Abstract
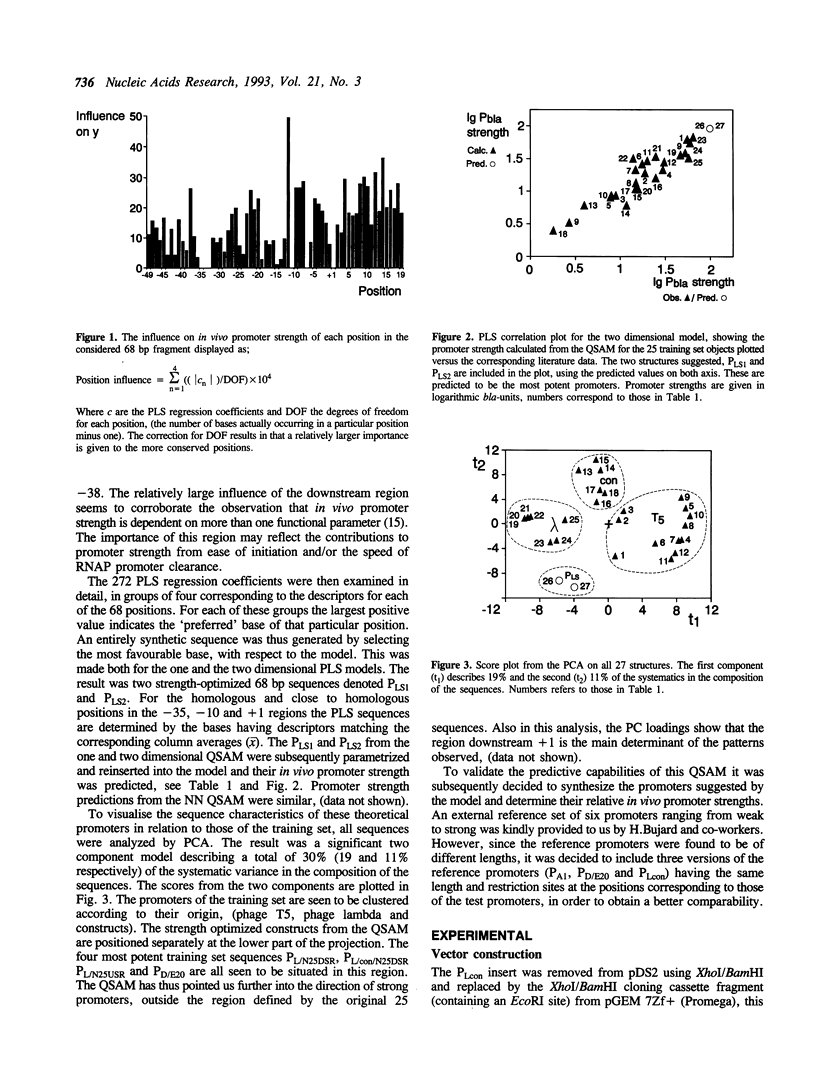
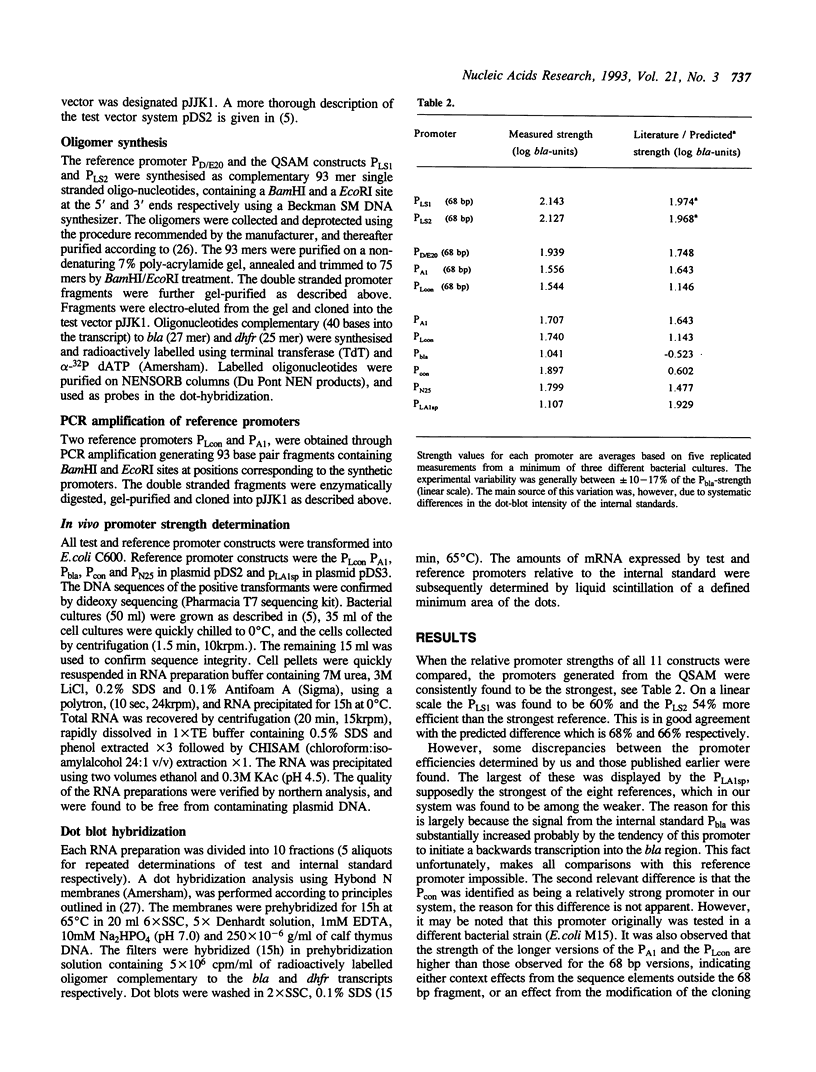
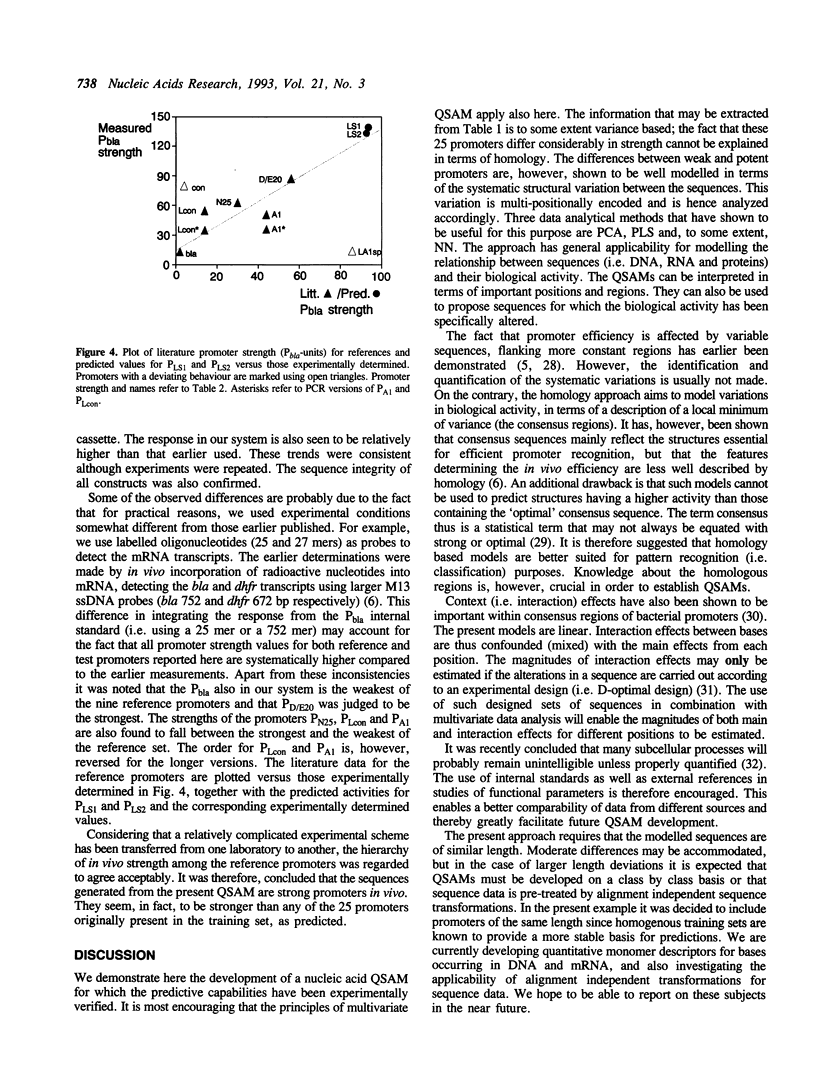
Models have been developed that allow the biological activity of a DNA segment to be altered in a desired direction. Partial least squares projections to latent structures (PLS) was used to establish a quantitative model between a numerical description of 68 bp fragments of 25 E.coli promoters and their corresponding quantitative measure of in vivo strength. This quantitative sequence-activity model (QSAM) was used to generate two 68 bp fragments predicted to be more potent promoters than any of those on which the model originally was based. The optimized structures were experimentally verified to be strong promoters in vivo.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunak S., Engelbrecht J., Knudsen S. Neural network detects errors in the assignment of mRNA splice sites. Nucleic Acids Res. 1990 Aug 25;18(16):4797–4801. doi: 10.1093/nar/18.16.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner M., Bujard H. Promoter recognition and promoter strength in the Escherichia coli system. EMBO J. 1987 Oct;6(10):3139–3144. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon L. R., Stormo G. D. Expectation maximization algorithm for identifying protein-binding sites with variable lengths from unaligned DNA fragments. J Mol Biol. 1992 Jan 5;223(1):159–170. doi: 10.1016/0022-2836(92)90723-w. [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Ray S. C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991 Jun 25;19(12):3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B., Zhou G. W. Neural network optimization for E. coli promoter prediction. Nucleic Acids Res. 1991 Apr 11;19(7):1593–1599. doi: 10.1093/nar/19.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrynin V. N., Korobko V. G., Severtsova I. V., Bystrov N. S., Chuvpilo S. A., Kolosov M. N. Synthesis of a model promoter for gene expression in Escherichia coli. Nucleic Acids Symp Ser. 1980;(7):365–376. [PubMed] [Google Scholar]
- Galas D. J., Eggert M., Waterman M. S. Rigorous pattern-recognition methods for DNA sequences. Analysis of promoter sequences from Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):117–128. doi: 10.1016/0022-2836(85)90262-1. [DOI] [PubMed] [Google Scholar]
- Graña D., Gardella T., Susskind M. M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988 Oct;120(2):319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg S., Eriksson L., Jonsson J., Lindgren F., Sjöström M., Skagerberg B., Wold S., Andrews P. Minimum analogue peptide sets (MAPS) for quantitative structure-activity relationships. Int J Pept Protein Res. 1991 May;37(5):414–424. doi: 10.1111/j.1399-3011.1991.tb00756.x. [DOI] [PubMed] [Google Scholar]
- Jacquet M. A., Reiss C. Transcription in vivo directed by consensus sequences of E.coli promoters: their context heavily affects efficiencies and start sites. Nucleic Acids Res. 1990 Mar 11;18(5):1137–1143. doi: 10.1093/nar/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J., Eriksson L., Hellberg S., Lindgren F., Sjöström M., Wold S. A multivariate representation and analysis of DNA sequence data. Acta Chem Scand. 1991 Feb;45(2):186–192. doi: 10.3891/acta.chem.scand.45-0186. [DOI] [PubMed] [Google Scholar]
- Kammerer W., Deuschle U., Gentz R., Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 1986 Nov;5(11):2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus R., Bujard H. PL of coliphage lambda: an alternative solution for an efficient promoter. EMBO J. 1988 Sep;7(9):2919–2923. doi: 10.1002/j.1460-2075.1988.tb03150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox J. Is molecular biology yet a science? Nature. 1992 Jan 16;355(6357):201–201. doi: 10.1038/355201a0. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M. C. Training back-propagation neural networks to define and detect DNA-binding sites. Nucleic Acids Res. 1991 Jan 25;19(2):313–318. doi: 10.1093/nar/19.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Van Dyke M. W. A rapid method for the purification of deprotected oligodeoxynucleotides. Nucleic Acids Res. 1991 Feb 11;19(3):674–674. doi: 10.1093/nar/19.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. A new family of powerful multivariate statistical sequence analysis techniques. J Mol Biol. 1991 Aug 20;220(4):877–887. doi: 10.1016/0022-2836(91)90360-i. [DOI] [PubMed] [Google Scholar]


