein (33). Notably, in our model the N-region interacts with the highly conserved RKTR tetrapeptide segment. The amino acid sequences of these regulatory motifs in A-RAF, B-RAF, and C-RAF are depicted in Fig. 1A. Contrary to the highly conserved phosphatidic acid binding region, which includes RKTR motif, the N-region of B-RAF reveals only 60% identity compared with the N-region of A-RAF and C-RAF (Fig. 1A). Of note, B-RAF Ser-446 (equivalent to C-RAF Ser-338 and A-RAF Ser-299) within the N-region was found to be constitutively phosphorylated. Together with aspartic acids at positions 448/449 (equivalent to Tyr-340/Tyr-341 in C-RAF and Tyr-301/Tyr-302 in A-RAF), the phosphorylated Ser-446 contributes to the high basal activity of B-RAF (30–32, 39). Having in mind these special features of B-RAF, we asked whether the putative interaction between the RKTR and the N-region in B-RAF may differ from those found in A-RAF and C-RAF. To address this question, we analyzed the spatial orientation of the B-RAF N-region relative to the RKTR peptide by molecular modeling and compared it with the model for C-RAF and A-RAF recently published by our group (33). As shown in the Fig. 1B, the spatial orientation of the N-region peptide relative to the RKTR segment differs significantly from that predicted for C-RAF and A-RAF (Fig. 1C (33)). In contrast to C-RAF and A-RAF, the orientation of the N-region in B-RAF is shifted relative to the RKTR motif. Moreover, as the number of contact points between the N-region and the catalytic domain in B-RAF is much larger than in C-RAF and A-RAF, a tighter binding between the N-region and the catalytic domain in B-RAF can be proposed. In our model the negatively charged 445DpSSDD449 motif of the N-region in B-RAF interacts with several basic amino acids from the αC helix: e.g. Asp-449 with Arg-506, Asp-448 with Lys-499, pSer-446 with Lys-507, and Asp-445 with Gln-496, whereas the positively charged Arg-443 of the N-region interacts with Asp-594 and the backbone carbonyl of Val-600 from the activation loop (Fig. 1B). Thus, the interactions that have been proposed to interfere with phosphorylation of Ser-338 in C-RAF and Ser-299 in A-RAF are lacking in B-RAF (33).
In this regard Garnett et al. (22) suggested that negatively charged N-region may stabilize the active conformation of the small lobe of the kinase domain. In fact, according to the steric constellation shown in Fig. 1B, an interaction between the N-region and the residues of the αC helix as well as the amino acids Asp-594 and Val-600 of the activation segment could be established. Both structural elements of the kinase domain are of essential importance for RAF kinase activity. Asp-594 and Val-600 are frequently mutated in cancer; thereby Val-600 is the most commonly mutated residue (3). Whereas Asp-594 is a part of the so-called DFG (Asp-Phe-Gly) motif and binds Mg2+, which coordinates β- and γ-phosphates of ATP, Val-600 is a part of the regulatory motif 599TVKS602 that has been found to become phosphorylated upon B-RAF activation (40). The αC helix is predicted to rotate and translate with respect to the rest of the lobe, making or breaking part of the active site (12). Given that the proposed mode of interaction between the N-region and the catalytic domain is valid for the full-length B-RAF, a stabilization of the active conformation by this interaction is indeed conceivable.
Single Substitutions of Basic Residues within the RKTR Motif Inhibit Catalytic Activity of C-RAF
The RKTR motif has been described previously as a part of the phosphatidic acid binding region and recently also as a part of the RAF dimerization surface (17, 26, 27, 34). As the importance of these basic residues within the RTKR motif for RAF activation has not been clearly defined yet, we examined the impact of substitutions for alanine on catalytic activity, subcellular localization, and dimerization of RAF kinases.
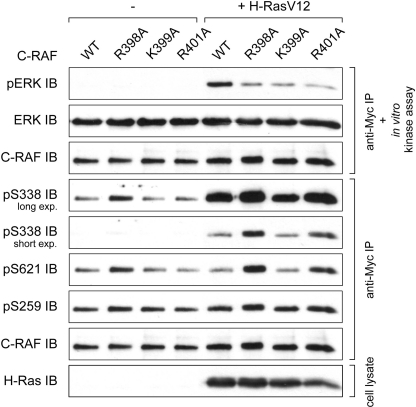
At first, we examined kinase activity of the C-RAF mutants R398A, K399A, and R401A and compared the activities with that of wild type RAF. To this end, Myc-tagged C-RAF cDNA was expressed in COS7 cells in the presence or absence of H-RasV12 as a cascade activator and isolated by anti-Myc immunoprecipitation. Subsequently, kinase activity was tested using purified MEK and ERK as coupled substrates. In all three mutants substitution of the basic residues for alanine resulted in considerable reduction of the kinase activity (Fig. 2). To address the question, whether the reduced kinase activity was due to the changes of the regulatory phosphorylation, we probed the phosphorylation status of the N-region Ser-338 and the 14-3-3 binding sites Ser-259 and Ser-621with appropriate antibodies. Surprisingly, phosphorylation status of the RKTR mutants was not consistent with the observed inhibition of their kinase activity, as phosphorylation of the N-region Ser-338 is thought to indicate the active state of C-RAF kinase. Whereas phosphorylation status of the C-RAF-K399A was almost unchanged, Ser-338 and Ser-621 phosphorylation was highly increased in C-RAF-R398A and to a lesser extent in C-RAF-R401A (Fig. 2). To test whether under physiological conditions the C-RAF mutants R398A, K399A, and R401A show similar effects as observed upon co-transfection with H-RasV12, we activated the cells with epidermal growth factor (EGF) as well. As depicted in supplemental Fig. S1., the impact of the substitutions on EGF-induced activation of C-RAF was similar to that found with H-RasV12-activated kinase (compare with Fig. 2). In contrast, the phosphorylation status of the RKTR mutants was almost unaltered (supplemental Fig. S1.).
FIGURE 2.
Single substitution of basic residues within the RKTR motif impairs catalytic activity of C-RAF. Kinase activity and phosphorylation status of the C-RAF RKTR mutants are shown. Myc-tagged C-RAF was expressed alone or together with H-RasV12 in COS7 cells. The kinase was immunoprecipitated by anti-Myc antibody, and catalytic activity was analyzed in an in vitro kinase assay using purified MEK and ERK as substrates. Phosphorylation status of C-RAF was analyzed by use of appropriate phosphospecific antibodies. Expression efficiency of H-RasV12 was determined by anti-H-Ras antibody.
In summary, these results suggest that (i) basic amino acids of the RKTR motif are crucial for the kinase activity of C-RAF, (ii) the low kinase activity of the RKTR mutants cannot be traced back to the changes in phosphorylation status, and (iii) different phosphorylation patterns of the RKTR mutants indicate alternative regulatory mechanisms for C-RAF-R398A and C-RAF-R401A versus C-RAF-K399A.
Mutation of the Putative PA Binding Sites Arg-398 and Arg-401 in C-RAF Results in Enhanced ERK-mediated Feedback Phosphorylation
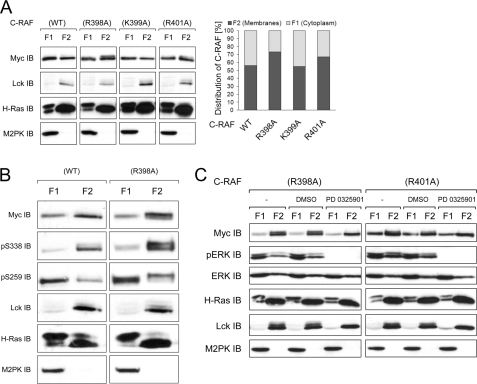
To test whether replacement of the basic residues within the C-RAF RKTR motif influences the subcellular distribution, we transfected COS7 cells with appropriate C-RAF constructs. The cells were activated by co-transfection with H-RasV12 and Lck. Subcellular fractions (cytosolic, membrane, nuclear, and cytoskeletal fractions) were collected 24 h after transfection. The results revealed that C-RAF proteins were absent from the nuclear fraction, and the cytoskeletal fraction contained only a small portion of the C-RAF protein (data not shown). Most of the C-RAF protein was distributed between the cytosol and the membrane fraction (Fig. 3A). Substitution of Lys-399 for alanine did not alter the distribution of activated C-RAF (about 56% of WT and K399A in the membrane faction), whereas exchange of Arg-398 or Arg-401 to alanine significantly increased the membrane-located portion of C-RAF up to 73% or 67%, respectively (Fig. 3A). This observation suggests that the residues Arg-398 and Arg-401 are involved in the membrane interaction and potentially also in the lipid binding of C-RAF.
FIGURE 3.
Membrane-located fraction of C-RAF-R398A and C-RAF-R401A display electrophoretic mobility shift. A, shown is subcellular distribution of the C-RAF RKTR mutants (immunoblot and quantification). Myc-tagged C-RAF WT and mutants were transfected together with H-RasV12 and Lck into COS7 cells. 24 h after transfection, cytoplasmic, membrane, nuclear (not shown), and cytoskeletal (not shown) fractions were collected. Data from three independent experiments were quantified by optical densitometry. Values represent the % ratio of C-RAF protein in each fraction relative to the total C-RAF protein detected in both cytosolic and membrane fractions together. B, phosphorylation status of the cytosol versus membrane-located C-RAF WT and C-RAF-R398A (close-up view) is shown. C, electrophoretic mobility of C-RAF-R398A and C-RAF-R401A upon treatment with the MEK inhibitor PD0325901 is shown. COS7 cells expressing Myc-tagged C-RAF-R398A and C-RAF-R401A together with H-RasV12 and Lck were treated with PD0325901 (10 μm) for 3 h before cytoplasmic, and membrane fractions were collected. PD0325901 mediated inhibition of ERK activity was confirmed by anti-pERK immunoblot. Although anti-ERK immunodetection was used as a loading control, anti-M2PK and anti-Lck were used as a control for cytosolic and membrane fraction, respectively. Expression efficiency of H-RasV12 and Lck were confirmed by appropriate antibodies.
The most intriguing finding disclosed by cell fractionation was that the RKTR mutants display differences in the electrophoretic mobility. A portion of the membrane-located mutant C-RAF-R398A and to a lesser extent the mutant C-RAF-R401A was shifted to higher molecular weight as detected by SDS-PAGE. This electrophoretic mobility shift was not found either in the sample of C-RAF WT or C-RAF-K399A. Numerous studies have reported a shift in electrophoretic mobility of RAF kinases upon multiple phosphorylation events (41–45). To find the cause of the mobility shift observed in Fig. 3A, we examined the phosphorylation status of activating Ser-338 and inhibiting Ser-259 in C-RAF-R398A compared with WT in cytosolic and membrane fractions. The immunoblotting analysis revealed that in both C-RAF WT and C-RAF-R398A, most of the protein phosphorylated at Ser-338 was located in the membranes fraction, whereas the cytosolic portion of C-RAF was preferentially phosphorylated at the inhibitory site Ser-259 (Fig. 3B). Interestingly, in the case of the mutant R398A, both the upper and the lower band of the membrane-located C-RAF was phosphorylated at Ser-338, whereas the inhibitory Ser-259 phosphorylation was present only in the protein located in the upper band. These results combined with the activity measurements (Fig. 2) suggest that the shifted portion of the C-RAF represents the kinase-inactivated form. As C-RAF has been shown to be a target for negative feedback hyperphosphorylation, which is mediated by ERK (41, 44, 46–48), we treated the cells with the highly potent MEK inhibitor PD0325901 before fractionation. The PD0325901-induced reduction of ERK activity was validated by use of an anti-phospho-ERK antibody. As shown in Fig. 3C, inhibition of the MEK/ERK pathway abolished the shifted portion of the C-RAF-R398A and C-RAF-R401A compared with untreated samples, suggesting that mobility shift observed for the mutants C-RAF-R398A and C-RAF-R401A was a result of the ERK-mediated feedback hyperphosphorylation.
RKTR Motif of A-RAF and C-RAF Shares Analogous Regulatory Functions
Previously, we and others reported that the RAF isoforms differ significantly in their regulation by the N-region (30, 32, 33, 49). In our recently published model, the residues of the kinase domain, which are involved in interaction with the N-region, are conserved in both A-RAF (Arg-359 and Lys-360) and C-RAF (Arg-398 and Lys-399) (33). Therefore, we assumed a similar regulation of A-RAF by Arg-359 and Lys-360 as found for analogous positions in C-RAF (see Fig. 2). To prove this assumption, we examined the catalytic activity, phosphorylation pattern, and subcellular distribution of A-RAF WT and A-RAF mutants R359A, K360A, and R362A. With respect to kinase activity, the results presented in Figs. 4A and S1B revealed that, similar to C-RAF, all three substitutions in A-RAF proved to be inhibitory upon activation with H-RasV12/Lck or EGF. Moreover, analogous to C-RAF-R398A, the phosphorylation of Ser-299 and Ser-582 of A-RAF-R359A was markedly increased compared with A-RAF WT (Fig. 4A). Remarkably, subcellular fractionation of the cells expressing A-RAF WT or A-RAF mutants with the modified RKTR structure also showed a shift in electrophoretic mobility for the A-RAF-R359A mutant (Fig. 4B). Furthermore, the membrane-located portion of A-RAF-R359A was increased compared with WT and other A-RAF mutants. These results suggest that similar to C-RAF, substitution of Arg-359 for alanine leads to hyperphosphorylation of the kinase upon activation with H-RasV12 and Lck. To test whether the mobility shift was result of the feedback phosphorylation by ERK, as observed for C-RAF-R398A and C-RAF-R401A, we treated the cells with the MEK inhibitor PD0325901 before fractionation. Surprisingly, inhibition of the MEK/ERK pathway did not abolish the shifted portion of the A-RAF-R359A, revealing that the origin of the mobility shift observed for A-RAF-R359A is not the same as for the C-RAF-R398A and C-RAF-R401A mutants. In summary, these data show that the regulation of A-RAF by the RKTR motif is similar to this of C-RAF.
FIGURE 4.
Regulation of A-RAF by RKTR motif is similar to C-RAF. A, shown are kinase activity and the phosphorylation status of the A-RAF RKTR mutants. Myc-tagged A-RAF WT and indicated mutants were expressed alone or together with H-RasV12 and Lck in COS7 cells. The A-RAF proteins were immunoprecipitated by anti-Myc antibody, and catalytic activity was analyzed in an in vitro kinase assay using purified MEK and ERK as substrates. Phosphorylation status of A-RAF was analyzed by use of appropriate phosphospecific antibodies. B, subcellular distribution of the A-RAF RKTR mutants (immunoblot and quantification) is shown. Myc-tagged A-RAF WT and mutants were transfected together with H-RasV12 and Lck into COS7 cells. 24 h after transfection, cytoplasmic, membrane, nuclear (not shown), and cytoskeletal (not shown) fractions were collected. Data from three independent experiments were quantified by optical densitometry. Values represent the % ratio of A-RAF protein in each fraction relative to the total A-RAF protein detected in both cytosolic and membrane fractions together. Anti-M2PK and anti-Lck immunodetection was used as a fractionation control for cytosolic and membrane fraction, respectively. Expression efficiency of H-RasV12 and Lck were confirmed by appropriate antibodies. C, electrophoretic mobility of A-RAF-R359A upon treatment with the MEK inhibitor PD0325901 is shown. COS7 cells expressing Myc-tagged A-RAF-R359A together with H-RasV12 and Lck were treated as described in Fig. 3C.
Mutation Analysis of the RKTR Motif in B-RAF Supports the Alternative Regulatory Mechanism of This Isoform
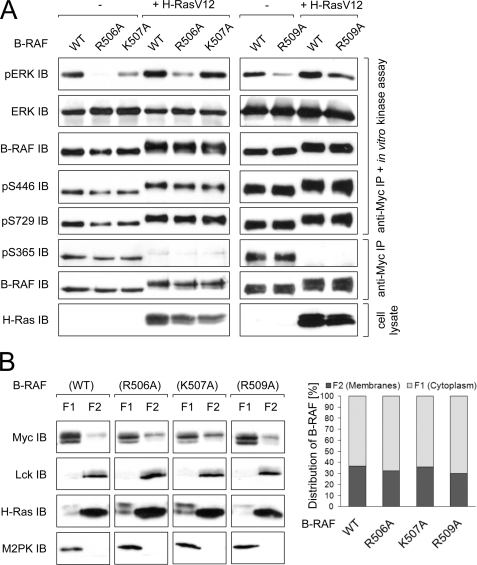
As the RKTR motif is highly conserved in all three RAF isoforms, it is very likely that this motif also plays an important role in the regulation of B-RAF. To address this issue, we tested the kinase activity of B-RAF proteins with modified RKTR sequence. We found that single substitutions of basic residues within the RKTR motif impair the kinase activity in all three cases (R506A, K507A, and R509A; see Fig. 5A), consistent with the results obtained for A-RAF and C-RAF (Figs. 2 and 4A). However, the inhibition pattern differed from that observed for A-RAF and C-RAF. Whereas in the case of A-RAF and C-RAF, the kinase activity was reduced to the similar extent in all three mutants, in B-RAF substitution of Arg-506 for alanine led to the highest reduction of activity. Furthermore, in contrast to A-RAF and C-RAF, neither phosphorylation of Ser-446 nor of Ser-729 was altered in any of the mutants (Fig. 5A). Also, the results of the subcellular fractionation revealed significant differences for B-RAF compared with A-RAF and C-RAF. Upon co-expression with H-RasV12 and Lck, B-RAF WT as well as all three B-RAF mutants were preferentially located in the cytosolic fraction (Fig. 5B). Furthermore, activation of B-RAF by H-RasV12 resulted in a shift of electrophoretic mobility compared with the non-activated kinase (Fig. 5, A and B). However, in contrast to C-RAF and A-RAF, this shift in mobility was observed for all B-RAF samples. Notably, the distribution of B-RAF between cytosol and membrane fractions was not altered by single amino acid exchanges within the RKTR motif, suggesting that in the case of B-RAF the basic residues within the RKTR motif play only a minor role with respect to their interaction with phospholipids or this interaction is not significant for subcellular distribution of this isoform. In summary, regulation of B-RAF by the RKTR motif differs fundamentally from that of A-RAF and C-RAF.
FIGURE 5.
Regulation of B-RAF by RKTR motif differs significantly from C-RAF and A-RAF. A, kinase activity and phosphorylation status of the B-RAF RKTR mutants are shown. Myc-tagged B-RAF WT and the indicated mutants were expressed alone or together with H-RasV12 in COS7 cells. The B-RAF proteins were immunoprecipitated by anti-Myc antibody, and catalytic activity was analyzed in an in vitro kinase assay using purified MEK and ERK as substrates. Phosphorylation status of B-RAF was analyzed by use of appropriate phosphospecific antibodies. B, shown is subcellular distribution of the B-RAF RKTR mutants (immunoblot and quantification). Myc-tagged B-RAF WT and mutants were transfected together with H-RasV12 and Lck into COS7 cells. 24 h after transfection, cytoplasmic, membrane, nuclear (not shown), and cytoskeletal (not shown) fractions were collected. Data from three independent experiments were quantified by optical densitometry. Values represent the % ratio of B-RAF protein in each fraction relative to the total B-RAF protein detected in both cytosolic and membrane fractions together. Anti-M2PK and anti-Lck immunodetection was used as a fractionation control for cytosolic and membrane fraction, respectively. Expression efficiency of H-RasV12 was confirmed by appropriate antibody.
RAF Dimer Formation Is Strongly Increased upon Mutation of the Second Arginine within the RKTR Motif
Basic residues of the RKTR motif have been reported to be part of the RAF dimer interface (26, 27). In the case of the isolated catalytic domain, the second arginine residue of this motif (Arg-362 in A-RAF, Arg-401 in C-RAF, and Arg-509 in B-RAF) has been suggested to be crucial for dimer formation due to the observation that mutation of this arginine abolished the homodimerization of this moiety (27). To investigate the dimerization properties of the kinase-impaired RAF mutants of the present study, we immunoprecipitated RAF dimers with an anti-Myc antibody after Myc-tagged full-length RAF proteins were expressed in COS7 cells. As Weber et al. (21) reported that dimer formation of RAF proteins is induced by active Ras, we additionally co-transfected the cells with H-RasV12 as well.
Homodimerization of C-RAF
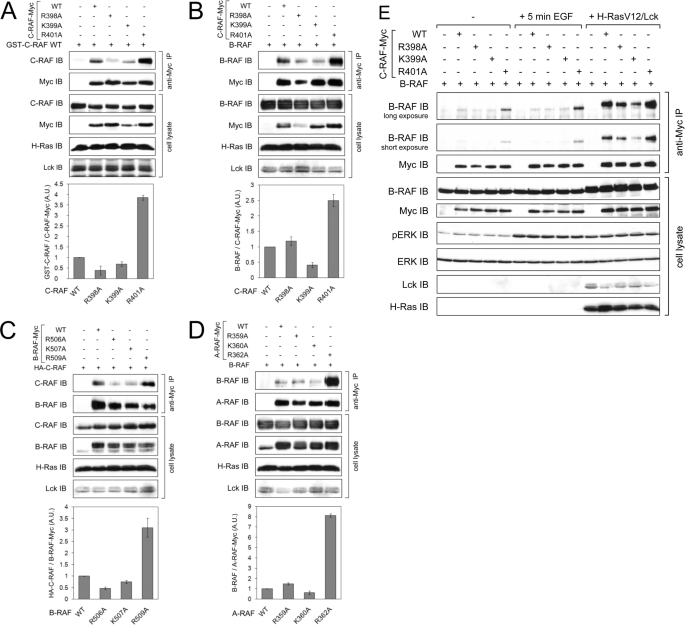
To monitor homodimer formation of C-RAF, C-RAF-Myc WT or its RKTR mutants were expressed together with a GST fusion of wild type C-RAF. As shown in Fig. 6A, full-length C-RAF WT dimerizes effectively upon activation with H-RasV12. However, dimerization was impaired when one of the protomers (C-RAF-Myc) was mutated at Arg-398 or Lys-399. Surprisingly, substitution of Arg-401 to alanine in one of the protomers led to a strong enhancement of homodimer formation (approximately up to 4-fold; see Fig. 6A).
FIGURE 6.
Substitution of the second arginine within the RKTR motif in one of the RAF protomers leads to a strongly increased homo- and heterodimer formation. Myc-tagged RAF WT or the indicated RKTR mutants were transfected into COS7 cells together with either untagged B-RAF WT, HA-tagged C-RAF WT, or GST-tagged C-RAF WT. Dimerization of RAF proteins was induced by co-transfection with H-RasV12 and Lck. RAF dimers were isolated from cell lysates by anti-Myc immunoprecipitation. Expression efficiency of H-RasV12 and Lck was confirmed by appropriate antibodies. For each part of the figure, data from three independent experiments were quantified by optical densitometry. The quantification results are expressed in terms of -fold dimer formation, where 1-fold represents the amount of immunoprecipitated dimer complexes for RAF WT. A, shown is homodimerization of Myc-tagged C-RAF WT and the indicated mutants with GST-tagged C-RAF WT. A.U., absorbance units. B, shown is heterodimerization of Myc-tagged C-RAF WT and the indicated mutants with untagged B-RAF WT. C, shown is heterodimerization of Myc-tagged B-RAF WT and the indicated mutants with HA-tagged C-RAF WT. D, shown is heterodimerization of Myc-tagged A-RAF WT and the indicated mutants with untagged B-RAF WT. E, shown is heterodimerization of Myc-tagged C-RAF WT and the indicated mutants with untagged B-RAF WT in non-stimulated cells, cells activated with EGF (100 ng/ml) for 5 min, and cells co-transfected with H-RasV12 and Lck.
Heterodimerization of C-RAF with B-RAF
To investigate heterodimer formation between C-RAF and B-RAF, we tried two different approaches; we expressed either Myc-tagged C-RAF mutants with untagged B-RAF WT or, vice versa, Myc-tagged B-RAF mutants with HA-tagged C-RAF WT. Both combinations yielded similar results with respect to C-RAF-K399A and C-RAF-R401A mutants; similar to homodimerization, dimer formation of C-RAF-K399A with B-RAF WT was reduced, whereas heterodimerization of C-RAF-R401 with B-RAF WT was about 3-fold the level observed for C-RAF WT (Fig. 6B). Similar results were obtained if C-RAF WT was precipitated with B-RAF-K507A and B-RAF-R509A (Fig. 6C). In contrast, the heterodimerization of C-RAF-R398A and B-RAF-R506A mutants was different. Whereas the ability of the mutant B-RAF-506A to dimerize with C-RAF-WT was decreased (Fig. 6C), dimer formation of the corresponding C-RAF-R398A with B-RAF WT was not impaired but, rather, slightly increased (Fig. 6B).
Heterodimerization of A-RAF with B-RAF
To complete our analyses, we tested the dimerization ability of A-RAF WT and its RKTR mutants. To this end we expressed Myc-tagged A-RAF mutants together with untagged B-RAF WT. The results of the anti-Myc immunoprecipitation revealed that dimer formation between A-RAF WT and B-RAF WT was much lower than observed between C-RAF WT and B-RAF WT (data not shown). Apart from that, the heterodimerization properties of A-RAF RKTR mutants were similar to C-RAF (compare Fig. 6, B and D). Again, the mutant A-RAF-R362A, which is analogues to C-RAF-R401A and B-RAF-R509A, displayed an extremely strong heterodimerization potency compared with A-RAF WT (Fig. 6D).
To investigate the dimerization properties of the RAF RKTR mutants under physiological conditions, we expressed C-RAF-Myc WT and mutants with untagged B-RAF WT in COS7 cells. The dimer complexes were isolated either from non-stimulated cells or cells activated with EGF for 5 min. Whereas dimerization of C-RAF-R398A and C-RAF-K399A under these conditions was similar to wild type, C-RAF-R401A mutant revealed strongly increased dimerization even in non-stimulated cells, and the dimerization of this mutant was more enhanced in cells activated with EGF (Fig. 6E).
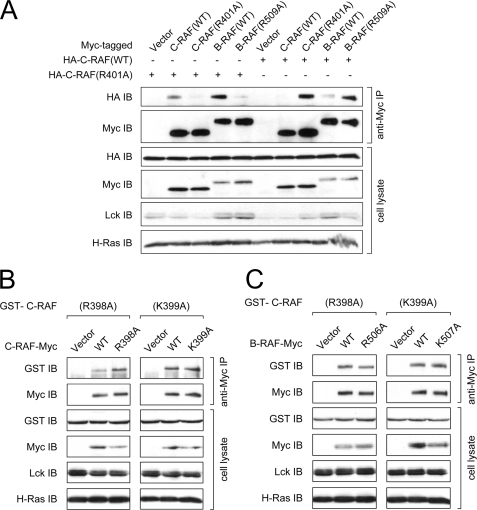
As the dimerization behavior of the C-RAF-R401A and the corresponding A-RAF and B-RAF was kind of unanticipated and in conflict with the published data (27), we tested homo- and heterodimer formation of C-RAF-R401A and B-RAF-R509A under conditions, where both protomers were mutated. The results of this experiment revealed that the strikingly increased dimerization ability of the mutant was reduced to the level of the wild type proteins if both promoters comprised the same mutation (Fig. 7A). Of note, the dimerization behavior of C-RAF mutants R398A and K399A and B-RAF mutants R506A and K507A under conditions where both protomers were mutated was similar to the situation observed with one mutated and one wild type protomer (Fig. 7, B and C). Together, these results indicate that under physiological conditions and using full-length RAF proteins, each basic amino acid of the RKTR motif influences the formation of RAF homo- and heterodimers and that the second arginine residue plays the most striking and controversial role in this process.
FIGURE 7.
Increased dimerization of the C-RAF-R401A and B-RAF-R509A was reduced to the level of the wild type if both protomers were mutated. A, homo- and heterodimerization of C-RAF-R401A mutant is shown. For homodimerization, Myc-tagged C-RAF WT or C-RAF-R401A mutant were transfected into COS7 cells together with HA-tagged C-RAF WT or C-RAF-R401A, whereas for heterodimerization, Myc-tagged B-RAF WT or B-RAF-R509A mutant were transfected together with HA-tagged C-RAF WT or C-RAF-R401A. B, homodimerization of C-RAF-R398A and C-RAF-K399A mutants is shown. For homodimerization, Myc-tagged C-RAF WT, C-RAF-R398A, or C-RAF-K399A mutants were transfected into COS7 cells together with GST-tagged C-RAF-R398A or C-RAF-K399A. C, for heterodimerization, Myc-tagged B-RAF WT, B-RAF-R506A, or B-RAF-K507A mutants were transfected into COS7 cells together with GST-tagged C-RAF-R398A or C-RAF-K399A. Dimerization of RAF proteins was induced by co-transfection with H-RasV12 and Lck. RAF dimers were isolated from cell lysates by anti-Myc immunoprecipitation. Expression efficiency of H-RasV12 and Lck were confirmed by appropriate antibodies.
DISCUSSION
Several groups previously reported that the highly conserved RKTR motif is involved in PA binding and dimerization of RAF proteins (16, 17, 26, 27, 34, 50). However, there are some inconsistencies and incompleteness of available information with respect to possible consequences of single amino acid exchanges within this motif for catalytic activity, subcellular distribution, and dimerization properties of the full-length RAF isoforms. Our study demonstrates that single substitutions of the basic residues in the RKTR motif result in impaired kinase activity of the full-length RAF isoforms (Figs. 2–5 and supplemental Fig. S1). However, these results are contradictory to the data published by Lu et al. (36), who tested the transformation capabilities of rat fibroblast cells transfected with N-terminal-truncated C-RAF constructs that correspond to isolated kinase domain and contain either C-RAF-R398A/K399A or C-RAF-K399A/R401A double amino acid exchanges. Analysis of focus formation and growth in soft agar revealed no differences between WT and the RKTR mutants (36). This suggests that either the regulation of C-RAF activity by the RKTR motif is not relevant for the isolated kinase domain-lacking N-terminal regulatory part as used in the study of Lu et al. (36) or the kinase domain per se (even activity-impaired) is sufficient for fibroblast transformation. The second explanation seems more plausible, as several recent studies reported that kinase-inactive C-RAF can still activate ERK signaling in cells via wild type C-RAF or B-RAF through homo- or heterodimer formation, respectively (24–29). Dimerization analysis in our study revealed that only C-RAF-K399A mutant was impaired in its ability to form heterodimers with wild type B-RAF, whereas C-RAF-R398A and C-RAF-R401A heterodimerized even better than wild type C-RAF (Fig. 7B). These observations suggest that mutations R398A and R401A may compensate for the dimerization defective substitution of Lys-399 in C-RAF-R398A/K399A and C-RAF-K399A/R401A, respectively. Consequently, it is more likely that the double mutants C-RAF-R398A/K399A and C-RAF-K399A/R401A are in fact impaired in their kinase activity, as also observed for the corresponding single mutants (Figs. 2, 4A, and 5A and supplemental Fig. S1); however, Lu et al. (36) did not observe any differences in the transforming ability of these mutants compared with wild type, possibly due to an unaltered dimerization with endogenous B-RAF. This consideration might be supported by the finding of the same study that the transforming ability of the C-RAF double mutant containing R401A/H402A substitutions was completely abrogated (36).
To address the question, whether the reduced catalytic activity of the RKTR mutants is caused by an altered phosphorylation status, we analyzed the activating phosphorylations of Ser-338 and Ser-621 and the inhibiting phosphorylation of Ser-259 in C-RAF as well as the effect of the corresponding sites in A-RAF and B-RAF. The most striking observation was a strongly increased phosphorylation of Ser-338 and Ser-621 in the mutant C-RAF-R398A, although the kinase activity was considerably reduced (Fig. 2 and supplemental Fig. S1A. Similarly, the corresponding A-RAF mutant R359A also showed increased phosphorylation of Ser-299 and Ser-582 (Fig. 4A). As the reduced kinases activity of the RKTR mutants could not be entirely explained by changes in the phosphorylation status, we analyzed their subcellular distribution. It has been shown that the membrane lipid PA is involved in endocytic trafficking as well as in the recruitment of proteins to endosomes (34). In addition, Rizzo et al. (16) reported that insulin stimulated the accumulation of C-RAF in endocytic vesicles containing insulin receptors, whereas substitution of Arg-398 for alanine inhibited insulin-dependent recruitment of this mutant to vesicles, possibly due to disruption of the C-RAF/PA interaction. Our results of cell fractionation revealed that the membrane-located portions of C-RAF-R398A and A-RAF-R359A were significantly elevated when compared with wild type (Figs. 3A and 4B). Membrane localization of C-RAF-R401A was also increased; however, to a lower extent (Fig. 3A). Of note, the early endosomes were detected exclusively in the cytosolic fraction, as verified by an anti-EEA-1 (an early endosome marker (51)) antibody (data not shown), suggesting that the increased membrane localization is not due to accumulation of the mutants in endocytotic vesicles. Moreover, as shown in Figs. 3 and 4B, we observed a shift in electrophoretic mobility in the SDS-PAGE pattern of the mutants with the increased membrane localization. The shift in mobility appears preferentially in the membrane-localized portion of the mutants. Several groups observed such a mobility shift of wild type C-RAF in response to stimulation by mitogens (41–45). In all cases, the decreased electrophoretic mobility was a result of hyperphosphorylation. Wartmann and Davis (43) and Dougherty et al. (41) reported that the mobility shift-associated hyperphosphorylation of C-RAF correlates with the reduction of the mitogen-stimulated C-RAF kinase activity. These data and the observation that C-RAF activation was prolonged by pharmacological inhibitors blocking ERK activation (46) or by an inhibitor of KSR that disrupts the RAF-MEK interaction (47) led to the conclusion that C-RAF is a target for ERK-mediated feedback inhibition. The mobility shift of C-RAF-R398A and C-RAF-R401A observed in our study is most likely a result of such a negative regulation through ERK-mediated phosphorylation, because treatment with the MEK inhibitor PD0325901 abolished the fraction of the C-RAF-R398A and C-RAF-R401A mutants that exhibits the mobility shift (Fig. 3C). A phosphorylation pattern of the mutant C-RAF-398A supported this assumption: whereas both the shifted and the non-shifted protein band was phosphorylated on the activating residue Ser-338, only the shifted protein band was phosphorylated on the inhibiting residue Ser-259, suggesting that the shifted portion of the protein represents the inactivated one (Fig. 3B). Considering the results published by Rizzo et al. (16) and the data of our study, we suggest that C-RAF-R398A as well as C-RAF-R401A are incapable to translocate to early endosome and, therefore, cannot be efficiently recycled. Consequently, these mutants accumulate at the plasma membrane in a hyperphosphorylated state, which is a result of the feedback phosphorylation by ERK and the enhance phosphorylation of residue Ser-259.
In the case of A-RAF-R359A, which is analogous to C-RAF-R398A, treatment with the MEK inhibitor did not prevent its mobility shift (Fig. 4C). This suggests that ERK is not the only kinase involved in hyperphosphorylation of A-RAF. Indeed, in our previous study we reported that the IH-region of A-RAF, which is target for ERK-mediated feedback phosphorylation, contains, besides putative ERK phosphorylation sites, several other sites that do not feature the ERK targeting motif SP/TP (52).
Remarkably, analysis of the B-RAF RKTR mutants did not reveal the mutant-specific shift in electrophoretic mobility and a similar accumulation in the membrane fraction as observed for A-RAF and C-RAF (compare Figs. 3A, 4B, and 5B). These results suggest that in the case of B-RAF, the basic residues within the RKTR motif either play a minor role for the interaction with PA or this interaction does not play an important role for regulation of B-RAF activation/deactivation. In fact, the RKTR motif has been implicated in binding to PA only for C-RAF and A-RAF (16, 17, 50) but not for B-RAF so far. Accordingly, endosome localization has been described for C-RAF and A-RAF (16, 53–55) but not for B-RAF. Moreover, our group has previously reported that C-RAF requires exclusively farnesylated H-Ras, whereas B-RAF associates effectively and with significantly higher affinity with both farnesylated and non-farnesylated H-Ras, suggesting that activation of B-RAF, in contrast to C-RAF, may take place both at the plasma membrane and in the cytosolic environment (56).
As the hyperphosphorylation of C-RAF-R398A and A-RAF-R359A was not found for the corresponding B-RAF mutant, although its kinase activity was similarly impaired, we concluded that an additional regulatory mechanism was involved. The recently published crystal structure of a C-RAF homodimer revealed that Arg-398, Lys-399, and Arg-401 are part of the dimerization interface, assigning a new function to the RKTR motif (26, 27). Supposing that the reduced kinase activity of the RKTR mutants may derive from an impaired dimer formation, we tested their dimerization ability in an co-immunoprecipitation assay. Surprisingly, only substitution of the lysine residue for alanine resulted in reduced heterodimerization if one of the protomers was wild type (Fig. 6, A–D). In contrast, heterodimer formation was slightly increased for the construct in which the first arginine was mutated, and it was exceedingly increased if the second arginine was exchanged in only one of the protomers (Fig. 6, A–D). At first glance these findings seem to be in conflict with the data of Rajakulendran et al. (27), who reported that substitution of Arg-509 for histidine resulted in a dimerization-deficient B-RAF mutant. However, the data of Rajakulendran et al. (27) and results of our study are not directly comparable due to the differences in experimental settings. (i) Whereas Rajakulendran et al. (27) tested dimerization of the truncated kinase domain, which lacks the N-terminal regulatory part as well as the C-terminal 14-3-3 binding site (the latter is reported to be important for RAF dimerization (21, 23, 57)) and contains numerous additional mutations that increase the level of soluble protein expression in Escherichia coli, we analyzed the dimerization properties of the full-length protein. (ii) Rajakulendran et al. (27) isolated RAF dimers from E. coli, which implicates a lack of phosphorylation. In contrast, in our study we isolated RAF dimers from mammalian cells. (iii) Finally, substitution for alanine is different compared with the exchange by histidine, as the latter can result in steric hindrance due to the larger side chain of histidine compared with alanine. On the other hand, Hatzivassiliou et al. (26) did not observe a significant inhibition of the heterodimer formation between B-RAF-R509H and C-RAF-R401H compared with wild type using full-length proteins expressed in HCT116 cells. An apparent inhibition of the heterodimerization appeared only if both wild type and mutant samples were treated with dimerization promoting drug GDC-0879 (26). In accordance with the data of Hatzivassiliou et al. (26), the results of our study demonstrate that the strikingly increased dimerization ability of the mutants B-RAF-R509A, C-RAF-R401A, and A-RAF-R362A was reduced to the level of wild type only if both protomers contained the same mutation (Fig. 7A). Thus, substitution for alanine in position Arg-509 in B-RAF, Arg-401 in C-RAF, and Arg-362 in A-RAF seems to mimic the situation that was observed for some structurally related ATP-competitive RAF kinase inhibitors; drug binding to one member of RAF homodimer or heterodimer inhibits one protomer but results in transactivation of the drug-free protomer (25–29). This suggests that substitution of the second arginine residue within the RKTR motif for alanine may result in a similar structural rearrangement of the RAF kinase domain, as was found for the C-RAF kinase domain co-crystallized with a GDC-0879 analog (26). Furthermore, the discrepancy between the data of Rajakulendran et al. (27) on the one hand and the data of Hatzivassiliou et al. (26) and data presented here on the other hand may indicate that severe dimerization problems of the isolated RAF kinase domains containing mutations within the RKTR motif and lacking C-terminal 14-3-3 binding site can be corrected by binding of 14-3-3 proteins to the full-length mutant and stabilizing the dimer. In fact, several groups reported that the presence of the C-terminal 14-3-3 binding sites surrounding Ser-621 in C-RAF and Ser-729 in B-RAF are indispensible for heterodimerization of C-RAF and B-RAF (21, 23, 57). Accordingly, the structural model of a RAF side-to-side dimer is compatible with the spatial requirements for binding to dimeric 14-3-3 proteins (27).
In conclusion, the results of the present study clearly show that the RKTR motif is a cross-point of multiple regulatory mechanisms. According to our model, this motif is located close to the regulatory N-region in all three RAF isoforms (Ref. 33 and Fig. 1B and C), and this close proximity may influence phosphorylation of Ser-338 in C-RAF and Ser-299 in A-RAF and activation of these kinases. Furthermore, due to the fact that the RKTR motif is a part of the αC helix, a key structural element whose conformation executes a regulatory function in several protein kinases (58), structural modifications within this motif per se may impair the catalytic efficiency of the RAF kinase domain. The proposed position of the N-region suggests that the RKTR motif is masked in the full-length RAF monomer, and intramolecular rearrangements are required to displace the N-region and to expose the RKTR tetrapeptide segment to exert its intrinsic functions in dimerization and PA binding. The mode of interactions between the N-region and the RKTR motif predicted for B-RAF may stabilize the active form of the monomeric RAF kinase, whereas in the RAF dimer the stabilizing function of the N-region would be adopted by the complimentary RKTR motif of the dimerization partner.
Supplementary Material
Acknowledgment
We thank Laura A. Goldberg (Lafayette College, PA) for proofreading.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 487, Projects C3 and B2.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PA
- phosphatidic acid
- COS7
- African green monkey SV40-transfected kidney fibroblast cell line
- IB
- immunoblot
- IP
- immunoprecipitation.
REFERENCES
- 1. Rapp U. R., Goldsborough M. D., Mark G. E., Bonner T. I., Groffen J., Reynolds F. H., Jr., Stephenson J. R. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 4218–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dhillon A. S., Kolch W. (2002) Arch. Biochem. Biophys. 404, 3–9 [DOI] [PubMed] [Google Scholar]
- 3. Wellbrock C., Karasarides M., Marais R. (2004) Nat. Rev. Mol. Cell Biol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 4. Rapp U. R., Götz R., Albert S. (2006) Cancer Cell 9, 9–12 [DOI] [PubMed] [Google Scholar]
- 5. Polzien L., Baljuls A., Rennefahrt U. E., Fischer A., Schmitz W., Zahedi R. P., Sickmann A., Metz R., Albert S., Benz R., Hekman M., Rapp U. R. (2009) J. Biol. Chem. 284, 28004–28020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonner T. I., Kerby S. B., Sutrave P., Gunnell M. A., Mark G., Rapp U. R. (1985) Mol. Cell. Biol. 5, 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daum G., Eisenmann-Tappe I., Fries H. W., Troppmair J., Rapp U. R. (1994) Trends Biochem. Sci. 19, 474–480 [DOI] [PubMed] [Google Scholar]
- 8. Huleihel M., Goldsborough M., Cleveland J., Gunnell M., Bonner T., Rapp U. R. (1986) Mol. Cell. Biol. 6, 2655–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ikawa S., Fukui M., Ueyama Y., Tamaoki N., Yamamoto T., Toyoshima K. (1988) Mol. Cell. Biol. 8, 2651–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. (2002) Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 11. Schreck R., Rapp U. R. (2006) Int. J. Cancer 119, 2261–2271 [DOI] [PubMed] [Google Scholar]
- 12. Roskoski R., Jr. (2010) Biochem. Biophys. Res. Commun. 399, 313–317 [DOI] [PubMed] [Google Scholar]
- 13. Hagemann C., Rapp U. R. (1999) Exp. Cell Res. 253, 34–46 [DOI] [PubMed] [Google Scholar]
- 14. Hekman M., Hamm H., Villar A. V., Bader B., Kuhlmann J., Nickel J., Rapp U. R. (2002) J. Biol. Chem. 277, 24090–24102 [DOI] [PubMed] [Google Scholar]
- 15. Prior I. A., Harding A., Yan J., Sluimer J., Parton R. G., Hancock J. F. (2001) Nat. Cell Biol. 3, 368–375 [DOI] [PubMed] [Google Scholar]
- 16. Rizzo M. A., Shome K., Watkins S. C., Romero G. (2000) J. Biol. Chem. 275, 23911–23918 [DOI] [PubMed] [Google Scholar]
- 17. Ghosh S., Strum J. C., Sciorra V. A., Daniel L., Bell R. M. (1996) J. Biol. Chem. 271, 8472–8480 [DOI] [PubMed] [Google Scholar]
- 18. Moodie S. A., Willumsen B. M., Weber M. J., Wolfman A. (1993) Science 260, 1658–1661 [DOI] [PubMed] [Google Scholar]
- 19. Zhang X. F., Settleman J., Kyriakis J. M., Takeuchi-Suzuki E., Elledge S. J., Marshall M. S., Bruder J. T., Rapp U. R., Avruch J. (1993) Nature 364, 308–313 [DOI] [PubMed] [Google Scholar]
- 20. Rajalingam K., Schreck R., Rapp U. R., Albert S. (2007) Biochim. Biophys. Acta 1773, 1177–1195 [DOI] [PubMed] [Google Scholar]
- 21. Weber C. K., Slupsky J. R., Kalmes H. A., Rapp U. R. (2001) Cancer Res. 61, 3595–3598 [PubMed] [Google Scholar]
- 22. Garnett M. J., Rana S., Paterson H., Barford D., Marais R. (2005) Mol. Cell 20, 963–969 [DOI] [PubMed] [Google Scholar]
- 23. Rushworth L. K., Hindley A. D., O'Neill E., Kolch W. (2006) Mol. Cell. Biol. 26, 2262–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hall-Jackson C. A., Eyers P. A., Cohen P., Goedert M., Boyle F. T., Hewitt N., Plant H., Hedge P. (1999) Chem. Biol. 6, 559–568 [DOI] [PubMed] [Google Scholar]
- 25. Heidorn S. J., Milagre C., Whittaker S., Nourry A., Niculescu-Duvas I., Dhomen N., Hussain J., Reis-Filho J. S., Springer C. J., Pritchard C., Marais R. (2010) Cell 140, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatzivassiliou G., Song K., Yen I., Brandhuber B. J., Anderson D. J., Alvarado R., Ludlam M. J., Stokoe D., Gloor S. L., Vigers G., Morales T., Aliagas I., Liu B., Sideris S., Hoeflich K. P., Jaiswal B. S., Seshagiri S., Koeppen H., Belvin M., Friedman L. S., Malek S. (2010) Nature 464, 431–435 [DOI] [PubMed] [Google Scholar]
- 27. Rajakulendran T., Sahmi M., Lefrançois M., Sicheri F., Therrien M. (2009) Nature 461, 542–545 [DOI] [PubMed] [Google Scholar]
- 28. Poulikakos P. I., Zhang C., Bollag G., Shokat K. M., Rosen N. (2010) Nature 464, 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cox A. D., Der C. J. (2010) Cancer Cell 17, 221–223 [DOI] [PubMed] [Google Scholar]
- 30. Marais R., Light Y., Paterson H. F., Mason C. S., Marshall C. J. (1997) J. Biol. Chem. 272, 4378–4383 [DOI] [PubMed] [Google Scholar]
- 31. Diaz B., Barnard D., Filson A., MacDonald S., King A., Marshall M. (1997) Mol. Cell. Biol. 17, 4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mason C. S., Springer C. J., Cooper R. G., Superti-Furga G., Marshall C. J., Marais R. (1999) EMBO J. 18, 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baljuls A., Mueller T., Drexler H. C., Hekman M., Rapp U. R. (2007) J. Biol. Chem. 282, 26575–26590 [DOI] [PubMed] [Google Scholar]
- 34. Andresen B. T., Rizzo M. A., Shome K., Romero G. (2002) FEBS Lett. 531, 65–68 [DOI] [PubMed] [Google Scholar]
- 35. Ghosh S., Moore S., Bell R. M., Dush M. (2003) J. Biol. Chem. 278, 45690–45696 [DOI] [PubMed] [Google Scholar]
- 36. Lu K. K., Bazarov A. V., Yoon L. S., Sedivy J. M. (1998) Cell Growth Differ. 9, 367–380 [PubMed] [Google Scholar]
- 37. Hekman M., Wiese S., Metz R., Albert S., Troppmair J., Nickel J., Sendtner M., Rapp U. R. (2004) J. Biol. Chem. 279, 14074–14086 [DOI] [PubMed] [Google Scholar]
- 38. Wan P. T., Garnett M. J., Roe S. M., Lee S., Niculescu-Duvaz D., Good V. M., Jones C. M., Marshall C. J., Springer C. J., Barford D., Marais R. (2004) Cell 116, 855–867 [DOI] [PubMed] [Google Scholar]
- 39. Fabian J. R., Daar I. O., Morrison D. K. (1993) Mol. Cell. Biol. 13, 7170–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang B. H., Guan K. L. (2000) EMBO J. 19, 5429–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dougherty M. K., Müller J., Ritt D. A., Zhou M., Zhou X. Z., Copeland T. D., Conrads T. P., Veenstra T. D., Lu K. P., Morrison D. K. (2005) Mol. Cell 17, 215–224 [DOI] [PubMed] [Google Scholar]
- 42. Morrison D. K., Kaplan D. R., Rapp U., Roberts T. M. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 8855–8859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wartmann M., Davis R. J. (1994) J. Biol. Chem. 269, 6695–6701 [PubMed] [Google Scholar]
- 44. Wartmann M., Hofer P., Turowski P., Saltiel A. R., Hynes N. E. (1997) J. Biol. Chem. 272, 3915–3923 [DOI] [PubMed] [Google Scholar]
- 45. Chen C., Sytkowski A. J. (2004) Blood 104, 73–80 [DOI] [PubMed] [Google Scholar]
- 46. Alessi D. R., Cuenda A., Cohen P., Dudley D. T., Saltiel A. R. (1995) J. Biol. Chem. 270, 27489–27494 [DOI] [PubMed] [Google Scholar]
- 47. Matheny S. A., Chen C., Kortum R. L., Razidlo G. L., Lewis R. E., White M. A. (2004) Nature 427, 256–260 [DOI] [PubMed] [Google Scholar]
- 48. Hekman M., Fischer A., Wennogle L. P., Wang Y. K., Campbell S. L., Rapp U. R. (2005) FEBS Lett. 579, 464–468 [DOI] [PubMed] [Google Scholar]
- 49. Ritt D. A., Zhou M., Conrads T. P., Veenstra T. D., Copeland T. D., Morrison D. K. (2007) Curr. Biol. 17, 179–184 [DOI] [PubMed] [Google Scholar]
- 50. Johnson L. M., James K. M., Chamberlain M. D., Anderson D. H. (2005) Biochemistry 44, 3432–3440 [DOI] [PubMed] [Google Scholar]
- 51. Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. (1995) J. Biol. Chem. 270, 13503–13511 [DOI] [PubMed] [Google Scholar]
- 52. Baljuls A., Schmitz W., Mueller T., Zahedi R. P., Sickmann A., Hekman M., Rapp U. R. (2008) J. Biol. Chem. 283, 27239–27254 [DOI] [PubMed] [Google Scholar]
- 53. Nekhoroshkova E., Albert S., Becker M., Rapp U. R. (2009) PLoS ONE 4, e4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pelkmans L., Fava E., Grabner H., Hannus M., Habermann B., Krausz E., Zerial M. (2005) Nature 436, 78–86 [DOI] [PubMed] [Google Scholar]
- 55. Pelkmans L., Zerial M. (2005) Nature 436, 128–133 [DOI] [PubMed] [Google Scholar]
- 56. Fischer A., Hekman M., Kuhlmann J., Rubio I., Wiese S., Rapp U. R. (2007) J. Biol. Chem. 282, 26503–26516 [DOI] [PubMed] [Google Scholar]
- 57. Ritt D. A., Monson D. M., Specht S. I., Morrison D. K. (2010) Mol. Cell. Biol. 30, 806–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huse M., Kuriyan J. (2002) Cell 109, 275–282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.