Abstract
The mammalian retina is a tractable model system for analyzing transcriptional networks that guide neural development. Spalt family zinc-finger transcription factors play a crucial role in photoreceptor specification in Drosophila, but their role in mammalian retinal development has not been investigated. In this study, we show that that the spalt homolog Sall3 is prominently expressed in developing cone photoreceptors and horizontal interneurons of the mouse retina and in a subset of cone bipolar cells. We find that Sall3 is both necessary and sufficient to activate the expression of multiple cone-specific genes, and that Sall3 protein is selectively bound to the promoter regions of these genes. Notably, Sall3 shows more prominent expression in short wavelength-sensitive cones than in medium wavelength-sensitive cones, and that Sall3 selectively activates expression of the short but not the medium wavelength-sensitive cone opsin gene. We further observe that Sall3 regulates the differentiation of horizontal interneurons, which form direct synaptic contacts with cone photoreceptors. Loss of function of Sall3 eliminates expression of the horizontal cell-specific transcription factor Lhx1, resulting in a radial displacement of horizontal cells that partially phenocopies the loss of function of Lhx1. These findings not only demonstrate that Spalt family transcription factors play a conserved role in regulating photoreceptor development in insects and mammals, but also identify Sall3 as a factor that regulates terminal differentiation of both cone photoreceptors and their postsynaptic partners.
Keywords: Cell fate, Evolution, Neural development, Photoreceptor, Transcription factor, Mouse
INTRODUCTION
The mammalian retina is composed of seven major cell classes, with intrinsic cues being sufficient for specification of the majority of cell types (Cayouette et al., 2003; Cepko, 1999; Hatakeyama and Kageyama, 2004; Livesey and Cepko, 2001). Retinal photoreceptors comprise two major classes, the rods and cones, which detect dim and bright light, respectively. Several transcription factors have been identified that regulate photoreceptor differentiation. Some, such as the homeodomain factors Otx2 and Crx, are required for the differentiation and survival of both rods and cones (Chen et al., 1997; Furukawa et al., 1997; Nishida et al., 2003). Others, including Rorβ, Nrl and Nr2e3, repress the expression of cone-specific genes in rod photoreceptor precursors while often also activating the expression of rod-specific genes (Akhmedov et al., 2000; Haider et al., 2000; Jia et al., 2009; Mears et al., 2001). A number of cone-expressed transcription factors have been identified, including Trβ (Thrβ), Rxrγ and Rorα, which together with the E3 SUMO ligase Pias3 simultaneously drive expression of medium wavelength-sensitive cone opsin (Mop; Opn1mw) while repressing the expression of short wavelength-sensitive cone opsin (Sop; Opn1sw) (Fujieda et al., 2009; Onishi et al., 2010a; Roberts et al., 2005; Roberts et al., 2006).
Photoreceptors synapse onto both bipolar interneurons and GABAergic interneurons known as horizontal cells. Horizontal cells send lateral inhibitory projections to multiple nearby photoreceptors through their large dendritic arbors, enhancing spatial resolution in light detection. Horizontal cells exit mitosis between embryonic day (E) 12 and E16 in mice, and initially migrate towards the ganglion cell layer, but then reverse direction and migrate towards their final position at the outer edge of the inner nuclear layer (Edqvist and Hallbook, 2004). Several transcription factors regulate horizontal cell differentiation, including Foxn4, Ptf1a and Prox1 (Dyer et al., 2003; Fujitani et al., 2006; Li et al., 2004). Prox1 regulates the expression of Lhx1, which is necessary for horizontal cells to adopt their appropriate laminar position. Horizontal cells of Lhx1−/− mice fail to undergo radial migration and instead adopt positions in the inner portion of the inner nuclear layer, resembling wide-field amacrine cells in their morphology and dendritic arborization, while continuing to express molecular markers of horizontal cells (Poche et al., 2007).
We have previously identified the zinc-finger transcription factor Sall3 as prominently and selectively expressed in developing mouse retina (Blackshaw et al., 2004). Sall3 is a homolog of the spalt gene of Drosophila, which plays a crucial role in multiple aspects of photoreceptor development (Domingos et al., 2004; Mollereau et al., 2001; Sprecher et al., 2007). To determine whether Sall3 might play an evolutionarily conserved role in mammalian retinal differentiation, we conducted both gain- and loss-of-function analyses of Sall3 in developing mouse retina.
MATERIALS AND METHODS
Animals
Timed pregnant CD-1 mice and adult C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA, USA). CB6-Tg(Gad1-EGFP)G42Zjh/J mice (Huckfeldt et al., 2009) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Sall3 knockout mice (Sall3−/−) (Parrish et al., 2004) were kindly provided by A. P. Monaghan (University of Pittsburgh, Pittsburgh, PA, USA) and Nrl knockout mice were kindly provided by A. Swaroop (National Institutes of Health, Bethesda, MD, USA). Sall3−/−; Gad1-EGFP mice were generated by breeding Sall3+/− mice into the Gad1-EGFP line and subsequent backcrossing. Six3-Cre mice were kindly provided by Yasuhide Furuta (M. D. Anderson Cancer Center, University of Texas, Houston, TX, USA) and Lhx1lox/lox mice were kindly provided by Richard Behringer (M. D. Anderson Cancer Center). All experimental animal procedures were pre-approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Immunohistochemistry and TUNEL staining
For immunohistochemical analysis of retinal cryosections, eyes were carefully dissected and intact retinas removed. The retinas were fixed in cold 4% paraformaldehyde (PFA) for 40 minutes (E16), 50 minutes [postnatal day (P) 0] or 1 hour (P7, P14) and then immersed in 30% sucrose in PBS overnight before embedding in O.C.T. compound (Sakura Finetek, USA). Sections (25 μm) of embedded retinas were cut on a Leica CM3050S cryostat and blocked with PBS containing 5% horse serum, 0.2% Triton X-100, 0.02% sodium azide and 0.1% BSA for 1 hour at room temperature and then subjected to immunohistochemistry as previously described (de Melo et al., 2003).
Primary antibodies used were goat anti-Brn3 (1:200; C-13, sc-6026, Santa Cruz Biotechnology), mouse anti-calbindin (1:200; C9848, Sigma-Aldrich), rabbit anti-calretinin (1:200; AB5054, Chemicon), goat anti-Chat (1:100; AB144P, Chemicon), sheep anti-Chx10 (1:200; X1180P, Exalpha Biologicals), mouse anti-cone arrestin [1:500; a generous gift of Cheryl Craft (Zhu et al., 2002)], rabbit anti-Dab1 (1:200; a generous gift of Brian Howell, SUNY Upstate Medical University, Syracuse, NY, USA), mouse anti-Gad1/2 [1:200; GAD-6, Developmental Studies Hybridoma Bank (DSHB), University of Iowa], goat anti-GFP (1:500; 600-101-215, Rockland Immunochemicals), rabbit anti-GFP (1:1000; A6455, Invitrogen), mouse anti-glutamine synthase (GS) (1:200; 610518, BD Biosciences), rabbit anti-Glyt1 (1:200; AB1770, Chemicon), mouse anti-Ki67 (1:200; 550609, BD Pharmingen), mouse anti-Lhx1 (1:200; 4F2, DSHB), rabbit anti-Lhx2 (1:1500; generated for this study by Covance), mouse anti-NF165 (1:200; 2H3, DSHB), chicken anti-M-opsin (1:500; a generous gift of Jeremy Nathans, Johns Hopkins University School of Medicine, Baltimore, MD, USA), mouse anti-P27kip1 (1:200; 18-2370, Invitrogen), mouse anti-Pax6 (1:200; DSHB), rabbit anti-parvalbumin (1:200; PV-28, Swant), mouse anti-PKCα (1:200; 05-154, Millipore), mouse anti-Prox1 (1:200; MAB5654, Millipore), mouse anti-rhodopsin (1:200; a generous gift of Robert Molday, University of British Columbia, Vancouver, Canada), rabbit anti-Rxrγ (1:200; SC-555, Santa Cruz Biotechnology), rabbit anti-Sall3 (1:500; HPA016656, Sigma), goat anti-S-opsin (1:500; N-20, sc-14363, Santa Cruz Biotechnology) and mouse anti-syntaxin (1:200; S0664, Sigma). Secondary antibodies (1:500) used were: Alexa 568 goat anti-chicken IgG (A11041, Invitrogen); FITC-conjugated donkey anti-goat IgG (705-095-147), anti-mouse IgG (715-095-150), anti-rabbit IgG (711-095-152) and anti-sheep IgG (713-095-147) and Texas Red-conjugated donkey anti-goat IgG (705-075-147), anti-mouse IgG (715-075-150) and anti-rabbit IgG (711-075-152) (all Jackson ImmunoResearch). All section immunohistochemical data shown were imaged and photographed on a Zeiss 510 confocal microscope.
TUNEL assays were performed using a TMRred In Situ Cell Death Detection Kit as per the manufacturer's instructions (Roche Applied Science). E16.5 and P0 retina sections were prepared and sectioned as described above. TUNEL assays were imaged and photographed on a Zeiss 510 confocal microscope.
Retinal explant culture preparation and flatmount immunohistochemistry
Retinas used for explant cultures were dissected at P0 as described above in sterile PBS. The dorsal pole was marked by notching the retinal tissue. Four cuts were made using microdissection scissors and the retinas were transferred to Nucleopore Track-Etch membrane filters (Whatman) and the tissue flattened out evenly. Filters were then floated on DMEM/F12 containing 10% FBS and 1× penicillin/streptomycin in 12-well plates and incubated at 37°C for 7 or 14 days. Explants were harvested and immersed in 4% PFA for 1 hour, then immersed in 30% sucrose in PBS overnight and embedded in O.C.T. compound. Immunohistochemistry was performed as described above.
Flatmount staining was performed by removing the sclera, choroid and cornea in PBS. Retinas are then fixed in 4% PFA for 30 minutes, washed three times in PBS and blocked for 1 hour. Retinas were then incubated in primary antibody for 48 hours at 4°C with gentle agitation, washed three times in PBS for 20 minutes each and incubated in secondary antibody for 24 hours. Retinas were then washed three times for 20 minutes each in PBS, dissected, flattened and mounted with Vectashield (Vector Labs).
In vivo electroporation
In vivo electroporation experiments were performed as previously described (Matsuda and Cepko, 2004). pCAG-Sall3 was constructed by cloning the full-length open reading frame of the human SALL3 gene (derived from BC148296) into the pCAGIG vector. Approximately 0.3 μl of 5 μg/μl DNA solution was injected into the subretinal space of P0 mouse retinas and square electric pulses were applied (100 volts, five 50-millisecond pulses at 950-millisecond intervals). Electroporated retinas were harvested at P14.
Microarray analysis
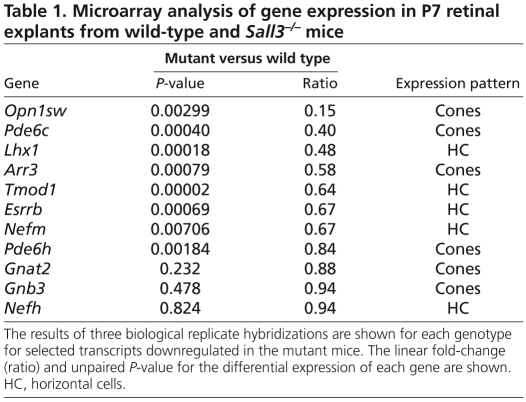
Sall3+/+ and Sall3−/− retinas were explanted as described for 7 days. Retinas were harvested and total RNA was extracted using the RNeasy Mini Kit (Qiagen). Two explants for each genotype were pooled and three replicates were prepared. The independent RNA preparations were labeled and hybridized essentially as previously described using the Affymetrix Mouse Exon 1.0 array platform (Onishi et al., 2010b). Data were analyzed using Spotfire (TIBCO). A list of previously identified cone-enriched genes was compiled from previously published microarray data (Corbo et al., 2007; Jia et al., 2009). The P-value for the cone enrichment of transcripts that were significantly downregulated (P<0.05) in the microarray data set relative to all detectable transcripts was calculated using one-way ANOVA. The sources for the horizontal cell-enriched genes shown in Table 1 are as follows: Nefm and Nefh (Chien and Liem, 1995), Esrrb (Onishi et al., 2010b), Tmod1 (Yao and Sung, 2009). Microarray data have been deposited in GEO under accession number GSE24083.
Table 1.
Microarray analysis of gene expression in P7 retinal explants from wild-type and Sall3−/− mice
Chromatin immunoprecipitation (ChIP)
ChIP was performed using six pooled retinas from P7 C57BL/6J mice as previously described (Peng and Chen, 2005). A rabbit antibody to Sall3 (3 μl of 1 mg/ml; ab41740, Abcam) and the normal rabbit IgG control (Santa Cruz Biotechnology) were used for immunoprecipitation (IP). The immunoprecipitated DNA and the input (without IP) and mock (no chromatin DNA) controls were analyzed by PCR using primers spanning the promoter or 3′ regions of each gene (Peng and Chen, 2005). Quantitative real-time PCR was performed using the SYBR Green Jumpstart Taq Readymix qPCR Kit (Sigma) and CFX96 Real-Time PCR System (BioRad) according to the manufacturers' protocols. The results of ChIP assays were analyzed by candidate gene-based PCR using primers spanning the promoter region of each gene. The data shown in Fig. 6 are representative of a minimum of three replicate experiments. Controls were performed using normal rabbit IgG (Santa Cruz Biotechnology) in IP reactions as negative controls and input without IP as positive controls for PCR reactions.
Fig. 6.
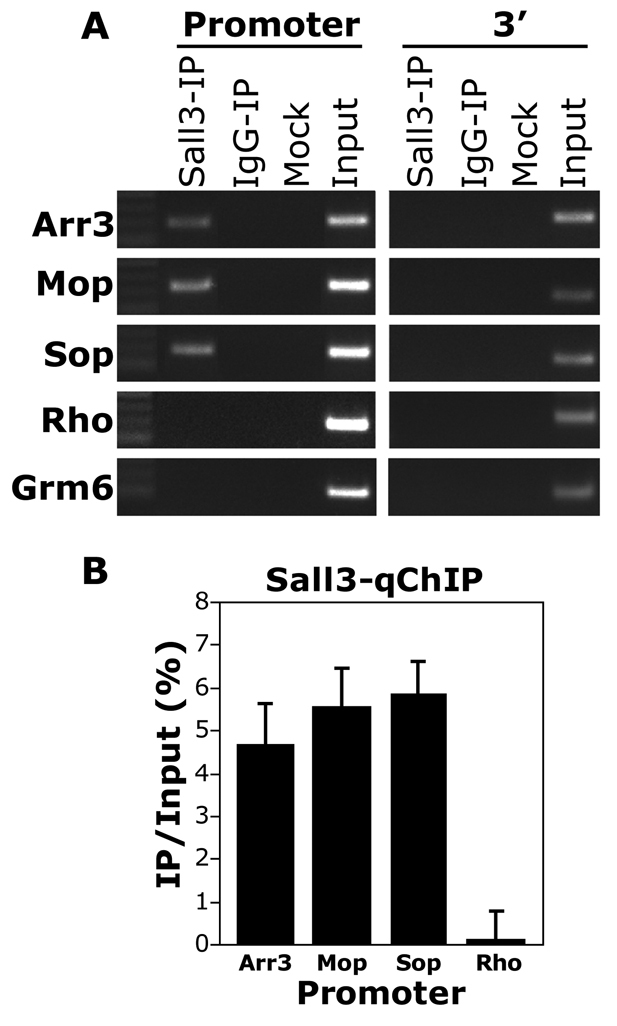
ChIP assay demonstrating direct binding of Sall3 to promoters of cone-specific genes. (A) Sall3 binds to the promoter of the cone-specific genes Arr3, Sop and Mop, but not to the promoter of the rod-specific gene rhodopsin (Rho) or the bipolar-specific gene Grm6. Sall3 does not bind to the 3′ regions of cone-, rod- or bipolar-specific genes. Sall3-IP, Sall3 immunoprecipitation; IgG-IP, non-specific rabbit IgG control; mock, no chromatin negative control; input, PCR positive control. (B) Quantitative real-time PCR experiment representing IP to input ratio (%) following normalization to the IgG control values. Arr3, 4.69%; Mop, 5.57%; Sop, 5.89%; Rho, 0.1%. Error bars represent s.e.m. from three independent experiments (n=3).
RESULTS
Sall3 is prominently expressed in differentiating horizontal interneurons and cone photoreceptors
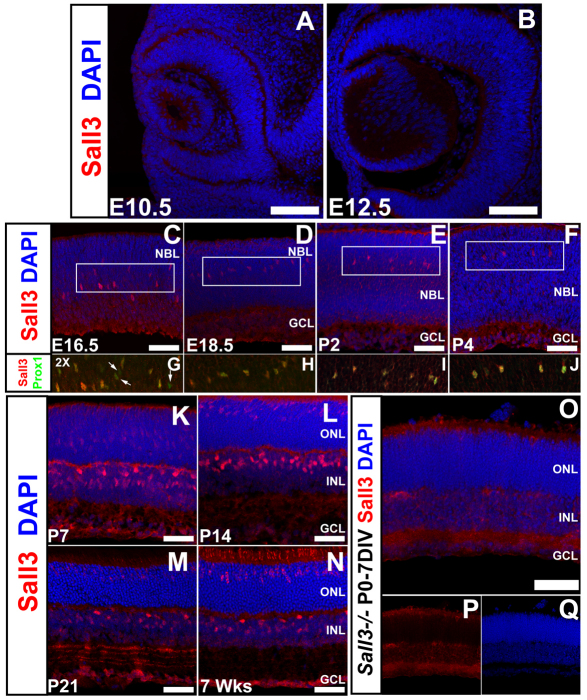
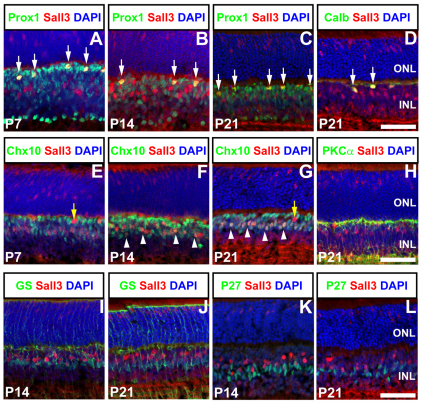
Our previous in situ hybridization data suggested that Sall3 RNA is expressed in developing horizontal cells beginning at ~E16, with expression later detected in a subset of photoreceptors, and in an uncharacterized cell population in the retinal inner nuclear layer (INL) (Blackshaw et al., 2004). To map Sall3 protein distribution during retinal development, we performed immunohistochemistry with antibodies to Sall3. Sall3 protein expression was first detected at E16.5 in developing horizontal cells, as determined by coexpression with the horizontal cell marker Prox1 (Fig. 1C-J). Prox1 protein expression is first detected in the retina at E14.5 (Dyer et al., 2003). Immunostaining for Sall3 at E14.5 demonstrated that Prox1 expression precedes that of Sall3 (see Fig. S1A-D in the supplementary material). Sall3 expression was confined to postmitotic cells, as no Sall3+ Ki67+ cells were detected at any time point analyzed (see Fig. S1E-M in the supplementary material). By P5, Sall3 protein was detected in a population of cells in both the medial INL and the outer nuclear layer (ONL) (see Fig. S1K,L and Fig. S2A,B in the supplementary material). This pattern of ONL, medial INL and horizontal cell staining persisted into adulthood (7 weeks postnatal) (Fig. 1N). Immunostaining for the horizontal cell-specific markers Lhx1 (Liu et al., 2000), NF165 (Nefm) (de Melo et al., 2003) and calbindin (Hamano et al., 1990) confirmed that Sall3 is expressed in differentiating horizontal cells (Fig. 2D; see Fig. S2A-C,G-I in the supplementary material). Sall3-expressing cells in the medial INL coexpressed the pan-bipolar marker Chx10 (Vsx2) from P7 onward (Fig. 2E-G), but not the rod bipolar marker PKCα (Prkcα) (Fig. 2H). Sall3-positive cells did not coexpress the Müller glia markers glutamine synthase or p27kip1 (Cdkn1b) (Fig. 2I-L), further suggesting that these cells are a cone bipolar subpopulation. Sall3 protein was absent at all times from amacrine cells and retinal ganglion cells (Fig. 1; data not shown).
Fig. 1.
Expression of Sall3 protein during mouse retinal development. (A-C) Expression of Sall3 was first identified at E16.5 in a population of cells scattered in the neuroblastic layer (NBL). (D-F) Expression of Sall3 remains restricted to cells in the medial NBL until P4. The boxed areas in C-F are magnified 2× in G-J. (G-J) These cells robustly coexpress the transcription factor Prox1, indicating that they are horizontal cells. Prox1-positive Sall3-negative cells (G, arrows) are present at E16.5 but not at E18.5-P4 (H-J). (K-N) Sall3 expression is upregulated in cells in the outer nuclear layer (ONL) and the medial inner nuclear layer (INL) at P7, and remains expressed in horizontal cells in the outer INL. This pattern is preserved throughout the remainder of retinal development. (O-Q) Antibodies to Sall3 do not stain cell bodies in the retinas of 7 day Sall3−/− retinal explants, demonstrating the specificity of the antibody. The weak staining observed in the inner plexiform layer (IPL) is a secondary antibody artifact (data not shown). GCL, ganglion cell layer. Scale bars: 100 μm in A,B; 50 μm in C-F,K-O.
Fig. 2.
Coexpression of Sall3 with markers of inner retinal cell types. (A-C) Sall3 is coexpressed in a subset of Prox1-expressing cells (arrows) in the outer INL adjacent to the outer plexiform layer (OPL) at P7, P14 and P21. Furthermore, Sall3-expressing cells in the medial INL do not coexpress Prox1. (D) Sall3 is coexpressed (arrows) with the horizontal cell marker calbindin (Calb) in cells in the outer INL at P21. (E-G) Sall3-positive cells (arrowheads) in the medial but not outer INL (yellow arrows) coexpress Chx10 after the first postnatal week, indicating that they are bipolar cells. (H) Sall3-positive bipolar cells do not coexpress PKCα, implying that they are not rod bipolar cells. (I-L) Sall3-positive cells do not express the Müller glia markers glutamine synthase (GS) (I,J) and P27kip1 (K,L). Scale bars: 50 μm.
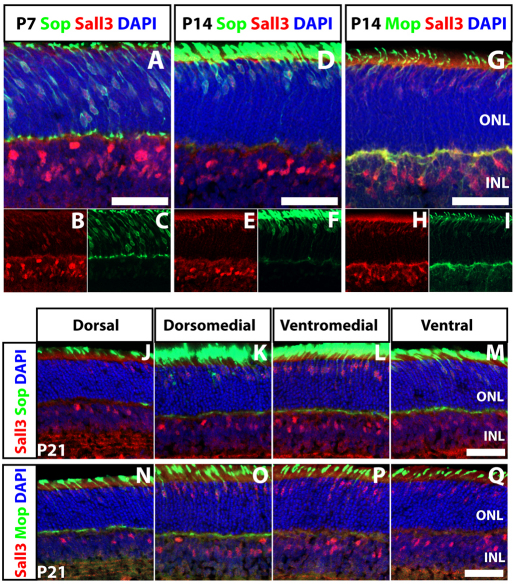
Sall3 and short wavelength-sensitive cone opsin (Sop) were coexpressed from P7 onward in the ONL, indicating that Sall3 is expressed in retinal cones (Fig. 3A-F,J-M). Notably, by P21, Sall3 immunostaining was more prominent in cones of the medial and ventral retina, where the great majority of cones express Sop, as compared with the dorsal retina (Haverkamp et al., 2005; Szel et al., 1993). Furthermore, expression of Sall3 was only detected in a subset of medium wavelength-sensitive cone opsin (Mop)-expressing cells (Fig. 3G-I,N-Q). Coexpression of Sall3 and Mop was most prominent in dorsomedial and ventromedial retina, where the overwhelming majority of cones coexpress both Mop and Sop (Onishi et al., 2010a; Szel et al., 1993). We thus conclude that Sall3 is selectively expressed in short wavelength-sensitive cone photoreceptors (S-cones). This finding accords with the analysis of retinas of mice mutant for Nrl and Rorβ, in which rod photoreceptors are converted to an S-cone fate, and which both show a substantial increase in Sall3 mRNA (Hsiau et al., 2007; Jia et al., 2009).
Fig. 3.
Coexpression of Sall3 with Sop and Mop in cone photoreceptors of the developing retina. (A-F) Sall3 is coexpressed with Sop in mouse cone photoreceptors at P7 and P14. (G-I) Sall3 is only coexpressed with a subset of Mop-expressing photoreceptors at P14. (J-M) Sall3 expression in cones correlates with levels of Sop expression along the dorsoventral retinal axis at P21. Sections are taken at the following positions along an axis running through the central retina, where 0% is most dorsal and 100% most ventral: dorsal (10%), dorsomedial (35%), ventromedial (60%) and ventral (85%). (N-Q) Limited Sall3 expression is observed in the dorsal retina, where most cones predominantly express Mop. Sall3 is coexpressed with Mop in the dorsomedial and ventromedial retina, where the majority of cells coexpress Sop and Mop. Strong expression of Sall3 is seen in ventral retina, where only Sop-positive and Sop+ Mop+ cones are observed. Scale bars: 50 μm.
Sall3 is necessary and sufficient for expression of S-cone-specific genes
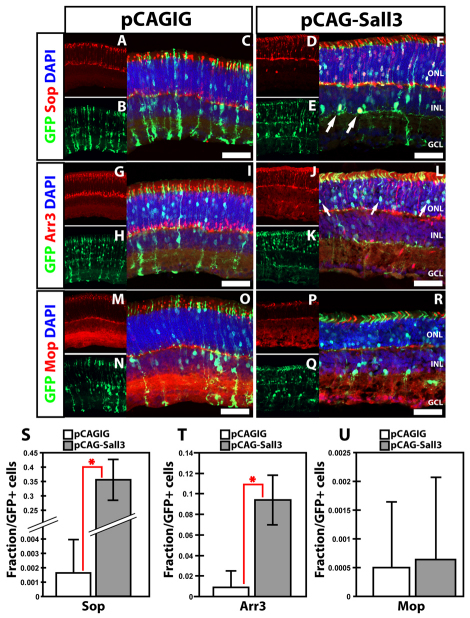
To determine whether Sall3 might be sufficient to drive other retinal cell types toward a cone fate, we used in vivo electroporation of plasmids expressing full-length Sall3 under the control of the ubiquitous CAG promoter to overexpress Sall3 beginning at P0.5 (Matsuda and Cepko, 2004; Onishi et al., 2009). Electroporation of CAG-Sall3 induced both Sop and cone arrestin (Arr3) (Zhu et al., 2002) expression in a large fraction of electroporated cells (Fig. 4A-L). Thirty-five percent of all pCAG-Sall3-electroporated cells coexpressed Sop, compared with 0.164% of cells electroporated with pCAGIG control construct (P<0.0005; n=5) (Fig. 4S). Furthermore, 9.5% of cells electroporated with pCAG-Sall3 expressed Arr3, whereas only 0.898% of GFP-positive controls were Arr3 positive (P<0.0005; n=5) (Fig. 4T). In sharp contrast, no induction of Mop was observed following Sall3 overexpression (P>0.05; n=5) (Fig. 4M-R,U), nor was rhodopsin expression repressed in electroporated cells (see Fig. S3 in the supplementary material). Electroporated photoreceptors showed no change in outer segment length or in the position of their cell bodies, nor did they show cone-like morphology. We also observed that overexpression of Sall3 induced Sop expression in a subset of electroporated cells that resembled wide-field amacrine cells, indicating that Sall3 is sufficient to induce cone opsin expression in non-photoreceptor cells (Fig. 4F, arrows).
Fig. 4.
Overexpression of Sall3 by in vivo electroporation of P0 retinas results in ectopic expression of S-cone markers at P14. (A-F) Expression of Sop in mouse retinas electroporated with control pCAGIG (A-C) and pCAG-Sall3 (D-F) constructs. Overexpression of Sall3 results in ectopic expression of Sop in the ONL (F) and in subsets of electroporated cells in the INL (F, arrows). (G-L) Expression of cone arrestin (Arr3) in retinas electroporated with control (G-I) and Sall3 (J-L) overexpression constructs. Overexpression of Sall3 results in the ectopic expression of Arr3 in the ONL of electroporated retinas (L). (M-R) Expression of Mop in retinas electroporated with control (M-O) and Sall3 (P-R) expression constructs. (S-U) Quantification of Sop (S), Arr3 (T) and Mop (U) expression in electroporated retinas at P14. Sop: 0.0016±0.003 control versus 0.35±0.067 Sall3; n=5, *P=0.0003. Arr3: 0.009±0.016 control versus 0.09436±0.0234 Sall3; n=5, *P=0.0003. Mop: 0.0005±0.0011 control versus 0.006±0.0014 Sall3; n=5, P=0.8677. Error bars indicate s.d. Scale bars: 50 μm.
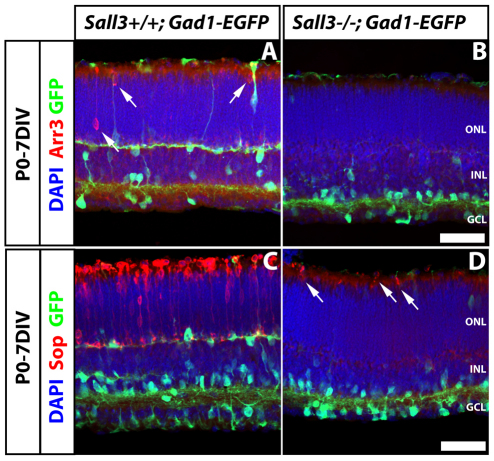
In light of the prominent expression of Sall3 in developing cones, we examined the expression of cone-specific markers in retinas of Sall3−/− mice. We observed no difference in the expression of the early cone marker Rxrγ in P0 retinas of wild-type and knockout animals, implying that cone specification occurs normally in Sall3−/− retinas (see Fig. S4 in the supplementary material). Owing to the neonatal lethality of the Sall3 knockout, retinal explant cultures were examined to investigate whether cone development proceeds normally. In P0 retinas that were explanted in vitro for 7 days (P0-7DIV), we observed a virtual elimination of the expression of a number of cone-specific genes, including both Arr3 (Fig. 5A,B) and Sop (Fig. 5C,D). Arr3 expression was absent in P0-7DIV Sall3−/− retinal explants (Fig. 5B), but a few Sop-positive cells were detected. However, these Sop-positive cells showed abnormal morphology, including truncated outer segments and the positioning of cell soma immediately adjacent to the outer limiting membrane (Fig. 5D, arrows). This drastic reduction in Sop expression was also observed in 14 day (P0-14DIV) Sall3−/− retinal explants (see Fig. S5F in the supplementary material, arrow). In P0-14DIV Sall3−/− explants, a few Arr3-positive cells were seen, although many fewer than in wild-type explants (see Fig. S5I,L in the supplementary material, arrow). Mop expression could not be examined because neonatal retinal explants do not express Mop protein (Soderpalm et al., 1994). Finally, rod numbers were unaffected in Sall3−/− P0-14DIV explants, with expression of rhodopsin expressed normally in rods (see Fig. S5M-R in the supplementary material).
Fig. 5.
Sall3+/+; Gad1-EGFP and Sall3−/−; Gad1-EGFP P0 retinas explanted for 7 days in vitro (P0-7DIV) demonstrate defects in cone photoreceptor development. (A,B) Arr3 expression in Sall3+/+ and Sall3−/− mouse retinal explants. Arr3 is expressed in the ONL in Sall3+/+ explants (A, arrows). Sall3−/− explants lack Arr3 expression (B). (C,D) Sop expression in Sall3−/− and Sall3+/+ retinal explants. Sop is robustly expressed in the ONL of Sall3+/+ explants (C), whereas Sall3−/− explants display severely reduced numbers of Sop-positive cells in the ONL (D, arrows). The Gad1-EGFP transgene was used to visualize horizontal cell morphology. Scale bars: 50 μm.
To determine whether Sall3 directly regulates the expression of Sop, Mop and/or Arr3, we performed chromatin immunoprecipitation (ChIP) from P7 C57BL/6J wild-type mouse retinas. We observed that Sall3 bound directly to the promoters of these genes, but did not bind to the 3′ regions of these genes or to the promoters of genes that are selectively expressed in rods or rod bipolar cells (Fig. 6). Interestingly, although our overexpression experiments did not reveal any significant changes in Mop expression, ChIP experiments suggested that Sall3 binds to the promoter of this gene in vivo. These data are in line with other studies that have reported a considerable degree of redundancy and crosstalk among transcription factors involved in the regulation of photoreceptor-specific gene expression (Onishi et al., 2009; Peng et al., 2005; Peng and Chen, 2005).
In vivo electroporation of Sall3 results in the generation of ectopic horizontal cells in the inner retina
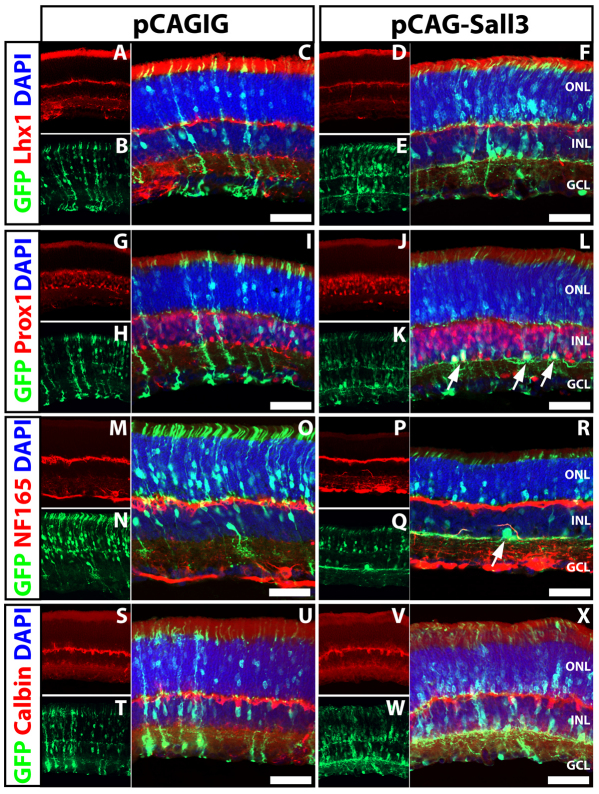
Since Sall3 is also expressed in retinal horizontal cells, we next examined the expression of horizontal cell markers in cells overexpressing Sall3 at P14. Overexpression of Sall3 altered the morphology of electroporated cells in the INL, which displayed a wide-field lateral dendritic morphology reminiscent of horizontal cells (Fig. 7). We hypothesized that these could be horizontal cells that were ectopically positioned adjacent to the inner plexiform layer (IPL). Overexpression of Sall3 did not induce expression of either of the horizontal cell markers Lhx1 and calbindin (Fig. 7A-F,S-X), but a robust induction of Prox1 was observed in electroporated horizontal-like cells localized adjacent to the IPL (Fig. 7L, arrows). Since Prox1 is expressed in AII amacrine cells in the mouse retina (Dyer et al., 2003), we investigated whether Prox1-expressing cells generated following Sall3 overexpression were abnormal AII amacrine cells, but did not detect expression of the AII amacrine markers Dab1, calretinin (Calb2), parvalbumin and Glyt1 (Slc6a9) (see Fig. S6 in the supplementary material). Furthermore, overexpression of Sall3 resulted in the expression of NF165 in the lateral dendrites of these same horizontal-like cells (Fig. 7R, arrow), which was never observed in cells electroporated with the pCAGIG control plasmid (Fig. 7M-O). These data imply that Sall3 overexpression can induce both the expression of a subset of horizontal cell markers and a horizontal cell-like morphology in late-born retinal cells, but cannot completely respecify these cells to a horizontal fate.
Fig. 7.
Overexpression of Sall3 by in vivo electroporation of P0 retinas results in expression of horizontal cell markers in the INL at P14. (A-F) Expression of Lhx1 in mouse retinas electroporated with control pCAGIG (A-C) and pCAG-Sall3 (D-F) plasmids. (G-L) Expression of Prox1 in retinas electroporated with control (G-I) and pCAG-Sall3 (J-L) constructs. Cells in the inner INL coexpress Prox1 in retinas electroporated with pCAG-Sall3 (L, arrows) but not with pCAGIG (I). (M-R) Expression of NF165 in retinas electroporated with pCAGIG (M-O) and pCAG-Sall3 (P-R). Cells in the inner INL electroporated with pCAG-Sall3 express NF165 (R, arrow) but not following electroporation with pCAGIG (O). (S-X) Expression of calbindin in retinas electroporated with control (S-U) or pCAG-Sall3 (V-X) plasmids. Scale bars: 50 μm.
Loss of function of Sall3 disrupts horizontal cell differentiation
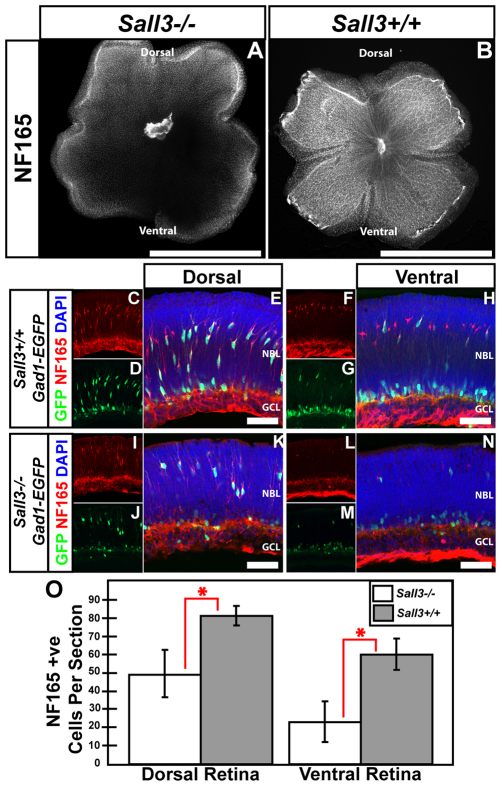
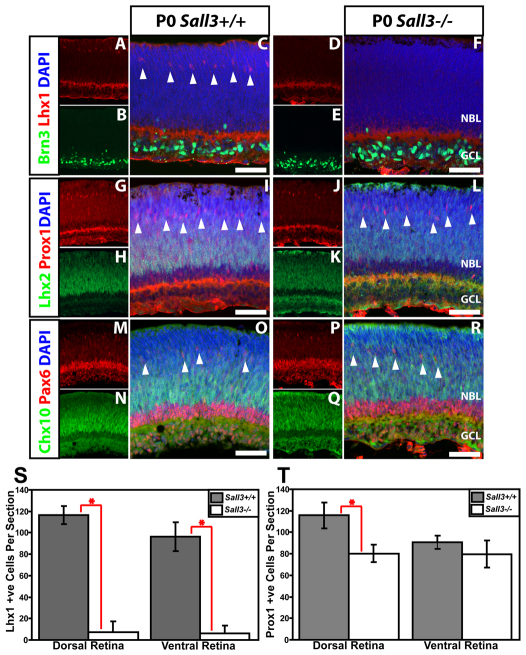
To examine the effects of Sall3 loss of function (LOF), we conducted immunohistochemical analysis of wild-type and Sall3−/− retinas using molecular markers of horizontal cells. At P0.5, both section and flatmount immunohistochemistry showed a dramatic reduction of NF165-positive horizontal cells (Fig. 8). This effect was especially pronounced in ventral retinas of Sall3−/− mice as compared with wild-type controls, with a less prominent reduction in the dorsal retina (Fig. 8A,K,N). A statistically significant 38% reduction in cell number was observed in dorsal retina (P<0.002), with cell number in the ventral retina reduced by two-thirds (P<0.0002) (Fig. 8O).
Fig. 8.
Sall3−/− mice show fewer horizontal interneurons in the ventral retina. (A,B) P0 flatmounts show fewer NF165-positive horizontal cells in Sall3−/− than Sall3+/+ retinas. Decreased NF165 expression is most prominent in the ventral portion of Sall3−/− retinas (A). (C-N) Localization of NF165 and GFP in dorsal and ventral P0 sections of Sall3+/+; Gad1-EGFP and Sall3−/−; Gad1-EGFP retinas. (C-E) Sall3+/+ dorsal retinas feature elaborate NF165+ GFP+ horizontal cells in the medial NBL. (F-H) Ventral Sall3+/+ retinas display NF165-positive horizontal cells; however, fewer horizontal cells coexpress GFP. (I-K) Expression of NF165 and Gad1-EGFP in dorsal Sall3−/−; Gad1-EGFP retinas reveals a decreased number of NF165+ GFP+ cells. (L-N) Expression of NF165 and GFP in ventral Sall3−/−; Gad1-EGFP retinas demonstrates substantially diminished numbers of NF165+ GFP+ cells. (O) Quantification of NF165-positive horizontal cells in P0 dorsal and ventral Sall3+/+ and Sall3−/− retinas: Dorsal retina: 49.33±13.03 Sall3−/− versus 81.32±4.32 Sall3+/+; n=5, *P=0.0011. Ventral retina: 23±11.33 Sall3−/− versus 60.17±8.66 Sall3+/+; n=5, *P=0.0001. Error bars indicate s.d. Scale bars: 2 mm in A,B; 50 μm in E,H,K,N.
To better visualize the morphology and position of horizontal cells, the Gad1-EGFP transgene was bred into the Sall3−/− line, as GFP fluorescence readily allows robust visualization of horizontal cell morphology (Huckfeldt et al., 2009). The number of GFP-positive cells was reduced in the medial neuroblastic layer (NBL) in Sall3−/− mice (Fig. 8C-N). However, compared with wild-type samples, cells within the medial NBL of Sall3−/− retinas that coexpressed NF165 and GFP did not demonstrate any clear difference in the relative intensity of either NF165 or GFP staining.
To determine whether Sall3 LOF results in increased apoptosis, we performed TUNEL assays on Sall3−/− and Sall3+/+ retinas at E16.5 and P0 (see Fig. S7 in the supplementary material). No difference in TUNEL labeling was observed in the Sall3−/− animals as compared with controls at either time point.
Loss of function of Sall3 results in downregulation of Lhx1 in horizontal cells
To investigate how Sall3 LOF disrupts horizontal cell development, we examined the expression of transcription factors that regulate horizontal cell development. At E16.5, expression of Prox1 and Lhx1 did not substantially differ between wild-type and Sall3−/− retinas (see Fig. S8 in the supplementary material). At P0, however, Lhx1 expression was almost completely absent from horizontal cells of Sall3−/− mice (Fig. 9A-F,S). Prox1 expression was not as severely affected as Lhx1 in Sall3−/− mice. There was a reduction in Prox1-positive cells in the dorsal retina, but this was very mild relative to that seen for Lhx1 (Fig. 9T). Expression of the retinal ganglion cell marker Brn3 (Pou4f1), the retinal progenitor, ganglion, amacrine and horizontal cell marker Pax6, and the progenitor markers Chx10 and Lhx2 was unaffected (Fig. 9A-L). This implies that Sall3 is required to maintain expression of Lhx1 in horizontal cells while they undergo radial migration, but is not necessary to initiate the expression of Lhx1 during outward radial migration. Using a Six3-Cre; Lhx1lox/lox line, we were able to selectively inactivate Lhx1 expression in retina. As previously described, Lhx1 immunoreactivity was eliminated in these mice (Poche et al., 2007). Expression of Sall3 in photoreceptors and putative cone bipolar cells was unaffected in these animals (see Fig. S2J-R in the supplementary material). Lhx1 LOF resulted in a substantial reduction in the number of horizontal cells in the outer INL (see Fig. S2J,M,P in the supplementary material). However, the remaining horizontal cells expressed Sall3, Prox1 and NF165 (see Fig. S2M,P in the supplementary material, arrows). Furthermore, ectopic horizontal cells could be identified in close proximity to the IPL, as determined by coexpression of Sall3 and Prox1 (see Fig. S2M in the supplementary material, arrowhead). These data suggest that, whereas Sall3 is required to maintain expression of Lhx1 postnatally, Lhx1 is not required for the expression of Sall3 in horizontal cells.
Fig. 9.
Expression of the horizontal cell markers Lhx1, Prox1 and Pax6, the retinal ganglion cell marker Brn3 and the retinal progenitor markers Lhx2 and Chx10 in P0 Sall3−/− and Sall3+/+ retinas. (A-F) Lhx1 and Brn3 immunostaining of Sall3+/+ and Sall3−/− mouse retinas at P0. Lhx1-expressing horizontal cells were present in Sall3+/+ (C, arrowheads) but not Sall3−/− (F) retinas. (G-L) Prox1 and Lhx2 immunostaining of Sall3−/− and Sall3+/+ retinas at P0. Prox1-expressing horizontal cells are present in Sall3+/+ (I, arrowheads) and Sall3−/− (L, arrowheads) retinas. (M-R) Chx10 and Pax6 immunostaining of Sall3+/+ and Sall3−/− retinas at P0. Pax6-expressing horizontal cells are present in Sall3+/+ (O, arrowheads) and Sall3−/− (R, arrowheads) retinas. (S) Quantification of Lhx1-positive horizontal cells in P0 dorsal and ventral Sall3−/− and Sall3+/+ retinas. Dorsal retina: 116.4±8.5 Sall3+/+ versus 7.3±9.9 Sall3−/−; n=3, *P=0.00015. Ventral retina: 96.3±13.3 Sall3+/+ versus 6.0±7.4 Sall3−/−; n=3, *P=0.0016. (T) Quantification of Prox1-positive horizontal cells in P0 dorsal and ventral Sall3+/+ and Sall3−/− retinas. Dorsal retina: 115.7±12.0 Sall3+/+ versus 80.3±8.1 Sall3−/−; n=3, *P=0.017. Ventral retina: 90.7±6.2 Sall3+/+ versus 79.7±12.5 Sall3−/−; n=3, P=0.27. Error bars indicate s.d. Scale bars: 50 μm.
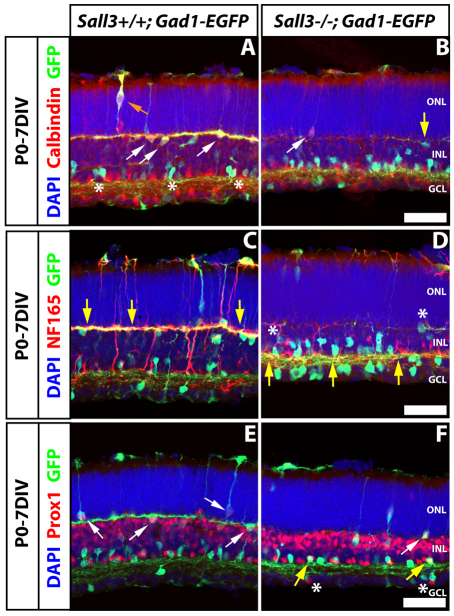
We next examined retinal explants at later developmental stages to further investigate the effects of Sall3 LOF on horizontal cell development. In P0-7DIV retinal explants of Sall3+/−; Gad1-EGFP mice, the overwhelming majority of GFP-positive horizontal cell bodies were found in the outer portion of the INL (Fig. 10A, white arrows). These cells expressed calbindin and the Gad1-EGFP transgene and extended GFP-positive dendrites tangentially through the outer plexiform layer (OPL). Coexpression of calbindin and GFP was specific to horizontal cells, with separate subpopulations of amacrine cells expressing calbindin or GFP (Fig. 10A, asterisks). A limited number of radially migrating calbindin+ GFP+ horizontal cell precursors were also observed in the ONL at this stage (Fig. 10A, orange arrow). In Sall3−/−; Gad1-EGFP mice, calbindin+ GFP+ horizontal cells were not detected. Robust calbindin expression was not observed in the OPL (Fig. 10B), although rare calbindin-positive soma were sometimes seen positioned either apical or basal to the OPL (Fig. 10B, white arrow), and a much reduced number of GFP-positive horizontal cell bodies was observed in the outer portion of the INL (Fig. 10B, yellow arrow).
Fig. 10.
Horizontal cell development is disrupted in P0-7DIV Sall3−/− retinas. (A,B) Calbindin and GFP expression. Calbindin+ GFP+ horizontal cells are localized in the outer INL (A, white arrows) in wild-type retinas, whereas GFP-positive cells in the inner INL are calbindin-negative (A, asterisks). Occasionally, calbindin+ GFP+ migrating horizontal cells are seen in the ONL (A, orange arrow). Sall3−/− retinas lack calbindin+ GFP+ horizontal cells; however, cells positive for either calbindin (B, white arrow) or GFP (B, yellow arrow) are still seen in the outer INL. (C,D) NF165 and Gad1-EGFP expression. NF165+ GFP+ horizontal cell processes are found in the OPL (C, yellow arrows) in Sall3+/+; Gad1-EGFP retinas. Sall3−/−; Gad1-EGFP retinas display few NF165+ GFP+ horizontal cell processes in the OPL (D, asterisks) and NF165+ GFP+ processes are localized in the IPL (D, yellow arrows). (E,F) Prox1 and GFP expression. Prox1+ GFP+ cells are localized adjacent to the OPL in Sall3+/+; Gad1-EGFP retinas (E, arrows). A reduced number of Prox1+ GFP+ cells is found adjacent to the OPL in Sall3−/− mice (F, white arrow), and Prox1+ GFP+ cells are now localized adjacent to the IPL (F, yellow arrows). Prox1-positive cells can also be found in the GCL of Sall3−/− retinas (F, asterisks). Scale bars: 50 μm.
To further investigate the fate of horizontal cells that would normally be present adjacent to the OPL, we analyzed NF165 immunofluorescence in both genetic backgrounds. The dendrites of horizontal cells in Sall3+/+; Gad1-EGFP mice were robustly positive for both NF165 and GFP, with intense colabeling observed throughout the OPL (Fig. 10C, arrows). Furthermore, little NF165 expression and no coexpression of NF165 and GFP was observed in the IPL of Sall3+/+; Gad1-EGFP retinas. In sharp contrast, Sall3−/−; Gad1-EGFP mice showed very weak expression of NF165 in the OPL (Fig. 10D); the few correctly positioned GFP-positive cell bodies extended grossly atrophic NF165-positive dendritic processes (Fig. 10D, asterisks). Significant populations of GFP-positive cells were observed in Sall3−/−; Gad1-EGFP retinas, with soma located in the inner INL and extending lateral NF165-positive dendrites along the scleral border of the IPL (Fig. 10D, arrows). All horizontal cells in wild-type retinas were robustly positive for both GFP and Prox1 (Fig. 10E, arrows), and Prox1+ GFP+ cells were not detectable in other regions of the INL or the ganglion cell layer (GCL). In Sall3−/−; Gad1-EGFP mice, however, in addition to the rare correctly positioned horizontal cell bodies that coexpressed Prox1 and GFP (Fig. 10F, white arrow), a number of cells in both the inner INL and GCL were now also seen to coexpress both markers (Fig. 10F, yellow arrows); such cells were not detected in Sall3+/+; Gad1-EGFP mice.
In P0-14DIV retinal explants, we likewise observed reduced NF165 expression in the OPL (see Fig. S9M-R in the supplementary material), but noted robust expression of NF165 in the IPL. Expression of calbindin remained virtually absent in the OPL, and appropriately positioned calbindin-positive horizontal cell soma were rarely seen (see Fig. S9S-X in the supplementary material). These data imply that Sall3 LOF results in the mislocalization of a subset of differentiating horizontal cells to the inner portion of the INL, similar to what is observed in Chx10-Cre; Lhx1lox/lox retinas (Poche et al., 2007). Sall3 LOF does not affect the initiation of Lhx1 expression in horizontal cells, and these cells appear to undergo outward radial migration normally, unlike Lhx1−/− horizontal cells, which generally do not undergo outward radial migration. However, most Sall3-deficient horizontal cells do not take up residence adjacent to the developing OPL but instead localize to the vitreal face of the INL, extending their dendrites laterally in the IPL.
Sall3 loss of function globally disrupts the expression of horizontal cell- and S-cone-specific genes
To determine whether additional developmental defects result from Sall3 LOF, we performed triplicate microarray analysis of P7 retinal explants obtained from wild-type and Sall3−/− mice. Through a survey of the literature, along with the previously published microarray analysis of Nrl−/− and Rorβ−/− mice, we were able to obtain a list of over 110 cone-enriched transcripts (Blackshaw et al., 2004; Hsiau et al., 2007; Jia et al., 2009) (see Table S1 in the supplementary material). We observed that cone-enriched transcripts were significantly downregulated in Sall3−/− retinas (P<2.0×10−4). Notably, we found that cone-specific components of the phototransduction pathway were particularly downregulated, with Sop (Opn1sw) being the second most highly downregulated transcript identified (Table 1 and see Fig. S10 in the supplementary material), although some cone-specific genes such as Cnga3 showed only modest changes in mutant retinas, and the expression of other cone-specific phototransduction components, such as Gnat2 and Gnb3, was unaffected (Table 1). In comparison, among horizontal cell-enriched genes, only Lhx1 showed a greater than 2-fold downregulation in expression that was statistically significant (P<0.0003). The horizontal cell-enriched intermediate filament proteins, such as NF165 and Tmod1, showed a less than 2-fold downregulation in expression (Table 1), whereas other horizontal cell-enriched genes, such as Nefh, Borg4 (Cdc42ep4), septin 4, Prox1 and calbindin showed little change in expression (Blackshaw et al., 2004). Transcripts that were upregulated in Sall3−/− mice did not show any significant enrichment for known retinal cell type-specific transcripts. Finally, despite the prominent expression of Sall3 in a subset of bipolar cells, known bipolar cell genes showed no significant changes in expression in the Sall3−/− retina (Kim et al., 2008) (see Table S1 in the supplementary material).
DISCUSSION
Our findings demonstrate that Sall3 plays a crucial role in regulating the development of both cone photoreceptors and horizontal interneurons. Our model of Sall3 action is summarized in Fig. 11. Sall3 is necessary for the expression of a range of cone-specific genes, with only a very small number of Sop-positive and Arr3-positive cells detectable in Sall3−/− retinas. Microarray experiments revealed that many other cone-enriched transcripts are selectively downregulated in Sall3−/− retinas, including Pde6c, Pde6h and Crb1, whereas some, such as Gnat2 and Gnb3, show little or no change. Strikingly, overexpression of Sall3 by electroporation was sufficient to induce the expression of both Sop and Arr3, not only in photoreceptors but also in a subset of cells in the INL, without affecting expression of Mop. Rod photoreceptors overexpressing Sall3 in electroporated retinas were morphologically indistinguishable from those of controls. These rod photoreceptors that overexpressed Sall3 retained their normal morphology, laminar position and pattern of expression of cell-specific markers.
Fig. 11.
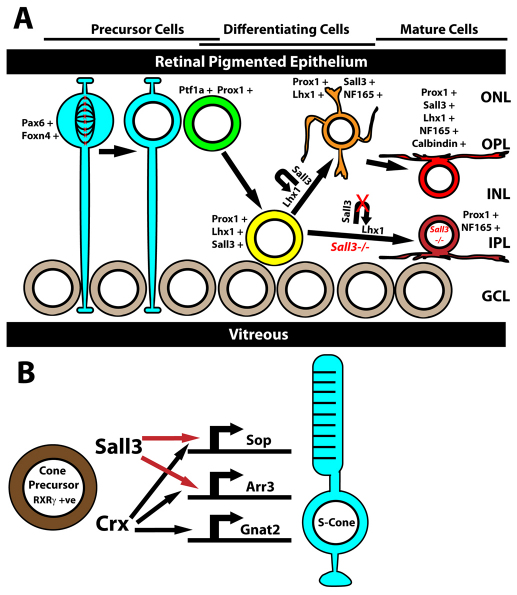
Role of Sall3 in horizontal cell and S-cone photoreceptor development. (A) Sall3 and Lhx1 expression is activated in postmitotic horizontal cell precursors, which express Ptf1a and Prox1. Expression of Sall3 maintains Lhx1 expression in differentiating horizontal cells. Loss of Sall3 expression results in horizontal cells being displaced radially to the IPL and losing expression of calbindin. Blue, M-phase retinal progenitor cell; green, newly postmitotic horizontal cell beginning inward radial migration; yellow, horizontal cell precursor beginning outward radial migration; orange, horizontal cell precursor that has completed outward radial migration; red, mature wild-type horizontal cell; maroon, displaced mature Sall3−/− horizontal cell. (B) Expression of Sall3 in Rxrγ-positive cone precursors directly activates the expression of Sop and Arr3 but not the cone alpha transducin subunit (Gnat2). ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
These data imply that Sall3 is activated in Sop-expressing photoreceptors undergoing terminal differentiation, and that Sall3 itself might directly activate a subset of cone-specific genes in a coordinated manner, analogous to the orphan nuclear hormone receptor Errβ (Esrrb) in rod photoreceptors (Onishi et al., 2010b). Several different nuclear hormone receptors, along with the transcriptional co-regulator Pias3, have been shown to be required for medium wavelength-sensitive cones to activate the expression of Mop while simultaneously repressing the expression of Sop. Sall3, however, represents the first cone-expressed transcription factor that selectively actives the expression of Sop.
The strong activation of S-cone-specific transcripts by Sall3 implies a surprising functional homology with the Spalt gene complex of Drosophila in the regulation of photoreceptor differentiation. In Drosophila, Spalt genes are necessary for specification of the inner R7 and R8 photoreceptors, which are responsible for color discrimination and in many respects form a cone-like photoreceptor class in the compound eye. Strikingly, loss of function of Spalt genes results in the ectopic expression of Rh1 opsin (NinaE), which is normally expressed in the outer R1-R6 photoreceptors, in the R7 and R8 cells, whereas expression of Rh3-Rh6 is lost (Domingos et al., 2004; Mollereau et al., 2001). However, axonal projections of inner photoreceptors are unaltered in Drosophila sal mutants. This partial shift in photoreceptor identity resembles that seen following Sall3 overexpression in rod photoreceptors. These cells appear morphologically normal and continue to express rhodopsin, but now robustly express cone-specific genes. Notably, Spalt genes are selectively expressed in blue-sensitive Rh5-positive R8 photoreceptors and are required for expression of Rh5 opsin (Sprecher et al., 2007).
In mice, we observe that Sall3 is both necessary and sufficient for the expression of blue-sensitive cone opsin but not green-sensitive cone opsin. Such a direct conservation of gene function in photoreceptor development is unusual, and even more surprising because the blue-sensitive visual opsins of insects and vertebrates evolved independently (Shichida and Matsuyama, 2009). Although this observation might represent evolutionary convergence, it could alternatively imply that ancestral bilateria possessed a dedicated short wavelength-sensitive photoreceptor cell type, the differentiation of which was guided by a Spalt family gene, with the blue-sensitive opsin gene expressed by this cell having changed in different lineages; photoreceptors might, at one point, have coexpressed both blue-sensitive ciliary and rhabdomeric opsins. This possibility is not without precedent, as both vertebrate and invertebrate photoreceptors are known to coexpress different opsin genes with similar spectral sensitivities (Mazzoni et al., 2008; Porter et al., 2009). In some cases, opsin genes with similar spectral sensitivity but which are nonetheless highly divergent at the molecular level are coexpressed, such as the blue-sensitive melanopsin and retinal cone opsins of the chick retina (Bailey and Cassone, 2005; Bellingham et al., 2006). Analysis of Spalt family gene expression in invertebrates from multiple phyla with well-characterized color vision should further clarify this finding.
Sall3 also plays a pivotal role in regulating the differentiation of horizontal cells, one of two cell types directly postsynaptic to cone photoreceptors, and might do so in part through maintenance of Lhx1 expression. We did not observe defects in the expression of other previously reported horizontal cell-expressed transcription factors in Sall3−/− retinas, including Pax6, Foxn4 and Ptf1a, although cell counts did reveal a reduction in the number of Prox1-positive cells in the dorsal retina at P0. However, the final laminar position of Sall3−/− horizontal cells closely resembles that seen in Lhx1 mutants. Horizontal cells of Chx10-Cre; Lhx1lox/lox mutants fail to undergo initial outward radial migration, resulting in ectopic localization to the inner INL and extension of their dendritic arbor within the IPL (Poche et al., 2007; Poche et al., 2008). Horizontal cells of Sall3−/− retinas appear to initiate Lhx1 expression and undergo outward radial migration normally. However, by P0, Lhx1 expression is drastically reduced, and at later ages the majority of horizontal cell bodies are found at the scleral border of the IPL, eventually taking up residence with amacrine cells and extending dendrites into the IPL, phenocopying the Lhx1 mutants. The phenomenon of ectopic inner nuclear/plexiform layer-associated horizontal cells was also seen in Sall3 overexpression experiments in which Sall3 appeared to be sufficient to at least partially specify horizontal cells, including activating Prox1 and NF165. The lack of coexpression with markers specific to AII amacrine cells indicated that the Prox1-expressing wide-field cells generated in Sall3 electroporation experiments were not a dysmorphic AII population. This does not exclude the possibility that Sall3 overexpression results in the generation of a rare Sall3+ Prox1+ wide-field amacrine subclass. Notably, Spalt genes also regulate prospero expression in developing Drosophila photoreceptors, which then acts to guide differentiation of the R7 photoreceptor subtype (Cook et al., 2003; Domingos et al., 2004). Overexpression of Sall3 in neonatal retinas was not sufficient to induce expression of Lhx1, and, consequently, the horizontal-like cells demonstrated the same ectopic positioning seen in Lhx1 and Sall3 mutants.
The crucial role of Sall3 in regulating Lhx1 expression is further underlined by our microarray analysis, which indicates that Lhx1 is one of the most strongly downregulated genes in Sall3−/− retinas. Taken together, these data suggest that sustained Lhx1 expression might regulate multiple stages of horizontal cell migration, and that Sall3 is necessary to maintain Lhx1 expression during the postnatal differentiation of horizontal cells. A limited number of Sall3+ NF165+ and Sall3+ Prox1+ horizontal cells are present in Six3-Cre; Lhx1lox/lox retinas, implying that Sall3 regulates aspects of horizontal cell development at least in part through an Lhx1-independent pathway and that expression of Sall3 itself might not require Lhx1.
A subset of bipolar interneurons selectively expressed Sall3. Dedicated S-cone-selective bipolar cells in the mouse retina that are analogous to primate S-cone-selective midget bipolar cells have been identified (Haverkamp et al., 2005). The tantalizing possibility exists that Sall3 might regulate the differentiation of S-cones and their dedicated bipolar interneuron. However, the microarray analysis did not reveal any significant changes in known mouse bipolar-expressed genes, and the role of Sall3 in bipolar interneuron development remains to be resolved.
Supplementary Material
Acknowledgements
We thank T. Shimogori, J. Nathans, W. Yap and members of the S.B. laboratory for comments on the manuscript. This work was supported by NIH EY0170015, a W. M. Keck Distinguished Young Scholar in Medical Research Award and a Sloan Scholars Award (to S.B.); and by NIH EY012543, NIH EY02687 and a Lew Wasserman Merit Award from Research to Prevent Blindness (to S.C.).
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Author contributions
J.d.M. co-designed and performed all the experiments described, with the exception of the ChIP, which was performed by G.-H.P and S.C. S.B. co-designed the experiments and directed the study. J.d.M. and S.B. wrote the paper.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.061846/-/DC1
References
- Akhmedov N. B., Piriev N. I., Chang B., Rapoport A. L., Hawes N. L., Nishina P. M., Nusinowitz S., Heckenlively J. R., Roderick T. H., Kozak C. A., et al. (2000). A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl. Acad. Sci. USA 97, 5551-5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. J., Cassone V. M. (2005). Melanopsin expression in the chick retina and pineal gland. Brain Res. Mol. Brain Res. 134, 345-348 [DOI] [PubMed] [Google Scholar]
- Bellingham J., Chaurasia S. S., Melyan Z., Liu C., Cameron M. A., Tarttelin E. E., Iuvone P. M., Hankins M. W., Tosini G., Lucas R. J. (2006). Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 4, e254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S., Harpavat S., Trimarchi J., Cai L., Huang H., Kuo W. P., Weber G., Lee K., Fraioli R. E., Cho S. H., et al. (2004). Genomic analysis of mouse retinal development. PLoS Biol. 2, E247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M., Barres B. A., Raff M. (2003). Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron 40, 897-904 [DOI] [PubMed] [Google Scholar]
- Cepko C. L. (1999). The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr. Opin. Neurobiol. 9, 37-46 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang Q. L., Nie Z., Sun H., Lennon G., Copeland N. G., Gilbert D. J., Jenkins N. A., Zack D. J. (1997). Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017-1030 [DOI] [PubMed] [Google Scholar]
- Chien C. L., Liem R. K. (1995). The neuronal intermediate filament, alpha-internexin is transiently expressed in amacrine cells in the developing mouse retina. Exp. Eye Res. 61, 749-756 [DOI] [PubMed] [Google Scholar]
- Cook T., Pichaud F., Sonneville R., Papatsenko D., Desplan C. (2003). Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev. Cell 4, 853-864 [DOI] [PubMed] [Google Scholar]
- Corbo J. C., Myers C. A., Lawrence K. A., Jadhav A. P., Cepko C. L. (2007). A typology of photoreceptor gene expression patterns in the mouse. Proc. Natl. Acad. Sci. USA 104, 12069-12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J., Qiu X., Du G., Cristante L., Eisenstat D. D. (2003). Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J. Comp. Neurol. 461, 187-204 [DOI] [PubMed] [Google Scholar]
- Domingos P. M., Brown S., Barrio R., Ratnakumar K., Frankfort B. J., Mardon G., Steller H., Mollereau B. (2004). Regulation of R7 and R8 differentiation by the spalt genes. Dev. Biol. 273, 121-133 [DOI] [PubMed] [Google Scholar]
- Dyer M. A., Livesey F. J., Cepko C. L., Oliver G. (2003). Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet. 34, 53-58 [DOI] [PubMed] [Google Scholar]
- Edqvist P. H., Hallbook F. (2004). Newborn horizontal cells migrate bi-directionally across the neuroepithelium during retinal development. Development 131, 1343-1351 [DOI] [PubMed] [Google Scholar]
- Fujieda H., Bremner R., Mears A. J., Sasaki H. (2009). Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J. Neurochem. 108, 91-101 [DOI] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R. J., Furukawa T., et al. (2006). Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development 133, 4439-4450 [DOI] [PubMed] [Google Scholar]
- Furukawa T., Morrow E. M., Cepko C. L. (1997). Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531-541 [DOI] [PubMed] [Google Scholar]
- Haider N. B., Jacobson S. G., Cideciyan A. V., Swiderski R., Streb L. M., Searby C., Beck G., Hockey R., Hanna D. B., Gorman S., et al. (2000). Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet. 24, 127-131 [DOI] [PubMed] [Google Scholar]
- Hamano K., Kiyama H., Emson P. C., Manabe R., Nakauchi M., Tohyama M. (1990). Localization of two calcium binding proteins, calbindin (28 kD) and parvalbumin (12 kD), in the vertebrate retina. J. Comp. Neurol. 302, 417-424 [DOI] [PubMed] [Google Scholar]
- Hatakeyama J., Kageyama R. (2004). Retinal cell fate determination and bHLH factors. Semin. Cell Dev. Biol. 15, 83-89 [DOI] [PubMed] [Google Scholar]
- Haverkamp S., Wassle H., Duebel J., Kuner T., Augustine G. J., Feng G., Euler T. (2005). The primordial, blue-cone color system of the mouse retina. J. Neurosci. 25, 5438-5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T. H., Diaconu C., Myers C. A., Lee J., Cepko C. L., Corbo J. C. (2007). The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE 2, e643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckfeldt R. M., Schubert T., Morgan J. L., Godinho L., Di Cristo G., Huang Z. J., Wong R. O. (2009). Transient neurites of retinal horizontal cells exhibit columnar tiling via homotypic interactions. Nat. Neurosci. 12, 35-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Oh E. C., Ng L., Srinivas M., Brooks M., Swaroop A., Forrest D. (2009). Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc. Natl. Acad. Sci. USA 106, 17534-17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. S., Ross S. E., Trimarchi J. M., Aach J., Greenberg M. E., Cepko C. L. (2008). Identification of molecular markers of bipolar cells in the murine retina. J. Comp. Neurol. 507, 1795-1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mo Z., Yang X., Price S. M., Shen M. M., Xiang M. (2004). Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43, 795-807 [DOI] [PubMed] [Google Scholar]
- Liu W., Wang J. H., Xiang M. (2000). Specific expression of the LIM/homeodomain protein Lim-1 in horizontal cells during retinogenesis. Dev. Dyn. 217, 320-325 [DOI] [PubMed] [Google Scholar]
- Livesey F. J., Cepko C. L. (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci. 2, 109-118 [DOI] [PubMed] [Google Scholar]
- Matsuda T., Cepko C. L. (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA 101, 16-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni E. O., Celik A., Wernet M. F., Vasiliauskas D., Johnston R. J., Cook T. A., Pichaud F., Desplan C. (2008). Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol. 6, e97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001). Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447-452 [DOI] [PubMed] [Google Scholar]
- Mollereau B., Dominguez M., Webel R., Colley N. J., Keung B., de Celis J. F., Desplan C. (2001). Two-step process for photoreceptor formation in Drosophila. Nature 412, 911-913 [DOI] [PubMed] [Google Scholar]
- Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., Furukawa T. (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6, 1255-1263 [DOI] [PubMed] [Google Scholar]
- Onishi A., Peng G. H., Hsu C., Alexis U., Chen S., Blackshaw S. (2009). Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron 61, 234-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A., Peng G. H., Chen S., Blackshaw S. (2010a). Pias3-dependent SUMOylation controls mammalian cone photoreceptor differentiation. Nat. Neurosci. 13, 1059-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A., Peng G. H., Poth E. M., Lee D. A., Chen J., Alexis U., de Melo J., Chen S., Blackshaw S. (2010b). The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc. Natl. Acad. Sci. USA 107, 11579-11584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish M., Ott T., Lance-Jones C., Schuetz G., Schwaeger-Nickolenko A., Monaghan A. P. (2004). Loss of the Sall3 gene leads to palate deficiency, abnormalities in cranial nerves, and perinatal lethality. Mol. Cell. Biol. 24, 7102-7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G. H., Chen S. (2005). Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis. Neurosci. 22, 575-586 [DOI] [PubMed] [Google Scholar]
- Peng G. H., Ahmad O., Ahmad F., Liu J., Chen S. (2005). The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 14, 747-764 [DOI] [PubMed] [Google Scholar]
- Poche R. A., Kwan K. M., Raven M. A., Furuta Y., Reese B. E., Behringer R. R. (2007). Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J. Neurosci. 27, 14099-14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche R. A., Raven M. A., Kwan K. M., Furuta Y., Behringer R. R., Reese B. E. (2008). Somal positioning and dendritic growth of horizontal cells are regulated by interactions with homotypic neighbors. Eur. J. Neurosci. 27, 1607-1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. L., Bok M. J., Robinson P. R., Cronin T. W. (2009). Molecular diversity of visual pigments in Stomatopoda (Crustacea). Vis. Neurosci. 26, 255-265 [DOI] [PubMed] [Google Scholar]
- Roberts M. R., Hendrickson A., McGuire C. R., Reh T. A. (2005). Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest. Ophthalmol. Vis. Sci. 46, 2897-2904 [DOI] [PubMed] [Google Scholar]
- Roberts M. R., Srinivas M., Forrest D., Morreale de Escobar G., Reh T. A. (2006). Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc. Natl. Acad. Sci. USA 103, 6218-6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichida Y., Matsuyama T. (2009). Evolution of opsins and phototransduction. Philos. Trans. R. Soc. Lond. B 364, 2881-2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderpalm A., Szel A., Caffe A. R., van Veen T. (1994). Selective development of one cone photoreceptor type in retinal organ culture. Invest. Ophthalmol. Vis. Sci. 35, 3910-3921 [PubMed] [Google Scholar]
- Sprecher S. G., Pichaud F., Desplan C. (2007). Adult and larval photoreceptors use different mechanisms to specify the same Rhodopsin fates. Genes Dev. 21, 2182-2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szel A., Rohlich P., Mieziewska K., Aguirre G., van Veen T. (1993). Spatial and temporal differences between the expression of short- and middle-wave sensitive cone pigments in the mouse retina: a developmental study. J. Comp. Neurol. 331, 564-577 [DOI] [PubMed] [Google Scholar]
- Yao W., Sung L. A. (2009). Specific expression of E-Tmod (Tmod1) in horizontal cells: implications in neuronal cell mechanics and glaucomatous retina. Mol. Cell. Biomech. 6, 71-82 [PubMed] [Google Scholar]
- Zhu X., Li A., Brown B., Weiss E. R., Osawa S., Craft C. M. (2002). Mouse cone arrestin expression pattern: light induced translocation in cone photoreceptors. Mol. Vis. 8, 462-471 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.