Abstract
Piwil2 (mili in mouse or hili in humans), a member of the PIWI/Argonaute gene family, plays important roles in stem cell self-renewal, RNA silencing, and translational regulation in various organisms. Recent demonstration of stable Piwil2 expression in pre-cancerous stem cells and in various human and animal tumor cell lines suggests its association in tumorigenesis. Here, we show that cisplatin induces chromatin relaxation in Mili-Wild type (WT) mouse embryonic fibroblasts (MEFs), but not in Mili-knockout (KO) MEFs. Moreover, in contrast to Mili-WT MEFs, Mili-KO MEFs showed a discernable H3 hypoacetylation response upon cisplatin treatment. Levels of the histone acetyltransferase (HAT), p300, were dramatically different due to a consistent cisplatin post-treatment decrease in Mili-WT and an increase in Mili-KO MEFs. Concomitant reduction of specific HAT activity of p300 could explain the decrease of H3 acetylation in Mili-KO MEFs. Our data also shows Mili is required for maintaining the euchromatic marks in MEFs upon cisplatin treatment. In addition, Mili-KO MEFs exhibited a significant deficiency in repairing cisplatin-induced DNA damage and displayed higher sensitivity to cisplatin. Further analysis revealed that Piwil2 was also enhanced in two completely different cisplatin-resistant ovarian cancer cell lines. Interestingly, knockdown of Piwil2 expression in these two cell lines also resulted in their enhanced sensitivity to cisplatin and decreased their efficiency for removing cisplatin-induced DNA intrastrand crosslinks (Pt-GG). The overall data showed that Piwil2 is a key factor in regulating chromatin modifications especially in response to cisplatin. To conclude, the overexpression of Piwil2 in some cancers could lead to cellular cisplatin resistance, possibly due to enhanced chromatin condensation affecting normal DNA repair.
Keywords: Piwil2, cisplatin, chromatin remodeling, DNA repair
1. Introduction
The piwi family genes are defined by highly conserved PAZ (Piwi/Argonaute/Zwille) and Piwi (P-element induced wimpy testis) domains and play important roles in stem cell self-renewal [1], spermatogenesis [2], RNA silencing [3], translational regulation [3] and chromatin remodeling [4, 5] in various organisms. Piwil2 (mili in mouse or hili in humans), a member of the PIWI/Argonaute gene family [6], is exclusively expressed in germ line stem cells of testis in normal adults [2], but is widely expressed in various types of tumors, including prostate, breast, gastrointestinal, ovarian and endometrial cancer of human and in breast tumors, rhabdomyosarcoma and medulloblastoma of mouse [7]. The findings that Piwil2 is widely expressed at early stages of various types of cancers [7] and can be detected in precancerous stem cells [8], indicate that it might play an important role in tumor initiation. It has also been reported that Piwil2 inhibits apoptosis and stimulates proliferation through activation of Stat3/Bcl-XL pathway. Inhibition of constitutive signaling pathways by repression of Piwil2 expression can inhibit tumor cell growth in vitro and in vivo, which provides a novel means for therapeutic intervention in human cancer [7].
Cisplatin is one of the most potent anti-tumor agents, which displays clinical activity against a wide variety of solid tumors. However, the effective use of cisplatin is limited by the development of cisplatin resistance in cancer cells. The anti-neoplastic activity of cisplatin results from cisplatin-induced DNA damage. Cisplatin forms primarily 1, 2-intrastrand crosslinks between adjacent purines in DNA, e.g. cis-Pt(NH3)2d(GpG) (Pt-GG) and cis-Pt(NH3)2d(ApG) (Pt-AG). These lesions contribute to 90% of total damage introduced by cisplatin [9]. The cisplatin-induced intrastrand crosslinks are mainly removed by nucleotide excision repair (NER). Thus, increase of this DNA repair capacity is believed to confer resistance to platinum-based chemotherapy, while decrease of NER efficiency is thought to enhance the sensitivity of cancer cells to cisplatin [10].
NER offers the most versatile choice among all repair systems operational in living cells in terms of lesion recognition. This highly conserved DNA repair system can eliminate a wide variety of helix-distorting lesions. In eukaryotic cells, the efficient repair of DNA damage is complicated by the fact that the genomic DNA is packaged through histone and non-histone proteins into chromatin, a highly condensed structure that hinders DNA accessibility and its subsequent repair. Therefore, the cellular repair machinery has to circumvent this barrier to gain access to the damaged site. In general, histone modifications, especially lysine acetylation, orchestrate the chromatin accessibility [11].
Early cytological studies have distinguished two types of chromatin: euchromatin and heterochromatin [12]. Heterochromatin was originally defined as the portion of the genome that remains condensed, relatively inaccessible, and harbors characteristic histone H3 lysine 9 hypoacetylation, H3 lysine 9 hypermethylation and H3 lysines 4 and 79 hypomethylation. In contrast, euchromatin is a lightly packed form of chromatin that is rich in gene concentration, and is often (but not always) under active transcription. Its histone modification marks are hyperacetylation of lysine 9 and hypermethylation of lysine 4 and 79 of H3. The DNA damage in heterochromatin is generally repaired more slowly than that in euchromatin [13]. Importantly, the euchromatin and heterochromatin states are dynamic and inter-convertible. For example, genes normally active in a euchromatin domain will typically be silenced when placed adjacent to or within a heterochromatic domain by chromosome rearrangement or transposition [12]. In S. cerevisiae, sub-telomeric silent chromatin (heterochromatin) is partially disrupted during the DNA damage response and reestablished following recovery [14, 15]. Therefore, the state of chromatin upon DNA damage plays a crucial role in the subsequent DNA repair process. Here, we showed that Piwil2 plays an important role in histone acetylation and sustainment of euchromatin decondensed state following cisplatin treatment in mammalian cells. Meanwhile, Piwil2-deficient cells exhibited reduced NER efficiency and enhanced sensitivity to cisplatin.
2. Materials and Methods
2.1. Cell lines, MEFs, and treatment
The human ovarian cancer cell line A2780 and its resistant subline CP70 were kindly provided by Dr. Paul Modrich (Duke University). Another A2780-derivative resistant subline CDDP was kindly provided by Dr. Karuppaiyah Selvendiran and Dr. Periannan Kuppusamy (The Ohio State University). Ovarian cancer cell line 2008 and its resistant cell line 2008C13 were kindly provided by Dr. Francois X. Claret (University of Texas - M. D. Anderson Cancer Center). The A2780-derivative and 2008-derivative cisplatin-resistant cell lines were produced by intermittent, incremental exposure of the sensitive parental cell line to various concentrations of cisplatin, as described before [16, 17]. Mili-WT and –KO MEFs were generated from 13.5-day-old embryos of mili WT and KO mice [2] and provided by Dr. Jianxin Gao (The Ohio State University) and were grown in DMEM containing 20% fetal bovine serum (FBS). For cisplatin treatment, cells were maintained in medium with the desired doses of cisplatin (Sigma, St. Louis, MO) for 1 h, and then washed with PBS and followed by incubation in fresh cisplatin-free medium for varying times post-treatment.
2.2. MNase digestion
MNase digestion assay, as described previously [18], was used to study chromatin relaxation. Briefly, MEFs were treated with cisplatin and lysed in hypotonic cell lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol, 25% glycerol, 0.2% Nonidet P-40, 0.5 μM spermidine, 0.15 μM spermine, and protease inhibitor mixture) for 10 min. Nuclei were pelleted by centrifugation and resuspended in 200 μl MNase buffer (10mM Tris-HCl, pH 8.0, 50 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 1 mM CaCl2) and digested with MNase (Sigma) at room temperature for 5 min. The reaction was then stopped by adding 5 X stop solution (0.1 M EDTA, 0.01 M EGTA, pH 8.0). DNA was isolated by phenol/chloroform and separated on a 1.8% agarose gel.
2.3. Western blot analysis
Whole cell lysates were prepared by boiling cell pellets for 10 min in lysis buffer, as described before [16]. After protein quantification, equal amounts of proteins were loaded, separated on a polyacrylamide gel, and transferred to a nitrocellulose membrane. Protein bands were immuno-detected with the following antibodies: Rabbit anti-XPC (1:5,000) and anti-DDB2 (1:1,000) antibodies were generated in our laboratory. Mouse anti-XPA (1:1,000), mouse anti-XPF (1:500), rabbit anti-ERCC1 (1:1,000), rabbit anti-p300 (1:1,000), and mouse anti-Tubulin (1:2,000) antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Rabbit anti-XPG (1:2,000) antibody was purchased from Bethyl Laboratory (Montgomery, TX). Rabbit anti-AcH3 (K9,14) (1:20,000), rabbit anti-AcH3 (K9) (1:1,000), rabbit anti-MeH3 (K4) (1:1,000), and rabbit anti-histone H3 (1:1,000) antibodies were purchased from Millipore (Billerica, MA). Rabbit anti-phosphorylated H3 (S10) (1:1,000), rabbit anti-AcH3 (K18) (1:1,000), rabbit anti-MeH3 (K9) (1:1,000), and rabbit anti-HP-1α (1:1,000) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Mouse anti-Piwil2 IgM antibody was generated provided by Dr. Jianxin Gao (The Ohio State University).
2.4. Immunoprecipitation and HAT assay
Whole cell extract was prepared from MEFs as described [19]. Briefly, cells were lysed in lysis buffer containing 50 mM Tris-HCl, pH7.5, 5 mM EDTA, 0.1% Nonidet P-40, and 250 mM NaCl supplemented with protease inhibitor and phosphorytase inhibitor cocktail (Roch Applied-Science) for 10 min at 4 °C, and sonicated for 5 seconds. The lysates were centrifuged at 18,000 g for 10 min. The supernatants were saved and protein concentration was determined using Bio-Rad protein assay kit. p300 proteins were immunoprecipitated from 1.5 mg of total cell lysats with 2 μg of the anti-p300 antibody and 30 μl of protein A/G beads (Calbiochem, Gibbstown, NJ) by incubating overnight with rotation at 4 °C. The beads were washed 4 times with lysis buffer, and used in HAT activity assay (kit from BioVision, Mountain View, CA). After the HAT assay, the p300 protein was eluted from beads and subjected to Western blotting to evaluate the p300 protein levels.
2.5. Cell survival assay
CDDP and 2008C13 cells were seeded in 60-mm plates after 48 h transfection with Piwil2 siRNA, and treated with cisplatin for 1 h. Cells were further cultured in drug-free medium for 10 days. Colonies of > 100 cells were counted after staining with methylene blue. MEFs were seeded in 96-well plates at an initial density of 1 × 103, incubated for 24 h, and treated with increasing doses of cisplatin for 1 h. Cultures were incubated for another 5 days, fixed with 3.7% formaldehyde for 30 min, and stained with 1.0% methylene blue for 30 min. 100 μl solvent (10% acetic acid and 50% methanol) was added to each well to dissolve the cells and, optical density (OD) of the released color was read at 660 nm. The relative cell survival was calculated with the values of mock-treated cells set as 100%.
2.6. Immuno-slot blot (ISB) assay
Cancer cells were pre-treated with hydroxyurea (HU) for 24 h, MEFs were grown to confluence and further kept in serum-free medium for 24 h to arrest cell growth. All cells were treated with cisplatin for 1 h, washed twice with PBS, and further cultured in HU containing medium (cancer cells) or HU-free medium (MEFs) for the desired time periods. The DNA was isolated, and the same amounts of denatured DNA were applied to nitrocellulose membranes. Cisplatin-induced DNA intrastrand cross-links (Pt-GG) were detected with anti-Pt-GG antibody as described before [16]. The intensity of each band was quantified, and the lesion concentrations were determined from a reference standards run in parallel to calculate the relative amounts of Pt-GG remaining at each time point.
2.7. RNAi
siRNA SMARTpools designed to target human Piwil2 were purchased from Dharmacon Inc (Denver, CO). A scramble non-targeting siRNA, synthesized by Dharmacon, Inc., was used in control experiments. Different siRNA (50 nM) were transfected into CDDP or 2008C13 cells using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacture’s instruction.
3. Results
3.1. Mili deficiency blocks DNA damage-induced chromatin relaxation and histone acetylation in MEFs
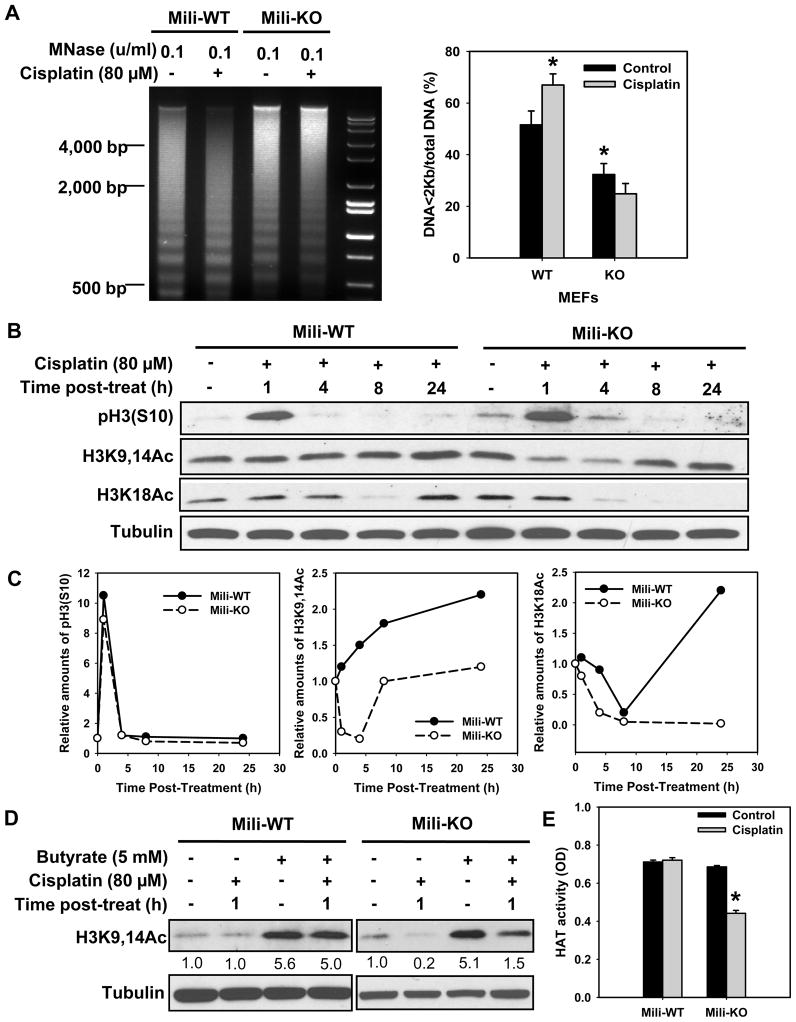
DNA lesions must be promptly recognized and repaired within the context of highly condensed chromatin fibers. Access to the DNA damage is facilitated by acetylation of histones H3 and H4, and critical involvement of chromatin remodeling factors to relax the chromatin structure around damaged sites [20]. To understand whether Piwil2 is involved in chromatin decondensation, especially in response to exposures of DNA damaging agents, we assessed the accessibility of chromatin to micrococcal nuclease (MNase) digestion in Mili-WT and -KO MEFs following cisplatin treatment. Fig. 1A shows that in Mili-KO MEFs chromatin exhibits a greater condensed state than Mili-WT MEFs, as reflected by the lesser MNase digestion in Mili-KO compared with Mili-WT MEFs. Accordingly, upon cisplatin treatment, WT MEFs exhibit significantly increased MNase susceptibility, while the extent of MNase digestion was not enhanced in cisplatin treated Mili-KO MEFs (Fig. 1A). These results indicated that both intrinsic and DNA damage-induced chromatin decondensation are compromised in the absence of Piwil2. We next assessed the acetylation of histone H3 in Mili-WT and -KO MEFs. As shown in Fig. 1B and C, cisplatin treatment caused a consistent increase of histone H3 acetylation at lysines 9,14 in WT cells up to 24 h. In contrast, H3K9,14Ac initially decreased significantly upon cisplatin treatment of Mili-KO MEFs at 1 and 4 h, and recovered later by 8 h time points. In addition, no obvious decrease of H3K18Ac was found in Mili-WT MEFs at 1 and 4 h upon cisplatin treatment. On the other hand, cisplatin treatment caused a continuous decrease of H3K18Ac that completely failed to recover by 24 h in Mili-KO MEFs. Phosphorylation of histone H3 at serine 10 (pH3S10) increased at 1 h and restored to its original level at 4 h time points in both Mili-WT and KO MEFs (Fig. 1B,C). These data indicated that Piwil2 is a key factor regulating histone H3 acetylation, but not phosphorylation upon DNA damage.
Fig. 1.
Mili is required for cisplatin-induced chromatin relaxation and histone acetylation. (A) Mili-WT and KO MEFs were treated with cisplatin for 1 h, further cultured in drug-free medium for another 1 h. The nuclei were isolated and subjected to MNase treatment. The genomic DNA was then isolated and resolved in agarose gel. The intensity of each lane was scanned and the ratio of DNA<2Kb to total DNA was calculated. The experiment was repeated three times. Bars represent SD. *: p < 0.05 compared with WT control. (B) Mili-WT and KO MEFs were treated with cisplatin for 1 h, further cultured in drug-free medium for various time periods. The phosphorylated and acetylated histone H3 was detected using Western blotting. (C) Quantitative analysis of modified histone H3 from the blot in (B), normalized to Tubulin. (D) MIli-WT and Mili-KO cells were pre-treated with Sodium butyrate for 0.5 h, before being treated with cisplatin for 1 h in the presence of butyrate. The cells were then cultured in the presence of butyrate for another 1 h. Acetylated H3 was detected using Western blotting. The band intensities were quantitatively assessed by scanning programs, and normalized to Tubulin levels. (E) Mili-WT and KO MEFs were treated with cisplatin for 1 h, further cultured in drug-free medium for another 2 h. The nuclear extract was prepared, the HAT activity was assayed and expressed with OD values. The HAT activity is an average of three independent repeats. Bars represent SD. *: p < 0.01 compared with control.
Since acetylation status is controlled by histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities, we wanted to investigate which process is affected by the absence of Piwil2. Sodium butyrate is a classical and specific inhibitor of HDAC. Treatment of both Mili-WT and Mili-KO MEFs with sodium butyrate showed the accumulation of total acetylated H3 (K9,14) in both cisplatin- and mock-treated cells. Nevertheless, cisplatin-induced reduction of H3K9,14Ac was still observed in Mili-KO MEFs (Fig. 1D), indicating that cisplatin-induced decrease of histone H3 acetylation in Piwil2-deficient cells cannot be attributed to enhanced HDAC activity. Thus, we assessed the effect of cisplatin on total HAT activity in both WT and Mili-KO MEFs. As shown in Fig. 1E, the HAT activity is comparable between WT and Mili-KO MEFs, and cisplatin treatment did not change the cellular HAT activity in WT MEFs, but significantly reduced the total HAT activity in Mili-KO MEFs, which is consistent with the overall change of histone H3 acetylation described above. Combined data suggest that Piwil2 deficiency blocks cisplatin-induced histone acetylation through the inhibition of cellular HAT activity.
3.2. Piwil2 increases the specific p300 HAT activity upon cisplatin treatment
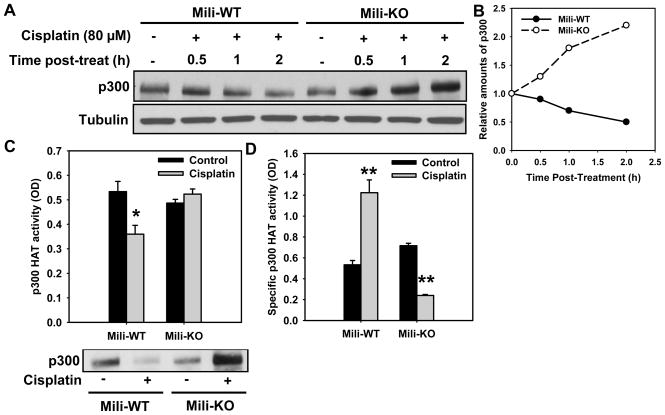
p300 is believed to be one of the key DNA damage-responsive HATs responsible for histone H3 acetylation [21]. We investigated whether Piwil2 has an influence on p300 in WT and Mili-KO MEFs. Surprisingly, we demonstrated that p300 decreased in WT MEFs, while increased in Mili-KO MEFs upon cisplatin treatment (Fig. 2A,B). Since a concomitant increase of HAT activity and degradation of p300 has been previously reported in mouse embryonal carcinoma cells treated with retinoic acid [19], we analyzed the p300 HAT activity in both cell types following cisplatin treatment. For this, p300 was immunoprecipitated from equal amounts of whole cell extracts of WT and KO MEFs with and without cisplatin treatment. HAT activity of immunoprecipitated p300 was determined in parallel with the quantitation of recovered protein by Western blotting. As shown in Fig. 2C, the total p300 HAT activity is comparable between WT and KO MEFs. However, cisplatin treatment decreased the total p300 HAT activity in WT MEFs but had no discernable effect on this activity in KO MEFs. We then calculated the specific HAT activity of p300 by normalizing the total p300 HAT activity to the amount of precipitated p300 protein. The results showed that the absolute specific HAT activity increased significantly in WT MEFs, and actually did decrease in KO MEFs upon cisplatin treatment (Fig. 2D). These data sets demonstrated that Piwil2 is influencing the selective depletion of p300 and concomitant increase in its HAT activity in cells following cisplatin treatment.
Fig. 2.
Mili is required for concomitant reduction of p300 protein and increase of specific HAT activity of p300 in response to cisplatin treatment. (A) Mili-WT and KO MEFs were treated with cisplatin for 1 h, and further cultured in drug-free medium for the indicated time periods. p300 was detected using Western blotting. (B) Quantitative analysis of p300 from the blot in (A), normalized to Tubulin. (C) Mili-WT and KO MEFs were treated with cisplatin for 1 h, washed and further cultured for another 2 h. p300 was immunoprecipitated from equal amount of whole cell extract, and assayed for HAT activity, which is expressed as the OD values. Results are the mean of three independent experiments. Bars represent SD. *: p < 0.05 compared with its corresponding control. After the HAT assay, the p300 immunoprecipitates were subjected to Western blot analysis to evaluate the p300 protein levels. (C), Specific activity of p300 HAT normalized to p300 protein level (Bars: SD, n=3). **: p < 0.01 compared with their corresponding control.
3.3. Mili maintains the euchromatic marks following cisplatin treatment of mammalian cells
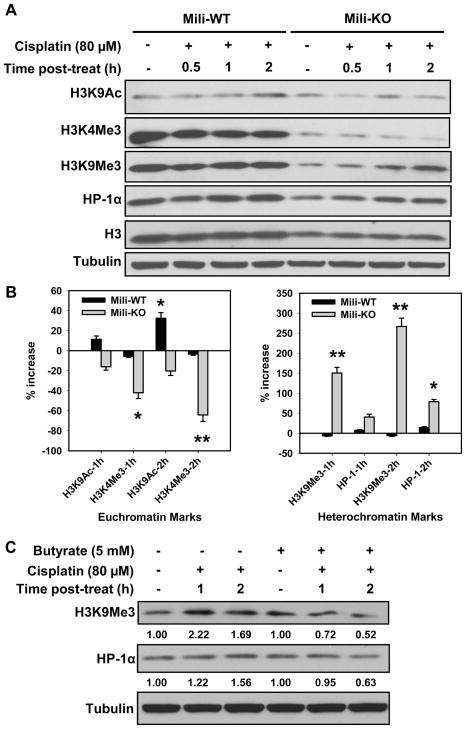
Piwi has been reported to associate with chromatin and promote the euchromatic character of sub-telomeric heterochromatin in Drosophila [4]. To explore whether Piwil2 plays a similar role in the equilibrium between euchromatin and heterochromatin in mammalian cells following DNA damage, we analyzed the euchromatic and heterochromatic marks in both Mili-WT and -KO MEFs treated with cisplatin. Since the most obvious change of histone acetylation was observed within 4 h of cisplatin treatment (Fig. 1B), we focused on the early alteration of euchromatic and heterochromatic marks in MEFs following cisplatin treatment. As shown in Fig. 3A, the steady-state levels of trimethylation of H3 at lysine 4 (H3K4Me3), trimethylation of H3 at lysine 9 (H3K9Me3), and HP-1α are lower in KO than those in WT MEFs. Considering the concomitant lower level of total histone H3 in KO MEFs, it seems that Mili deficiency reduces the abundance of histones and histone H3 methylation. Despite the different levels of methylated histone H3 and HP-1α in WT and KO MEFs, the dynamics of euchromatic and heterochromatic marks upon cisplatin treatment are distinct between WT and KO MEFs (Fig. 3A, B). For examples, the euchromatic mark H3K9Ac increased significantly in WT MEFs at 2 h following cisplatin treatment, but had no obvious change in KO MEFs at the same time point. In addition, both euchromatic marks, i.e., H3K4Me3, and heterochromatic marks, i.e., H3K9Me3 and HP-1α, did not exhibit a notable change upon cisplatin treatment of WT MEFs at various time points. On the contrary, the euchromatic mark H3K4Me3 decreased, while the heterochromatic marks H3K9Me3 and HP-1α increased significantly in Mili-KO MEFs at 2 and 4 h upon cisplatin post-treatment. These results indicated that Piwil2 is integral to the maintenance of euchromatic marks in mammalian cells following cisplatin treatment.
Fig. 3.
Mili is required for the maintenance of euchromatic marks in MEFs following cisplatin treatment. (A) Mili-WT and KO MEFs were treated with cisplatin for 1 h and further cultured in drug-free medium for indicated time periods. The different marks of euchromatin and heterochromatin were detected using Western blotting. (B) The band intensities were quantitatively assessed by scanning programs, normalized to Histone levels, and the changes of various marks in MEFs were plotted at 1 and 2 h post-treatment (Bars: SD, n=2). *: p < 0.05, **: p < 0.01 compared with their corresponding protein level changes in Mili-WT. (C) Mili-KO MEFs were pretreated with Sodium butyrate for 0.5 h, and then treated with cisplatin for 1 h followed by culturing in Butyrate-containing medium for 1 and 2 h. The heterochromatic marks were determined through Western blotting. Relative amount of H3K9Me3 and HP-1α at various times post-cisplatin treatment were quantified relative to the respective untreated levels and normalized to Tubulin controls.
The transition from euchromatin to heterochromatin is supposedly initiated by histone deacetylation [22]. Therefore, we wanted to understand whether HDAC is also involved in cisplatin-induced formation of heterochromatin in Mili-KO MEFs. Again, we used the specific HDAC inhibitor Sodium butyrate to inhibit the cellular HDAC activity. We found that treatment with Sodium butyrate reversed the increase of heterochromatic marks H3K9Me3 and HP-1α in KO MEFs following cisplatin treatment (Fig. 3C). Given the finding that HDAC does not contribute to cisplatin-induced hypoacetylation of histone H3 K9,14 in Mili-KO MEFS (Fig. 1D), we speculate that HDAC may enhance heterochromatic marks through deacetylating other acetylated histones, and Piwil2 can interfere with HDAC for maintaining the euchromatic marks in mammalian cells.
3.4. Piwil2-deficient cells exhibit high sensitivity to cisplatin and reduced NER for removing cisplatin-induced intrastrand crosslinks
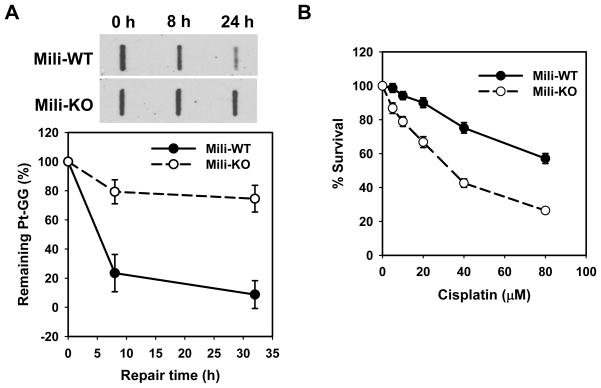
Given the role of Piwil2 in chromatin modifications upon DNA damage described above, we reasoned that Piwil2 may ultimately be acting in facilitating DNA repair. To test this hypothesis, we determined the removal rates of cisplatin-induced intrastrand crosslinks (Pt-GG) in WT and Mili-KO MEFs by using immuno-slot blotting (ISB) assay with anti-Pt-GG antibody. As shown in Fig. 4A, WT MEFs removed cisplatin-induced Pt-GG much rapidly; about 80% and 90% Pt-GG were lost at 8 and 24 h, respectively. In contrast, Mili-KO MEFs showed attenuated removal of Pt-GG; less than 20 and 30% Pt-GG was lost at 8 and 24 h, respectively. We next assessed the cisplatin sensitivity of WT and Mili-KO MEFs after 1 h cisplatin treatment with clonogenic survival assay. As shown in Fig. 4B, Mili-KO MEFs are more sensitive to cisplatin treatment compared with WT MEFs. Collectively, these data indicated that the absence of Piwil2 significantly affects the cells’ ability to remove cisplatin-induced DNA intrastrand crosslinks. As a result, the cells deficient in Piwil2 exhibit the higher sensitivity to cisplatin treatment.
Fig. 4.
Mili-KO MEFs exhibit high sensitivity to cisplatin and deficient NER for removing cisplatin-induced intrastrand crosslinks. (A) Mili-WT and KO MEFs were treated with cisplatin for 1 h, further cultured for the indicated time periods. The initial and remaining Pt-GG level were detected by ISB assay with anti-Pt-GG antibody (Bars: SD, n=3). (B) WT and Mili-KO MEFs were treated with cisplatin for 1 h. The cells were further cultured in drug-free medium for 5 days. The relative survival of cells was determined by methylene blue staining (Bars: SD, n=4).
Since NER is the main pathway to remove cisplatin-induced DNA intrastrand crosslinks, we determined the expression levels of various NER factors, at both transcript and protein levels, in Mili-WT and KO MEFs. Although Mili-KO MEFs exhibited deficient NER, the mRNA levels of XPA, XPF, XPG and ERCC1 in these cells are relatively higher than those in WT cells (Supplementary Fig. S1A). Moreover, protein levels of XPC, XPG, ERCC1 and DDB2 are also higher in Mili-KO MEFs than WT MEFs (Supplementary Fig. S1B). These results indicated that the cellular levels of NER factors are unlikely to be responsible for the decreased NER capability of Piwil2-deficient cells, and further support that loss of Piwil2-mediated chromatin remodeling as the main contributor to reduced NER capacity in these cells.
3.5. Enhanced expression of Piwil2 is one of the key factors contributing to the cisplatin resistance in human ovarian cancer cell lines
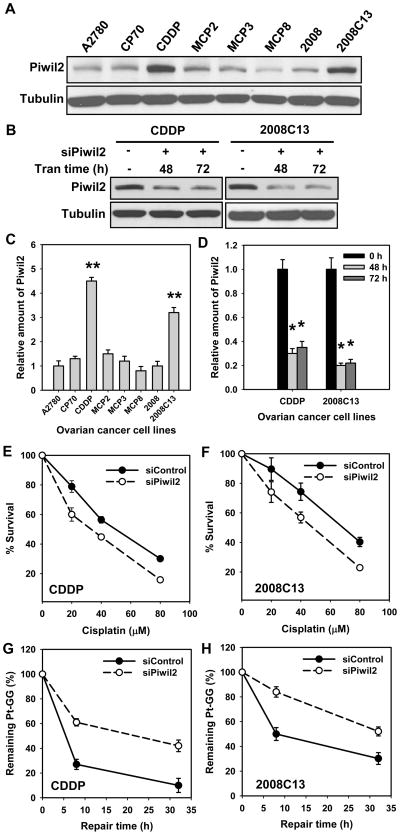
Piwil2 is widely expressed in tumors, including human ovarian cancer [7]. Since Piwil2 can modulate chromatin remodeling and DNA repair upon cisplatin treatment, as described above, we wanted to explore whether Piwil2 expression is related to cisplatin resistance in human ovarian cancer cell lines. We first assessed the level of Piwil2 expression in two cisplatin-sensitive ovarian cancer cell lines and their resistant variants. We found that cisplatin-resistant lines, CDDP and 2008C13, derived from A2780 and 2008 ovarian cancer cell lines, respectively, display dramatically higher level of Piwil2 protein compared with their corresponding parental cell lines (Fig. 5A,C). We then knocked down the expression of Piwil2 with siRNA and determined its influence on cisplatin sensitivity. Piwil2 siRNA transfection can knock down about 70% of Piwil2 in CDDP cells and 80% Piwil2 in 2008C13 cells (Fig. 5B,D). Clonogenic survival analysis showed that Piwil2 knockdown sensitized both CDDP and 2008C13 cells to cisplatin, with IC50 decreasing from 49.2 μM to 32.5 μM in CDDP cell line (Fig. 5E), and 70.8 μM to 50.4 μM in 2008C13 cell line (Fig. 5F), indicating that enhanced expression of Piwil2 is one of the contributing factors in cisplatin resistance of some cancer cells. We further determined the effect of Piwil2 knockdown on the removal of cisplatin-induced DNA damage in these human cancer cells. CDDP and 2008C13 cells, transfected with Piwil2 or control siRNA for 48 h, were subjected to cisplatin treatment. The amount of initially formed Pt-GG in the genomic DNA and the removal of Pt-GG were detected. A significant decrease in the repair of Pt-GG was observed at 8 and 24 h in both CDDP and 2008C13 cells transfected with Piwil2 siRNA as compared with the cells transfected with control siRNA (Fig. 5G & H). These results indicated that increased Piwil2 expression in human ovarian cancer cell lines CDDP and 2008C13 contributes to the cisplatin resistance, probably by enhancing the repair of cisplatin-induced DNA damage.
Fig. 5.
Enhanced Piwil2 protein level in cisplatin-resistant ovarian cancer cell lines is responsible for their low sensitivity to cisplatin and high DNA repair capacity. (A) The protein level of Piwil2 in human ovarian cancer cell line A2780, 2008 and their corresponding variants were detected by Western blotting. (B) Piwil2 was knocked down with target specific siRNA in CDDP and 2008C13 cells followed by Western blotting of protein extracts. (C) Quantitative analysis of Piwil2 from the blot in (A). The experiments were repeated twice, Bars: SD, **: p < 0.01 compared with A2780 cell line. (D) Quantitative analysis of Piwil2 from the blot in (B). The experiments were repeated twice, Bars: SD, *: p < 0.01 compared with 0 h group. (E, F) Piwil2 or control siRNA was transfected into CDDP and 2008C13 cells for 48 h, cells were treated with cisplatin for 1 h, and the relative survival was determined by colony forming assay after 10 days culture (average values of three experiments). Bars represent standard deviation (SD). (G, H) CDDP and 2008C13 cells were treated with cisplatin for 1 h after Piwil2 or control siRNA transfection. The cells were further cultured for the indicated time periods. The initial and remaining levels of Pt-GG were detected using ISB assay. The relative percentage of Pt-GG remaining at different time points is an average of three independent repeats. Bars represent SD.
4. Discussion
4.1. Piwil2 and histone acetylation upon cisplatin treatment
Histone acetylation functions in the relaxation of chromatin structure, which occurs in transcriptionally active regions during gene expression, and also in response to DNA damage [18, 20, 21, 23–25]. In terms of DNA repair, the modulation of chromatin structure caused by acetylation is thought to facilitate the access of DNA damage repair proteins to the embedded lesions [26, 27]. We clearly show that in the absence of Piwil2 histone H3 in MEFs is hypoacetylated in response to cisplatin treatment, indicating the importance of Piwil2 in histone acetylation upon DNA damage. Additionally following cisplatin treatment, H3K9,14Ac showed an initial decrease followed by a recovery at 8 hour, while the H3K18Ac displayed a consistent discernable decrease. These results indicated that acetylation of histone H3 at different residues may have different function(s). Given histone acetylation/deacetylation is a dynamic process, the acetylation status of H3, which changes by cisplatin treatment in Mili-KO MEFs, can be restored back at specific times corresponding to the completion of its putative function.
p300 is a general transcriptional co-activator that controls many biological activities. Its HAT activity endows p300 with the capacity to activate transcription by influencing chromatin structure and function through acetylation of nucleosomal histones and other transcription factors [28–30]. The HAT activity of p300 is upregulated through phosphorylation by protein kinase A (PKA) [19]. Moreover, p300 can also be inactivated by degradation through ubiquitin-proteasome pathway [31, 32]. Cisplatin treatment is reported to strongly reduce the expression of p300 protein in human ovarian cancer cell lines, and this reduction results in its repressed transcription [33]. Here, we show that cisplatin significantly reduces the expression of p300 in WT MEFs, while increases it in the absence of Piwil2. Interestingly, the reduction of p300, which might affect the global repression of transcription, was accompanied with the increase of specific HAT activity in WT MEFs upon cisplatin treatment, and Piwil2 is required in these processes. It is noteworthy that the concomitant increase of HAT activity and degradation of p300 was also reported previously during retinoic acid-induced differentiation of mouse embryonic carcinoma cells [19], and suggested that there are at least two subpopulations of p300, one having HAT activity and persisting during DNA repair, whereas the other is devoid of this activity and is degraded. Therefore, we posit that the Piwil2-mediated increase of specific HAT activity of p300 is instrumental in the acetylation of histones around the cisplatin-induced DNA lesions. Piwil2 deficiency in Mili-KO MEFs halts the increase of p300 specific HAT activity, and consequently inhibits the histone acetylation in response to cisplatin.
In this study, we also noted that Mili-KO MEFs exhibit intrinsic resistance to MNase digestion. It has been established that chromatin structure can be altered by two main mechanisms. One is histone modifications [34], and another is chromatin remodeling by multi-protein complexes that utilize the energy derived from ATP hydrolysis [35]. We did not find obvious difference in the steady-state level of histone H3 acetylation between Mili-KO and WT MEFs. Although our data show a reduced steady-state level of euchromatin mark H3K4Me3 in Mili-KO MEFs compared with WT MEFs, we also found the reduced heterochromatin mark H3K9Me3 in Mili-KO MEFs compared with WT MEFs. Thus, the difference in chromatin structure between Mili-KO and WT MEFs is hard to explain by only the differences of histone methylation status found in this study. The other histone modifications, e.g., acetylation, phosphorylation, and ubiquitylation of histone H3 and H4, as well as the ATP-dependent chromatin remodeling may contribute to the different intrinsic chromatin structure between Mili-KO and WT MEFs.
4.2. Piwil2 and euchromatin/heterochromatin dynamics
A considerable body of literature indicates equilibrium between active euchromatin and silent heterochromatin in eukaryotic cells. We show that cisplatin treatment does not disrupt this equilibrium in normal MEFs, as reflected by unchanged euchromatic/heterochromatic marks. However, cisplatin treatment resulted in increased heterochromatic marks and concomitant global decrease of euchromatic marks in MEFs deficient in Piwil2. Piwil2 has already been linked to epigenetic code for heterochromatin formation [5]. Although Piwi in Drosophila has a global function in heterochromatic silencing, Piwi promotes the euchromatic character of sub-telomeric region in Drosophila and its transcriptional activity [4]. Our findings in this study indicated that Piwil2 also plays an important role in maintaining the dynamic equilibrium between euchromatin and heterochromatin following cisplatin treatment in mammalian cells. It is well documented that DNA damage can induce global repression of transcription in cells [36–38]. The cisplatin-induced increase of heterochromatin formation in Mili-KO MEFs may reflect the global transcriptional repression, whereas Piwil2-mediated maintenance of euchromatic features counteracts the global silence and promotes the transcription of some DNA repair genes, such as XPC, (as documented in Supplementary Fig. S2). On the other hand, euchromatic marks relax chromatin and facilitate the repair of DNA damage, while heterochromatin halts the repair of DNA lesions [13]. Thus, Piwil2-mediated maintenance of euchromatic marks could be one of the mechanisms contributing to the enhanced DNA repair capacity.
The mechanism through which Piwil2 prevents heterochromatization is still unclear. Yin and Lin [4] have proposed a heterochromatin/euchromatin counter-balance model, according to which Piwi associates with piRNAs, a novel class of small RNAs that interact with Piwi proteins. The Piwi-piRNA complex then binds to piRNA-corresponding genomic sequences, and an unknown component in the complex prevents heterochromatization. As our results demonstrate that inhibition of HDAC could block the cisplatin-induced increase of heterochromatic marks in Mili-KO MEFs, we speculate that the cisplatin-activated HDAC, responsible for heterochromatization, may be inhibited by the presence of Piwil2. Consequently, the euchromatin status is maintained.
4.3. Piwil2 and cisplatin resistance of cancer cells
Although both mouse and human Piwil2 genes are specifically expressed in testis, an enhanced expression of Piwil2 also occurs in various cancer cell lines and tumors [7], indicating a relationship between Piwil2 and tumor development. In this study, we have not only found that Piwil2 is expressed in various human ovarian cancer cell lines, but also demonstrated its role in the cisplatin resistance of cancer cells. Piwil2 has been reported to inhibit apoptosis through activation of Stat3/Bcl-XL pathway [7], and this is one of the mechanisms contributing to the cisplatin resistance in the cancers with enhanced Piwil2 expression. In this study, we have now identified another mechanism of Piwil2-mediated cisplatin resistance. We show that both Piwil2 knockdown in human cancer cells and Mili-KO in MEFs compromised the removal of Pt-GG following cisplatin treatment, indicating that Piwil2 can facilitate the repair of cisplatin-induced DNA intrastrand crosslinks to help cells survive the DNA damage inflicted by platinum-based chemotherapeutic agents. Evidence for increased repair of DNA damage in resistant cancer cells has been demonstrated by many groups [see the review, [10]], and increased DNA repair is believed to be one of the mechanisms for developing cisplatin resistance. Therefore, repression of Piwil2 expression to curtail repair in tumor cells provides a novel means for therapeutic intervention in human cancer.
In summary, we have demonstrated that Piwil2 plays a role in the cellular chromatin acetylation and relaxation following treatment with DNA damaging agents. Piwil2 is also required for the maintenance of the equilibrium between euchromatin and heterochromatin in response to cisplatin treatment in mammalian cells. Moreover, we indicated that Piwil2 is critical to the functional NER to remove cisplatin-induced DNA damage, and enhanced Piwil2 is one of the mechanisms contributing to the development of cisplatin resistance in human ovarian cancer cells. Given the established relationship between DNA repair and chromatin remodeling, we speculate that Piwil2 promotes chromatin relaxation through preventing heterochromatization, and consequently enhances DNA repair. As a result, cisplatin resistance develops in Piwil2-overexpressed tumors.
Supplementary Material
Acknowledgments
We would like to thank Dr. Karuppaiyah Selvendiran (The Ohio State University) for providing CDDP cells, Dr. Tim Huang (The Ohio State University) for providing MCP2, MCP3 and MCP8 cells, Dr. Francois X. Claret (University of Texas-M.D. Anderson Cancer Center) for providing 2008C13 cells, Dr. Paul Modrich (Duke University) for providing A2780 and CP70 cells, Dr. Jianxin Gao (The Ohio State University) for providing MEFs and anti-Piwil2 antibody, and Dr. Jürgen Thomale (Institut für Zellbiolgoie, Germany) for providing Pt-GG antibody. This work was supported by NIH grants CA93413, ES2388 and ES12991 to AAW.
Footnotes
Conflict of interest.
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 3.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 4.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 5.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15:201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Shen R, Ye Y, Pu XA, Liu X, Duan W, Wen J, Zimmerer J, Wang Y, Liu Y, Lasky LC, Heerema NA, Perrotti D, Ozato K, Kuramochi-Miyagawa S, Nakano T, Yates AJ, Carson WE, III, Lin H, Barsky SH, Gao JX. Precancerous stem cells have the potential for both benign and malignant differentiation. PLoS One. 2007;2:e293. doi: 10.1371/journal.pone.0000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 10.Saldivar JS, Wu X, Follen M, Gershenson D. Nucleotide excision repair pathway review I: implications in ovarian cancer and platinum sensitivity. Gynecol Oncol. 2007;107:S56–S71. doi: 10.1016/j.ygyno.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Q, Wani AA. Histone modifications: crucial elements for damage response and chromatin restoration. J Cell Physiol. 2010;223:283–288. doi: 10.1002/jcp.22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 13.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 15.Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 16.Barakat BM, Wang QE, Han C, Milum K, Yin DT, Zhao Q, Wani G, Arafa ES, El-Mahdy MA, Wani AA. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int J Cancer. 2010;127:977–988. doi: 10.1002/ijc.25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansouri A, Ridgway LD, Korapati AL, Zhang Q, Tian L, Wang Y, Siddik ZH, Mills GB, Claret FX. Sustained activation of JNK/p38 MAPK pathways in response to cisplatin leads to Fas ligand induction and cell death in ovarian carcinoma cells. J Biol Chem. 2003;278:19245–19256. doi: 10.1074/jbc.M208134200. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, Barakat BM, Qin S, Ray A, El-Mahdy MA, Wani G, Arafa e, Mir SN, Wang QE, Wani AA. The p38 mitogen-activated protein kinase augments nucleotide excision repair by mediating DDB2 degradation and chromatin relaxation. J Biol Chem. 2008;283:32553–32561. doi: 10.1074/jbc.M803963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brouillard F, Cremisi CE. Concomitant increase of histone acetyltransferase activity and degradation of p300 during retinoic acid-induced differentiation of F9 cells. J Biol Chem. 2003;278:39509–39516. doi: 10.1074/jbc.M307123200. [DOI] [PubMed] [Google Scholar]
- 20.Costelloe T, Fitzgerald J, Murphy NJ, Flaus A, Lowndes NF. Chromatin modulation and the DNA damage response. Exp Cell Res. 2006;312:2677–2686. doi: 10.1016/j.yexcr.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 21.Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS One. 2009;4:e4864. doi: 10.1371/journal.pone.0004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Lippard SJ. Cisplatin-induced post-translational modification of histones H3 and H4. J Biol Chem. 2004;279:20622–20625. doi: 10.1074/jbc.M402547200. [DOI] [PubMed] [Google Scholar]
- 24.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci U S A. 2008;105:10320–10325. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng Y, Liu H, Gill HW, Yu Y, Waters R, Reed SH. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 2008;9:97–102. doi: 10.1038/sj.embor.7401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Downey M, Durocher D. Chromatin and DNA repair: the benefits of relaxation. Nat Cell Biol. 2006;8:9–10. doi: 10.1038/ncb0106-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Jones K, Gong F. The molecular basis of chromatin dynamics during nucleotide excision repair. Biochem Cell Biol. 2009;87:265–272. doi: 10.1139/O08-101. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 30.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 31.Poizat C, Sartorelli V, Chung G, Kloner RA, Kedes L. Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol Cell Biol. 2000;20:8643–8654. doi: 10.1128/mcb.20.23.8643-8654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poizat C, Puri PL, Bai Y, Kedes L. Phosphorylation-dependent degradation of p300 by doxorubicin-activated p38 mitogen-activated protein kinase in cardiac cells. Mol Cell Biol. 2005;25:2673–2687. doi: 10.1128/MCB.25.7.2673-2687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duyndam MC, van Berkel MP, Dorsman JC, Rockx DA, Pinedo HM, Boven E. Cisplatin and doxorubicin repress Vascular Endothelial Growth Factor expression and differentially down-regulate Hypoxia-inducible Factor I activity in human ovarian cancer cells. Biochem Pharmacol. 2007;74:191–201. doi: 10.1016/j.bcp.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Verger A, Crossley M. Chromatin modifiers in transcription and DNA repair. Cell Mol Life Sci. 2004;61:2154–2162. doi: 10.1007/s00018-004-4176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 36.Proietti-De-Santis L, Drane P, Egly JM. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25:1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rockx DA, Mason R, van HA, Barton MC, Citterio E, Bregman DB, Van Zeeland AA, Vrieling H, Mullenders LH. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc Natl Acad Sci U S A. 2000;97:10503–10508. doi: 10.1073/pnas.180169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda S, Naruse S, Yatani R. Effects of ultra-violet microbeam irradiation of various sites of HeLa cells on the synthesis of RNA, DNA and protein. Nature. 1967;213:696–697. doi: 10.1038/213696a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.