Abstract
Background and Aims
The cell cycle is controlled by cyclin-dependent kinases (CDKs), and CDK inhibitors are major regulators of their activities. The ICK/KRP family of CDK inhibitors has been reported in several plants, with seven members in arabidopsis; however, the phylogenetic relationship among members in different species is unknown. Also, there is a need to understand how these genes and proteins are regulated. Furthermore, little information is available on the functional differences among ICK/KRP family members.
Methods
We searched publicly available databases and identified over 120 unique ICK/KRP protein sequences from more than 60 plant species. Phylogenetic analysis was performed using 101 full-length sequences from 40 species and intron–exon organization of ICK/KRP genes in model species. Conserved sequences and motifs were analysed using ICK/KRP protein sequences from arabidopsis (Arabidopsis thaliana), rice (Orysa sativa) and poplar (Populus trichocarpa). In addition, gene expression was examined using microarray data from arabidopsis, rice and poplar, and further analysed by RT-PCR for arabidopsis.
Key Results and Conclusions
Phylogenetic analysis showed that plant ICK/KRP proteins can be grouped into three major classes. Whereas the C-class contains sequences from dicotyledons, monocotyledons and gymnosperms, the A- and B-classes contain only sequences from dicotyledons or monocotyledons, respectively, suggesting that the A- and B-classes might have evolved from the C-class. This classification is also supported by exon–intron organization. Genes in the A- and B- classes have four exons, whereas genes in the C-class have only three exons. Analysis of sequences from arabidopsis, rice and poplar identified conserved sequence motifs, some of which had not been described previously, and putative functional sites. The presence of conserved motifs in different family members is consistent with the classification. In addition, gene expression analysis showed preferential expression of ICK/KRP genes in certain tissues. A model has been proposed for the evolution of this gene family in plants.
Keywords: Arabidopsis thaliana, cell cycle, cyclin-dependent kinase inhibitor, ICK, exon–intron organization, gene expression, KRP, Oryza sativa, phylogeny, Populus trichocarpa, sequence motif
INTRODUCTION
In eukaryotes, the cell division cycle is strictly regulated by cyclin-dependent kinases (CDKs) together with specific cyclin partners (Morgan, 1997; Dewitte and Murray, 2003). The activity of CDKs can be regulated positively or negatively by transcriptional regulation, binding by other proteins, phosphorylation/dephosphorylation and proteolysis of the cyclin partner (Pines, 1995). While binding by cyclins activates CDKs, CDK inhibitors (CKIs), generally low-molecular-weight proteins, bind to CDKs and inhibit their activities (Sherr and Roberts, 1999; Mendenhall, 1998).
In mammals, there are two families of CKIs: KIP/CIP and INK4 (Sherr and Roberts, 1999). In yeast, three CKIs have been identified in the budding yeast Saccharomyces cerevisiae while only one in the fission yeast Schizosaccharomyces pombe (Mendenhall, 1998). In plants, two families of CKIs are now known. The first has a conserved domain at the C terminus that shows limited similarity to the mammalian KIP/CIP inhibitor p27Kip1 (Wang et al., 1997). This family of proteins from arabidopsis is referred to as ICKs (interactors/inhibitors of cyclin-dependent kinase; Wang et al., 1997; Lui et al., 2000), and also as KRPs (Kip-related proteins; De Veylder et al., 2001). Revised nomenclature refers to them as the ‘ICK/KRP family’ (Wang et al., 2006, 2007). Apart from the conserved domain at the C terminus, ICK/KRP proteins are very different from the mammalian and yeast CKIs (Wang et al., 2006). Recently, a second family of CKIs was described, represented by SIAMESE (SIM) from arabidopsis (Churchman et al., 2006) and EL2 from rice (Peres et al., 2007). They will be referred to as the SIM/EL2 family, based on the two initial members reported. SIM/EL2s are small proteins (about 14 kDa) and in the C-terminal region share a conserved EIEDFF sequence with the ICK/KRP proteins (Churchman et al., 2006; Peres et al., 2007).
ICK/KRP members were isolated either by the yeast two-hybrid approach from arabidopsis (Wang et al., 1997; Lui et al., 2000; Zhou et al., 2002), tobacco (Jasinski et al., 2002a, 2003) and alfalfa (Pettko-Szandtner et al., 2006), or by data mining from arabidopsis (De Veylder et al., 2001), maize (Coelho et al., 2005), rice (Barroco et al., 2006) and tomato (Bisbis et al., 2006). In vitro CDK inhibition activity has been demonstrated using recombinant ICK/KRP proteins to inhibit CDK complexes pulled down with p13Suc1-conjugated beads for arabidopsis ICK/KRP proteins (Wang et al., 1997, 1998; Lui et al., 2000; Nakai et al., 2006; Verkest et al., 2005), as well as ICK/KRP proteins from tobacco (Jasinski et al., 2002b), maize (Coelho et al., 2005), alfalfa (Pettko-Szandtner et al., 2006) and tomato (Bisbis et al., 2006). Domain-mapping studies showed that the interaction of ICK/KRPs with A-type CDKs and D-type cyclins is through the CDK/cyclin interacting/inhibiting domain (referred to hereafter as CID) located at the C-terminal region (Wang et al., 1998; Lui et al., 2000; Jasinski et al., 2002b). A tomato ICK/KRP protein SlKRP1 was shown recently to have another motif in the central region that is able to interact with CYCD3 as the SlKRP1 variant lacking the C-terminal conserved region still interacted with SlCYCD3 (Nafati et al., 2010).
Studies have shown that plants overexpressing an ICK/KRP gene display some common phenotypes, including reduced plant size, serrated leaves, reduced cell number and enlarged cells (Wang et al., 2000; De Veylder et al., 2001; Zhou et al., 2002; Jasinski et al., 2002a, 2003; Barroco et al., 2006; Kang et al., 2007; Bemis and Torii, 2007). At the cellular level, microinjection of recombinant ICK1 protein into Tradescantia virginiana stamen cells slowed mitosis by increasing the metaphase transit time (Cleary et al., 2002). While transgenic plants with a strong phenotype had a reduced ploidy level due to the inhibition of endoreduplication (De Veylder et al., 2001; Zhou et al., 2002; Jasinski et al., 2002a, 2003; Barroco et al., 2006), in transgenic plants weakly overexpressing ICK1 or ICK2, cells were induced to enter endoreduplication earlier, resulting in a higher level of ploidy (Verkest et al., 2005; Weinl et al., 2005). These observations suggest that strong overexpression of ICK1 or ICK2 inhibits the cell cycle progression at both G1/S and G2/M transitions, resulting in a reduced ploidy level and cell number, while weak overexpression of ICK1 or ICK2 preferentially inhibits mitosis and promotes the entry into endoreduplication.
Overexpression of an ICK/KRP gene also has specific effects on plant morphology and cellular differentiation. In addition to leaf serrations consistently observed in several studies (Wang et al., 2000; De Veylder et al., 2001; Zhou et al., 2002; Jasinski et al., 2002b), reduction in fertility was also observed in arabidopsis lines expressing ICK1 under the control of a pollen-specific promoter (Zhou et al., 2002) or in rice lines overexpressing the rice KRP1 (Barroco et al., 2006). Targeted expression of ICK1 in trichomes of arabidopsis plants under the control of the GL2 promoter caused reductions in both cell size and trichome branches (Schnittger et al., 2003). Interestingly, these trichomes collapsed and died earlier compared with the wild-type plants. Furthermore, the phenotypic effects in ICK/KRP-overexpressing lines could be attenuated by co-expressing a D-type cyclin, which interacts with ICK/KRPs (Jasinski et al., 2002a; Schnittger et al., 2003; Zhou et al., 2003a).
Despite significant progress made in understanding the functions and regulation of ICK/KRPs, many important questions remain. For instance, information is very limited regarding functional differences among different members of the ICK/KRP family in the same plant. The lack of a clear phylogenetic relationship hinders such studies. Apart from the conserved CID region, ICK/KRP proteins are very different from the mammalian CKIs and also show considerable sequence variability among themselves. In addition, although the function of CDK/cyclin interaction has been assigned to the CID region, specific sequences remain to be identified for possible biochemical functions such as interactions with other proteins, phosphorylation sites and motifs for regulating protein stability. In this study, a large number of new ICK/KRP sequences were identified and used for phylogenetic analysis, resulting in the classification of ICK/KRP proteins into three classes. Conserved sequences and putative functional motifs including novel motifs not known previously were identified, and they should be useful for further functional studies of these proteins. Furthermore, family-wide gene expression was also analysed using microarray and RT-PCR results.
MATERIALS AND METHODS
Sequence annotation and nomenclature
Using seven arabidopsis ICK/KRP protein sequences, ICK/KRP-related sequences were identified by the BLAST algorithm (Altschul et al., 1990) from the public databases indicated in Supplementary Data Table S1 (available online). Actual cDNA sequence entries (i.e. expressed sequence tags, ESTs) were preferred over predicted cDNA sequences (i.e. from genomic sequence). Sequences with p values higher than 1e-8 in terms of similarity to the zrabidopsis ICK/KRPs were excluded from the analysis. During our searches, some inconsistencies in sequence information were detected in the ICK/KRP data and they are explained in Supplementary Data Table S2. These inconsistencies were corrected manually using the ‘BCM Search Launcher’ tool: Sequence Utilities facility (http://searchlauncher.bcm.tmc.edu/seq-util/seq-util.html) (Smith et al., 1996). ESTs with >95 % identity in the coding region were considered alleles. The names for the seven arabidopsis, six rice and published ICK/KRPs follow their published annotations.
Alignments and phylogenetic analysis
Protein alignments were made with clustalw and a neighbour-joining tree was constructed with mega 3·1 (http://www.megasoftware.net) (Kumar et al., 2004). Poisson correction was used to calculate protein distances using a gap opening penalty of 10, a gap extension penalty of 0·1, a gap separation distance of 4, Blosum weight matrix and no residue-specific or hydrophilic penalties (Thompson et al., 1994). Bootstrap values are based on 1000 iterations for testing the significance of nodes.
Gene/genome organization and block duplication analysis
Exon–intron organization and chromosome position of ICK/KRP genes were both obtained by blasting the cDNA sequences against the arabidopsis genome sequence database in the Arabidopsis Information Resource (TAIR), the poplar JGI genome assembly v1·1 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) and the Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp). ICK/KRP genes on duplicated chromosomal blocks were determined using the ‘Paralogons in the Arasbidopsis thaliana’ database (http://wolfe.gen.tcd.ie/athal/dup) (Blanc et al., 2003) and by mapping rice and poplar ICK/KRPs in the duplicated blocks described in Guyot and Keller (2004) and Tuskan et al. (2006), respectively.
Protein properties and domain organization
Molecular weight (Mw) and isoelectric point (pI) were computed via Swiss-Prot/TrEMBL (http://ca.expasy.org/tools/pi_tool.html). The CID region was searched by using the smart program (http://smart.embl-heidelberg.de/; Schultz et al., 2000). ScanProsite (http://au.expasy.org/tools/scanprosite/; Sternberg and Islam, 1991) was used for detecting destruction box (R-X-X-[LM]-X-X-X-X-N) and phosphorylation site of p34Cdc2 ([TS]-P-X-[RKQSL]) patterns. p34Cdc2 phosphorylation sites were also detected by PhosphoBase v. 2·0 (http://www.cbs.dtu.dk/databases/PhosphoBase/ was used; current version available at http://phospho.elm.eu.org/) (Kreegipuu et al., 1999). Significant PEST regions were predicted by the ‘PEST finder’ program (https://embl.bcc.univie.ac.at/toolbox/pestfind/pestfind-analysis-webtool.htm; Rogers et al., 1986). Coiled-coil conformations were estimated by the Lupas algorithm program (http://www.ch.embnet.org/software/COILS_form.html; Lupas et al., 1991). Cell sorting signals (i.e. NLS) were obtained by the psort program (http://psort.ims.u-tokyo.ac.jp/) (Nakai and Horton, 1999).
Motif identification
Conserved motifs in ICK/KRP proteins were identified statistically by the MEME-MAST program, version 3·0 (http://meme.sdsc.edu/meme4_6_0/intro.html; Bailey and Elkan, 1994; Bailey and Gribskov, 1998) using motif setting as 6 to 200 length, 2 to 100 sites, and ‘p’ and ‘e’ values lower than 1e-8 and 1e-10, respectively. The resulting motifs were checked manually.
Microarray data
Expression values of ICK/KRPs were downloaded from publicly available Affymetrix array databases. For arabidopsis, ID and probe sequences were obtained at the Arabidopsis ATH1 Genome Array (http://www.affymetrix.com/products/arrays/specific/arab.affx), while expression values were obtained at the NASC's International Affymetrix Service (http://affymetrix.arabidopsis.info/; see Supplementary Data Tables S3 and S4, available online, for more detailed information on these arrays). For rice, ID and probe sequences were obtained at the ‘Rice Multi-platform Microarrary Search’ (http://www.ricearray.org/matrix.search.shtml) and at the ‘Plant Expression Database’ (PLEXdb) (http://www.plexdb.org/modules/PD_probeset/annotation.php?genechip=Rice57k), respectively. Rice microarrays corresponding to 48 GeneChips described by Jain et al. (2007) (Supplementary Data Table S5) were obtained at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) (accession no. GPL2025). The poplar ID and probe sequences were from the GPL2618 platform at the Gene Expression Omnibus. Poplar expression values were obtained from 24 GeneChips (accession no. GSE6422). The Pearson correlation coefficient, r, for arabidopsis ICK/KRP genes was calculated using the Expression Angler tool at the Botany Array Resource ‘BAR’ (http://www.bar.utoronto.ca/ntools/cgi-bin/ntools_expression_angler.cgi) (Toufighi et al., 2005) using the Atgenexpress tissue set with r-value cutoff range from 1·0 to 0·7. All microarray data were presented (heat maps) by using the Genesis software (Sturn et al., 2002).
RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) for most tissues, except germinating seeds for which total RNA was isolated as described by Vicient and Delseny (1999). First-strand cDNA synthesis was performed using 5 µg of total RNA and the Superscript RT-PCR III System kit (Invitrogen). RT-PCR was performed as previously described (Wen et al., 2008). cDNA inputs were standardized using a reference gene, At4g33380 (accession NM_119492), which is one of the most uniformly expressed genes (Czechowski et al., 2005). Gene-specific primers used are listed in Supplementary Data Table S6.
RESULTS
Analysis of ICK/KRP family in arabidopsis, rice and poplar
To obtain ICK/KRP-related sequences from different plant species, seven arabidopsis ICK/KRP sequences were used to search the databases of poplar (Populus trichocarpa) and two rice (Oryza sativa L.) subspecies (japonica and indica) (see ‘Materials and Methods’). Blast searches of the poplar genome database (Poplar v 1·0: http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) yielded seven significant matches with arabidopsis ICK/KRPs. Nomenclature of the poplar ICK/KRPs follows the phylogenetic relationship with arabidopsis ICK/KRPs. For indica rice (O. sativa subspecies indica), seven sequences related to arabidopsis ICK/KRPs were identified from the BGI rice genome database (http://rice.genomics.org.cn/rice/link/cite.jsp) (Zhao et al., 2004). For japonica rice (O. sativa subspecies japonica), Barroco et al. (2006) identified five genes designated as Orysa;KRP1 to Orysa;KRP5 by screening the database with the ‘GRYEW and KYNFD’ conserved sequences. However, Guo et al. (2007) reported two additional genes designated as Orysa;KRP6 (Os09g28580) and Orysa;KRP7 (Os01g37740). Orysa;KRP6 encodes a protein with the conserved CID region, while Orysa;KRP7 encodes a protein that lacks the CID, but shows sequence similarities to the Orysa;KRP4 and Orysa;KRP5 at the N-terminal region. Our analysis indicates that the protein encoded by Orysa;KRP7 is unlikely to be an alternative-splicing variant of a longer mRNA (which includes the CID region) as no match with the conserved CID region could be found in the intergenic region for this gene (data not shown). As the conserved CID region is the hallmark for the ICK/KRP family of plant CDK inhibitors (Wang et al., 1998; Lui et al., 2000; Jasinski et al., 2002b; Zhou et al., 2003b; Schnittger et al., 2003), we consider the presence of the CID region necessary for a sequence to be included in this family. Therefore, we suggest that there are six ICK/KRP genes in the rice genome (Supplementary Data Table S7, available online). The six rice ICK/KRP proteins from the two rice subspecies match each other well. To follow the initial name given to this gene family and also to distinguish the sequences from the japonica subspecies, ICK/KRP genes from Oryza sativa subspecies indica are referred to as ICKs.
ICK/KRP families in these model plants have similar numbers (six to seven). Excluding the small Orysa;KRP6, arabidopsis, poplar and rice ICK/KRP genes encode polypeptides ranging from 189 (Arath;ICK3/KRP5) to 289 (Arath;ICK7/KRP4) amino acid residues, with a molecular mass from 21·3 to 32·1 kDa (Supplementary Data Table S7).
ICK/KRP genes from other plant species
Using two criteria, a blast search ‘p’ value (the probability of such a match between random sequences) lower than 1e-8 and presence of the CID region, 133 non-redundant sequences related to ICK/KRPs were found from 63 plant species. Thirty-two of the sequences were not used in the final data set because they were not the complete coding sequences. Eight full-length ESTs were found to be mis-annotated (e.g. with incorrect start codon predictions, splicing errors and missing or additional exons) after comparing them with genomic DNA sequences. They were manually corrected (Supplementary Data Table S2). A total of 101 full-length sequences (from 40 species) were considered as members of the ICK/KRP family and used for phylogenetic analysis. Interestingly, ICK/KRP-related sequences were found only in vascular plants (two gymnosperm and 38 angiosperm species), while no sequences were found from algae (Chlamydomonas reinhardtii, Volvox carteri and Ostereococcus tauri) or bryophyte (Physcomitrella patens) genome databases.
Phylogenetic analysis of the ICK/KRP family
Based on sequence alignment and construction of a neighbour-joining phylogenetic tree, the putative proteins encoded by the 101 unique sequences were classified into three major classes (Fig. 1). The A-class contains 38 sequences only from dicotyledonous plants, while the B-class contains 21 sequences only from monocotyledonous plants. The C-class has 27 sequences from dicotyledons, 13 from monocotyledons and two from gymnosperms (Fig. 1). The A-class (dicot-specific) is represented by Arath;ICK1/KRP1, Arath;ICK2/KRP2, Arath;ICK4/KRP6 and Arath;ICK5/KRP7 and related sequences from poplar (Poptr;ICK1-2 and Poptr;ICK4-5); and the B-class (monocot-specific) is represented by Orysa;KRP1, Orysa;KRP2, Orysa;KRP3 and Orysa;KRP6. The C-class contains the remaining arabidopsis, poplar and rice ICK/KRPs. The tree topology is predominantly influenced by the core CID sequence as little difference was observed in several trees constructed using partial sequences containing the CID (data not shown). For instance, Arath;ICK4/KRP6 is related to the Arath;ICK5/KRP7, Poptr;ICK4 and Poptr;ICK5, but is grouped in the clade containing ICK1s and ICK2s from arabidopsis and poplar. When the E156, L158, L162, F178 and K191 residues in the core CID of Arath;ICK4/KRP6 are manually substituted with the conserved residues (D, F, A, Y and Q, respectively) derived from Ara;KRP7, Poptr;ICK4 and Poptr;ICK5, Arath;ICK4/KRP6 is re-grouped with them (data not shown). Using the consensus criteria of Joshi et al. (1997) and based on the alignment of the 93 sequences, a single conserved signature sequence, E-i-E-E-F-F-A-a-A-E-k-x-q-Q-k-r-F-x-E-K-Y-N-F-D-x-V-x-D-x-P-L-x-G-R-Y-E-W-V-r-l (see explanation in Supplementary Data Fig. S1, available online), is identified in the conserved CID. This signature can thus be used to define new ICK/KRP members.
Fig. 1.
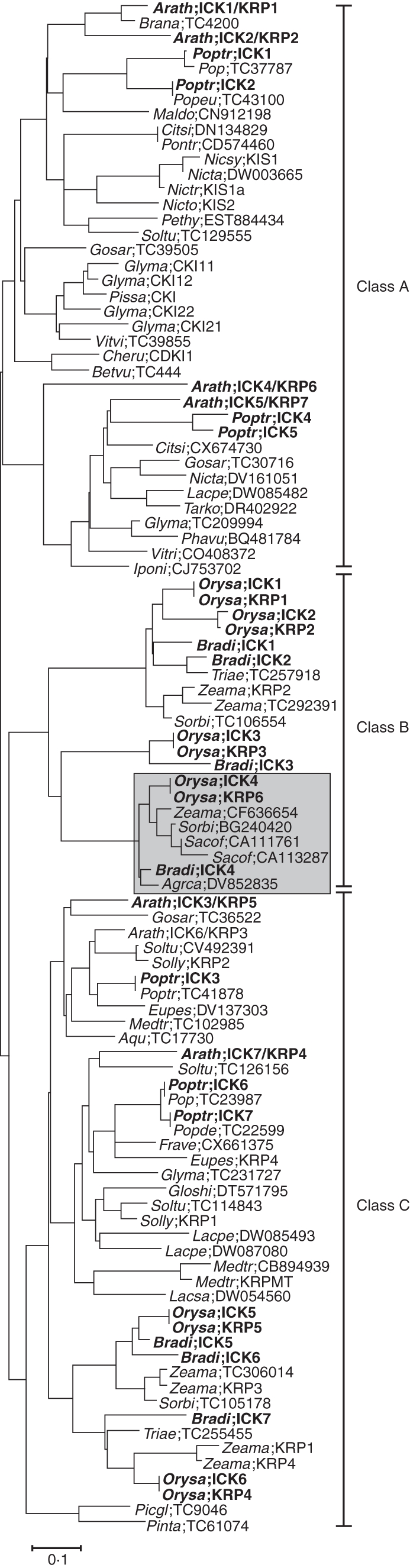
Phylogenetic tree of the plant ICK/KRP family. The tree was generated by aligning 93 ICK/KRP amino acid sequences by neighbour-joining distance, using clustalw and mega 3·1. ICK/KRPs from arabidopsis, poplar and rice are highlighted (bold). A sub-group of atypical ICK/KRPs (about 90 amino acid residues) is indicated by the shaded box. Classes are indicated to the right. The bar represents 0·1 amino acid substitutions per site in the primary structure. See Supplementary Data Table S2 (available online) for abbreviations.
Sub-groups in each class can also be distinguished, due to the use of a relatively large number of sequences and presumably also the quality of these sequences. Each class contains at least two sub-classes (clades) with sequences from diverse species, showing that they are more related to each other than to other ICK/KRP members of the same plant species. For instance, the A-class contains members from Leguminosae (soya, alfalfa, bean and pea), Solanaceae (tobacco, potato and petunia) and Crassulaceae (arabidopsis) as well as sequences from woody plants (poplar, lemon and ipomoea). Within the B-class, there is a group of atypical ICK/KRP sequences with about 90 amino acids in length from maize, sugar cane, barley and bent grass, which therefore constitute a new subgroup of ICK/KRPs exclusive to monocotyledons (Fig. 1). Interestingly, the more ancient ICK/KRP members from two conifer species, Pinus taeda (Pinta;TC61074) and Picea glabra (Picgl;TC9046), are classified within the C-class. Proteins of these two sequences (295 and 283 amino acids for Picgl;TC9046 and Pinta;TC61074, respectively) are longer than the other ICK/KRPs. The observation that the C-class has sequences from more ancient conifer species as well as monocotyledonous and dicotyledonous species suggests that the C-class might have existed first, before the emergence of the other two classes.
Genomic structure of ICK/KRP genes of arabidopsis, poplar and rice
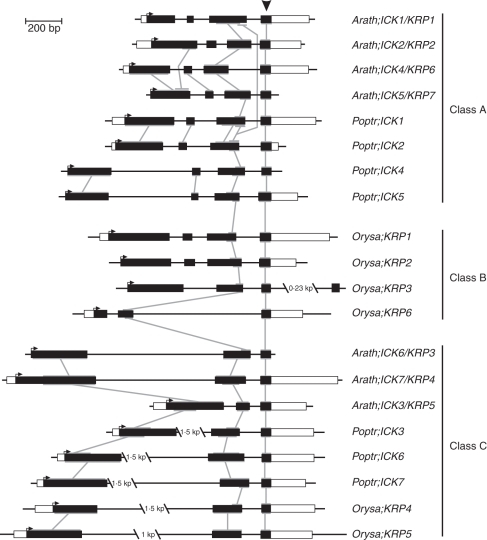
The exons and introns in arabidopsis, poplar and rice ICK/KRP genes were analysed by comparing the cDNA sequences with the genomic sequences. Overall, ICK/KRP genes are divided into two groups, one group with three exons (Arath;ICK3,6,7, Poptr;ICK3,6,7 and Orysa;KRP4-6) and the other group with four exons (Arath;ICK1,2,4,5, Poptr;ICK1,2,4,5 and Orysa;KRP1-3; Fig. 2). The first exon is generally larger than the last one in all ICK/KRPs. The second exon in the four-exon group is generally the shortest among all exons (except the one in Orysa;KRP3). The coding region corresponding to the CID is split into the last two exons in all sequences analysed except Orysa;KRP3, which has an unusual fourth exon (Fig. 2). The last exon is the most conserved in terms of size and sequence similarity and encodes the C-terminal portion of the CID, while the second last exon encodes the N-terminal portion of the CID. The exon–intron organization shows sequence relatedness that is consistent with the phylogenetic tree based on sequence analysis (Fig. 2). For instance, the exon and intron organizations of the arabidopsis and poplar A-class ICK/KRP genes are more similar among themselves than to the genes in the other two classes. In the C-class, the last exon is similar for eight ICK/KRPs from arabidopsis, poplar and rice. Interestingly, exon 2 of Arath;ICK3/KRP5 is smaller than other members in the C-class, indicating a possible shortening of this exon in Arath;ICK3/KRP5 during evolution. Comparison of introns revealed that the introns of rice ICK/KRPs in general are longer than arabidopsis ICK/KRPs whereas the exon lengths are similar (Fig. 2).
Fig. 2.
Exon–intron organization of the ICK/KRP genes from arabidopsis, rice and poplar. Exons and introns are represented by boxes and black lines, respectively. White boxes, when full-length cDNA sequences are known, correspond to untranslated regions. The genes are drawn to scale and are aligned by the most conserved exon in all genes (indicated by an arrowhead). Horizontal arrows indicate start codons. Interconnected grey lines correspond to related regions in exons. Sequences are organized according to the phylogenetic classes.
Duplication of ICK/KRP genes in arabidopsis, poplar and rice genomes
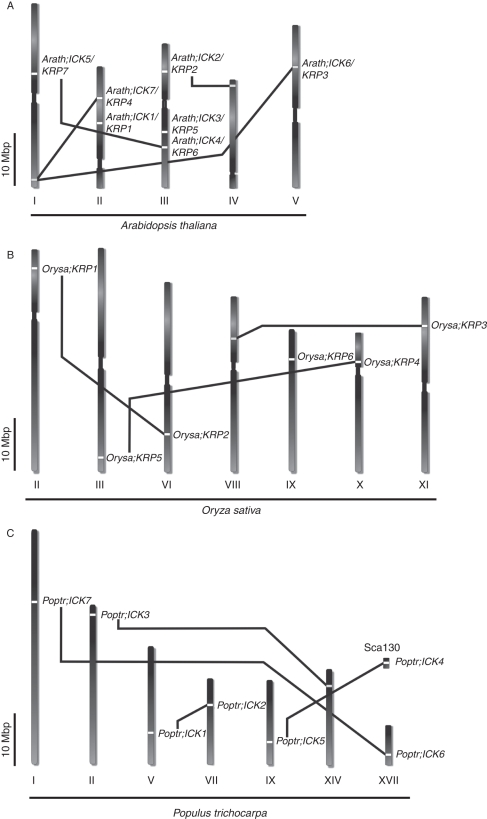
The phylogenetic and gene organization analyses on ICK/KRPs suggest that consecutive gene duplication would explain a progressive expansion of the family. To test this hypothesis we mapped the chromosome positions of arabidopsis, poplar and rice ICK/KRP genes, and traced their potential duplicated blocks using available bioinformatics tools (Blanc et al., 2003; Guyot and Keller, 2004; Tuskan et al., 2006) to determine possible gene duplications. Our analysis indicates that five ICK/KRP genes from both arabidopsis and rice and the seven from poplar are mapped to duplicated blocks (Fig. 3). In arabidopsis, Arath;ICK4/KRP6 and Arath;ICK5/KRP7 co-align in duplicated blocks on chromosomes III and I, while Arath;ICK2/KRP2, Arath;ICK6/KRP3 and Arath;ICK7/KRP4 are mapped to duplicated blocks, which do not co-align with any ICK/KRP-containing blocks (Fig. 3A). Furthermore, loci of Arath;ICK6/KRP3 (chromosome V) and Arath;ICK7/KRP4 (chromosome II) co-align with the same duplicated region free of ICK/KRP on chromosome I (Fig. 3A), suggesting that gene deletion or translocation might have occurred.
Fig. 3.
Genome localization of ICK/KRP genes in duplicated blocks. Chromosomal distribution of ICK/KRP genes in: (A) arabidopsis, (B) rice and (C) poplar. Chromosomes are presented as vertical bars and identified using roman numerals. In poplar, one unassembled scaffold, Sca130, contains the Poptr;ICK4 gene. Duplicated blocks related to ICK/KRPs are linked by black lines. A scale bar is included in each genome.
In contrast to arabidopsis ICK/KRPs, rice and poplar ICK/KRPs have more duplicated pairs (Fig. 3B, C). In rice, Guyot and Keller (2004) reported 52 duplicated blocks that cover 65·7 % of the genome. Five of the six rice ICK/KRP genes are in duplicated blocks. Orysa;KRP1 and Orysa;KRP2 are in a pair of duplicated blocks on chromosomes II and VI, while Orysa;KRP4 and Orysa;KRP5 are in another pair on chromosomes X and III. Orysa;KRP3 is located on chromosome XI in a duplicated segment that co-aligns with another block free of ICK/KRP on chromosome VIII. In poplar, Poptr;ICK1-2, Poptr;ICK4-5 and Poptr;ICK6-7 pairs co-align to common duplicated blocks, although the exact location of Poptr;ICK4 is not known as it belongs to a scaffold fragment that has not been assigned to a chromosome (Tuskan et al., 2006). The rice Orysa;KRP3 is not located in a known duplicated block, but sequence-wise, it is similar to Orysa;KRP4 and Orysa;KRP5.
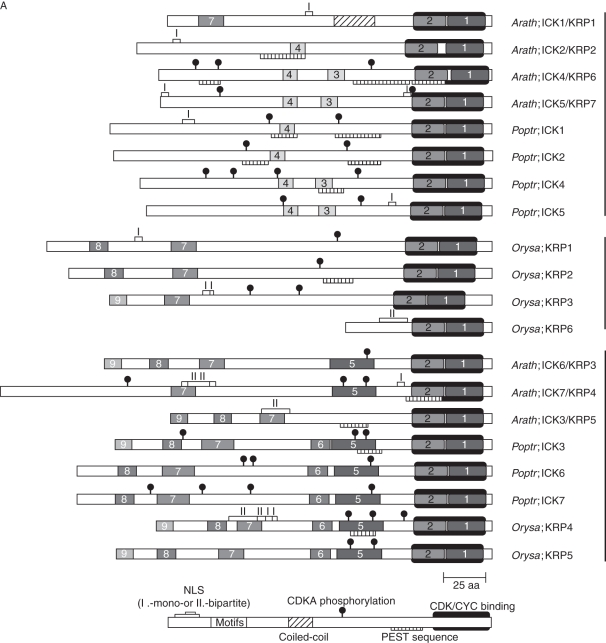
Conserved domains and motifs in ICK/KRP proteins of arabidopsis, poplar and rice
Using the protein sequences of ICK/KRPs from the model plants, we searched for the presence of putative functional domains and motifs: the conserved CID, nuclear localization signal (NLS: monopartite, type I; and bipartite, type II), protein degradation signal (PEST), coiled-coil domain and CDK phosphorylation site. CID is uniformly present at the C terminus in all ICK/KRP proteins (Fig. 4A). Our analysis shows a moderate frequency of putative NLSs (12 of 20 sequences in Fig. 4A). Although a putative NLS is not detected in Arath;ICK6/KRP3 or Arath;ICK4/KRP6, experimentally all seven arabidopsis, two tobacco and two tomato ICK/KRPs are shown to be localized in the nucleus (Jasinski et al., 2002b, 2003; Zhou et al., 2003b; Bird et al., 2007; Nafati et al., 2010). Ten of the 20 proteins are found to contain putative PEST sequences (Fig. 4A and Supplementary Data Table S8, available online), which may be involved in protein degradation (Rogers et al., 1986). Rice ICK/KRP appears to have a lower frequency with only two members containing the PEST domain (Orysa;KRP2 and Orysa;KRP4). Potential CDK phosphorylation sites were found in 16 of 20 ICK/KRPs (Fig. 4A and Supplementary Data Table S8), indicating a possible high prevalence of phosphorylation in this family of proteins. Experimentally, in vitro phosphorylation has been demonstrated for Arath;ICK2/KRP2 (Verkest et al., 2005) and alfalfa KRPMt (Pettko-Szandtner et al., 2006). In contrast to the frequent occurrence of putative NLSs, PEST and CDK phosphorylation sites, a coiled-coil domain, often involved in protein–protein interactions (Lupas, 1996), is only present in Arath;ICK1/KRP1.
Fig. 4.
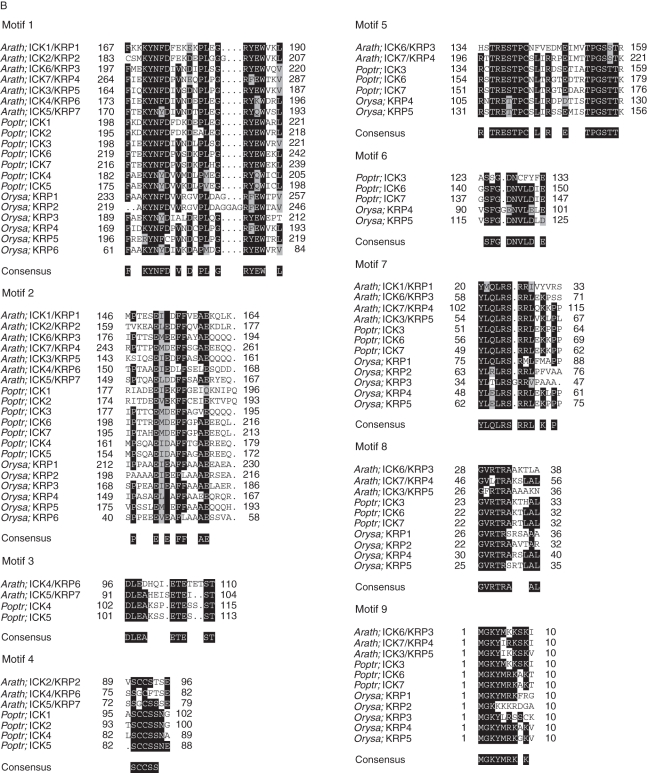
Putative functional and conserved motifs in plant ICK/KRP proteins. (A) Putative functional motifs are represented by symbols, and conserved motifs are indicated in boxes (with explanations given). Sequences are organized according to the phylogenetic relationship. (B) Sequences of conserved motifs in ICK/KRP proteins of arabidopsis, rice and poplar. Amino acid residues with at least 40 % identity are shaded.
Identification of conserved sequence motifs can help functional studies of proteins. In our analysis, nine consensus sequence motifs were identified by the MEME/MAST program (Bailey and Elkan, 1994; Bailey and Gribskov, 1998) (http://meme.nbcr.net/meme4_4_0/cgi-bin/meme.cgi) in the ICK/KRP sequences from the three model plants (Fig. 4). To identify conserved motifs, we excluded regions which are homologous only between recently duplicated genes (i.e. pairs of arabidopsis ICK4/KRP6 and ICK5/KRP7, and poplar ICK6/ICK7, respectively). The distribution of the motifs follows the phylogenetic classification. Motifs 1 and 2 are present in all ICK/KRP proteins; motifs 3–4 and motifs 5–6 are only present, respectively, in members of the A- and B-class, while motifs 7, 8 and 9 are present in members of both the B- and C-class (Fig. 4A). Conserved motifs were reported previously among seven arabidopsis and one Chenopodium ICK/KRP (De Veylder et al., 2001). Motifs 1–2, 3, 4, 5 and 6 described by De Veylder et al. (2001) correspond to motifs 1, 2, 7, 8 and 9 described here (Fig. 4B). During submission of this manuscript, Nafati et al. (2010) reported six motifs from analysis of ICK/KRP sequences, with motifs 1, 2, 3, 4, 5 and 6 corresponding to motifs 1, 2, 7, 5, 8 and 9 described here, respectively.
The functions of a few major sequence motifs are known. Motif 1 in ICK1 is required for interacting with CDKA (Wang et al., 1998) and for inhibiting CDK activity in plants (Zhou et al., 2003b). The functional importance of this motif for CDK interaction and inhibition may explain why this motif is the most conserved among all ICK/KRP proteins. Motif 2 is in a region suggested to be important for the interaction with CYCD3 (Wang et al., 1998). The recently described SIM/EL2 family of CKIs also contains a shorter version of this motif, and its function in interacting with D-type cyclins has been established (Peres et al., 2007). Interestingly, motif 2 of the tomato SlKRP1 was identified to be responsible for the interaction with CSN5A, a subunit of the COP9 signalosome (Nafati et al., 2010). Motif 7 of the arabidopsis ICK/KRP proteins confers nuclear localization of the fusion proteins with green fluorescent protein (GFP) (Zhou et al., 2006; Bird et al., 2007). This motif also specifies a punctate sub-nuclear localization pattern (Jakoby et al., 2006; Zhou et al., 2006). Most recently, Nafati et al. (2010) showed that another motif in the tomato SlKRP1 (corresponding to motif 8 here) functions redundantly with the motif corresponding to motif 7 here in conferring the punctate nuclear localization. In addition, motif 5 contains at least one putative CDK phosphorylation site in all sequences containing this motif, although phosphorylation with any of the sites is yet to be shown. The functional significance of motifs 3–6 and 9 (Fig. 4B) remains unknown.
Expression analysis of ICK/KRP genes
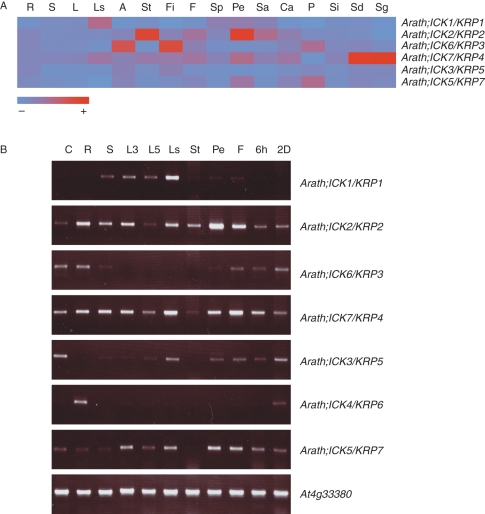
We analysed the expression of arabidopsis ICK/KRP genes for 73 different tissues/developmental conditions using microarray data from the NASC's International Affymetrix Service (Supplementary Data Tables S3 and S4). Expression of Arath;ICK4/KRP6 was not analysed as it was absent in the ATH1 GeneChip used. In general, arabidopsis ICK/KRPs were expressed at low levels but showed preferential expression in specific tissue/developmental conditions (Fig. 5A). To examine the expression patterns of ICK/KRP genes further, we also performed RT-PCR for the seven arabidopsis ICK/KRPs using selected tissues, mostly chosen to be similar to those used for the microarray experiments (Fig. 5B). For ease of comparison, the microarray and RT-PCR results are described together.
Fig. 5.
Expression of arabidopsis ICK/KRP genes. (A) ATH Affymetrix expression average values of six arabidopsis ICK/KRP genes from 16 tissues. Codes are: R, roots; S, seedling; L, leaf; Ls, senescing leaf; A, shoot apex; St, stem; Fi, shoot apices after plants were induced to flower (see Schmid et al., 2003); F, flower; Sp, sepal; Pe, petal; Sa, stamen; Ca, carpel; P, pollen; Si, silique; Sd, developing seed in silique; Sg, germinating seed. Bottom bar indicates expression level in mean-normalized values from null (light blue) to high (dark red). (B) RT-PCR expression of the seven arabidopsis ICK/KRP genes in different tissues. Codes are: C, cell suspension; R, root from 13-d seedlings; S, shoot from 13-d seedlings; 3L, leaf from 3-week plants; 5L, leaf from 5-week plants; Ls, senescing leaf from 5-week plants; St, stem from 5-week plants; Pe, petal; F, flower from 5-week plants; 6h and 2d, seeds after 6 h and 2 d of stratification, respectively. Expression of the At4g33380 gene (Czechowski et al., 2005) was used as input control. Pictures were taken under identical conditions (acquisition time and loading volumes) using a UV transilluminator (BioDoc-It System, UVP, www.uvp.com).
Similar patterns of expression were observed from both microarray and RT-PCR data for several tissues at least in terms of the prominent ICK/KRP members expressed, including: Arath;ICK1/KRP1 and Arath;ICK7/KRP4 in old and senescing leaves; Arath;ICK2/KRP2 in stems; Arath;ICK2/KRP2, Arath;ICK7/KRP4 and Arath;ICK5/KRP7 in flowers and petals (with Arath;ICK2/KRP2 most strongly expressed in petals); and Arath;ICK7/KRP4 in germinating seeds (Fig. 5A; compare with Fig. 5B). The expression of Arath;ICK1/KRP1 in old leaves and Arath;ICK2/KRP2 in stem and floral tissues has been reported previously (Wang et al., 1998; Lui et al., 2000; De Veylder et al., 2001). There are some differences between the microarray and RT-PCR results; RT-PCR results show relatively higher levels of Arath;ICK2/KRP2 and Arath;ICK7/KRP4 in roots and shoots (Fig. 5B), while there was no significant expression for either of the two genes in these tissues by microarray data (Fig. 5A). Similarly, clear increases in the levels of expression were observed by RT-PCR for Arath;ICK5/KRP7 in senescing leaves and flowers (Fig. 5B), but not by microarray data. One reason for the differences may be that RT-PCR is more sensitive to low levels of transcripts due to the power of PCR amplification. Also, the signal intensities in microarray analysis are not a measure of the absolute transcript abundance (Vigneault et al., 2007). In addition, differences in tissues and experimental conditions used for the microarray and RT-PCR analyses could contribute to the variation. Given these differences, it is unlikely that the patterns of expression based on microarray analysis would match exactly the patterns of expression based on RT-PCR analysis. We focus our attention on the relatively strong and clear patterns of expression.
For several tissue samples, only data from microarray or RT-PCR are available. Microarray data show that in the shoot apex, the expression of Arath;ICK6/KRP3 was strongest (Fig. 5A). By contrast, RT-PCR results show that in germinating seeds 2 d after imbibition, all ICK/KRPs except Arath;ICK1/KRP1 were weakly and almost evenly expressed (Fig. 5B). The RT-PCR analysis showed that Arath;ICK4/KRP6 (not present in the ATH1 chip) had a higher expression level in roots among different tissue samples used in the analysis (Fig. 5B).
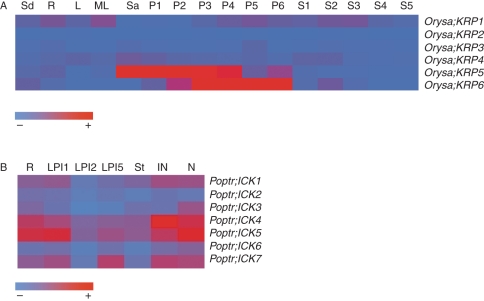
Expression of rice ICK/KRPs was analysed using the microarray data reported by Jain et al. (2007) (Supplementary Data Table S5). Little expression was observed for Orysa;KRP2 and Ory;KRP3 (Fig. 6A). Interestingly, Orysa;KRP1 was expressed at a relatively high level in mature leaf, and in this regard resembles Arath;ICK1/KRP1, suggesting that both may have a role in leaf differentiation or leaf senescence (Fig. 6A). Orysa;KRP1 is also the predominant member of rice ICK/KRPs to be expressed during seed germination (Fig. 6A) (Barroco et al., 2006). Like arabidopsis ICK/KRPs, some rice members are associated with proliferating tissues. For instance, Orysa;KRP5 and, to a lesser extent, Ory;KRP4 were expressed in the shoot apex while Orysa;KRP5 and Ory;KRP6 were expressed in early stages of panicle development (Fig. 6A).
Fig. 6.
Expression of rice and poplar ICK/KRP genes. GeneChip expression values for rice and poplar ICK/KRP genes in different tissues and at different developmental stages are used. (A) Rice. Codes are: Sd, seedling; R, root; L, young leaf; ML, mature leaf; Sa, shoot apex; inflorescence stages P1–P6 (P1, up to 3 cm; P2, 3–5 cm; P3, 5–10 cm; P4, 10–15 cm; P5, 15–22 cm; P6, 22–30 cm); seed stages S1–S5 [S1, 0–2 d after pollination (dap); S2, 3–4 dap; S3, 5–10 dap; S4, 11–20 dap; S5, 21–29 dap]. (B) Poplar. Codes are: R, root; LPI1, leaf plastochron index 1; LPI2, leaf plastochron index 2; LPI5, leaf plastochron index 5; St, stem; IN, internode; N, node. The bar at the bottom of each heat map indicates the absolute expression values from null (light blue) to high (dark red).
We analysed the microarray data of poplar (Ramírez-Carvajal et al., 2008) and the expression patterns are shown in Fig. 6B. Several observations are worth noting. Poptr;ICK4 and Poptr;ICK5 are the highest ICK/KRP expressed in poplar, followed by Poptr;ICK1 and Poptr;ICK7. More poplar ICK/KRP members are expressed in nodes than other tissues (Fig. 6B). In general, no ICK/KRP gene from poplar seems to be significantly expressed in stems.
DISCUSSION
Classification of plant ICK/KRP proteins into three major classes
ICK/KRP plant CKIs are considered a family of core cell cycle regulators in plants (Verkest et al., 2005; Wang et al., 2006, 2007). It is important to understand how widespread they are among diverse species. In the present study, ICK/KRP-related sequences were found in more than 60 vascular plant species by searching all the common public EST and genomic databases. Interestingly, ICK/KRP-related sequences were not found in algae and bryophytes. Searching the databases of several model animals and human using arabidopsis ICK sequences, we were able to identify p27Kip1 proteins from mouse and human but failed to identify any sequence from Drosophila or Caenorhabditis elegans (data not shown), suggesting that the ICK/KRP and KIP sequences became more similar among species which appeared later in the plant and animal lineages. These observations suggest that the ICK/KRP family in plants and the KIP/CIP family in animals might have evolved independently. The fact that the conserved sequences in plant ICK/KIP proteins and animal KIP proteins are structured differently (Wang et al., 1997) is also consistent with this suggestion. Thus, plant ICK/KRP genes might have appeared either after plants colonized continental lands or after vascular plants emerged. The emergence of ICK/KRP genes could serve the needs of more complex multi-tissue land plants. In this regard, the name ICK may be better suited to describe this family of genes.
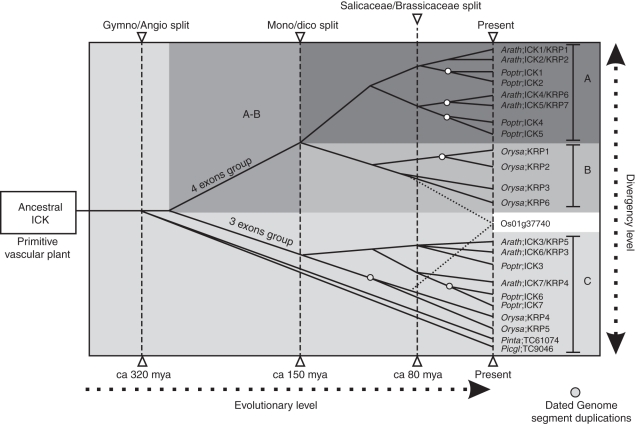
Based on the phylogenetic analysis of 101 unique ICK/KRP sequences, ICK/KRP proteins are classified into three classes. We were able to construct a clear phylogenetic tree for this family of proteins using a relatively large set of full-length protein sequences. Based on the distribution of sequences and sequence similarity, we propose a model to explain the evolution of the ICK/KRP family (Fig. 7). Early in the evolution of the family, two branches were formed consisting of the C-class and the A/B-classes. As the C-class contains members from dicotyledonous, monocotyledonous and gymnosperm plants while the A/B-class contains only sequences from dicotyledonous or monocotyledonous species, we propose that the C-class was present before the emergence of angiosperms. The A- and B-classes would have emerged from the C-class or from a common ancestral class (Fig. 7). During speciation, the C-class might have diverged but retained sufficient conservation to allow it to be distinguished based on the sequence phylogenetic analysis (lower part of Fig. 7). It appears that the A- and B-classes split prior to the speciation of dicotyledons and monocotyledons so that each of the two classes is now either monocot- or dicot-specific (Fig. 7). The gene number of the ICK/KRP family increased through segmental duplication in the plant genomes. We suggest the following duplication events. The first duplication probably occurred before or around the time of the angiosperm/gymnosperm split (320 Mya; Bowe et al., 2000), therefore dividing the family into A/B- and C-classes. A second duplication could have occurred before the monocotyledons and dicotyledons separated (150 Mya; Chaw et al., 2004) resulting in distinct A- and B-classes. The last and most evident duplication occurred during the specialization of monocotyledons and dicotyledons, probably even after the arabidopsis/poplar split (80 Mya; Wikstrom et al., 2001), resulting in the sub-groups observed in each class (i.e. the pairs Arath;ICK4/KRP6 and Arath;ICK5/KRP7, Poptr;ICK1/Poptr;ICK2 and Poptr;ICK4/Poptr;ICK5, and Orysa;KRP1/Orysa;KRP2 and Orysa;KRP4/Orysa;KRP5; Fig. 3).
Fig. 7.
A proposed model for the evolution of the ICK/KRP family. Based on sequence similarity and exon–intron organization, ICK/KRP genes are grouped into three major classes. While genes in the C-class have three exons, genes in the A- and B-classes have four exons. The C-class contains members from dicotyledons, monocotyledons and gymnosperms, suggesting a common ancestral class before the emergence of angiosperm plants. The A- and B- classes have only members from dicotyledons and monocotyledons, respectively, suggesting their split before the speciation of these two groups of plants. The number of family members increased through more recent segmental duplications in the plant genomes. Circles describe dated duplication events according to the literature. Potential origins for the Os01g37740 are marked with dotted lines. See text for more details.
The exon–intron organization of ICK/KRP genes supports the phylogenetic classification based on sequence analysis. Analysis of arabidopsis and rice genes shows that genes in the A- and B-classes (except Orysa;KRP6) have four exons, while genes in the C-class have only three (Fig. 2). It is tempting to speculate that one exon from a primitive gene in the C-class underwent a split, becoming the ancestor of the A- and B-classes. Moreover, within each class, members have similar exon–intron organization, probably due to more recent segment duplications giving rise to paralogues. To validate our phylogenetic analysis, we produced several trees with different numbers of sequences. When more than 80 sequences were used, the phylogenetic tree showed the same three major classes (data not shown). Therefore, the general phylogenetic topology consisting of the three classes will probably remain similar with additional sequences.
The ICK/KRP genes in different plants have been named by different laboratories chronologically. The phylogenetic relationships for ICK/KRP genes from different plants were not clear due to the sequence diversity and incomplete genome sequence information, making it difficult to compare genes from different plants. The present analysis has resulted in the clearest phylogenetic classification of ICK/KRP genes from different species so far. This classification makes it possible to number genes from other plants which have not been named, based on the alignment with the corresponding genes of model species such as arabidopsis and rice, thus avoiding further confusion by numbering genes in different species independently. In this regard, if the ICK number system is used, it should be easier to relate the gene numbers with the phylogenetic classification, facilitating future functional comparisons. Therefore, we have proposed ICK designations for the model species poplar, indica rice and Brachypodium distachyon. For arabidopsis and poplar, ICK1/2/4/5 belong to Class A and ICK6/7/3 to Class C (Fig. 1). For indica rice, ICK1–4 belong to Class B and ICK5 and 6 to Class C. Similarly, for Brachypodium distachyon ICK1–4 belong to Class B and ICK5–7 to Class C.
Analysis of conserved motifs and putative functional sites
Identification of conserved sequences in a family of proteins provides useful hints for functional characterization and for determining functional differences among different members. Analysis of 20 ICK/KRP protein sequences representing three model plants (arabidopsis, rice and poplar) identified nine conserved motifs (Fig. 4). The presence of the conserved motifs conforms in general to the phylogenetic classification. Rice members of the B-class contain motifs that are more common with motifs in members of the C-class (motifs 7–9), but not in members of the A-class. Other motifs seem to be restricted to a particular lineage. As the conserved motifs are shared by members from different plant species, these motifs probably have functions associated with them.
The best conserved sequence among ICK/KRP proteins is motif 1. This is not surprising given its role in interacting with CDK and inhibiting CDK activity (Wang et al., 1998; Zhou et al., 2003b). Despite the relatively high level of conservation for this motif, arabidopsis ICK/KRP proteins show some differences in their ability to interact with Arath;CDKA (Zhou et al., 2002). It is not known whether the differences are due to the variation in the consensus sequence or in the surrounding sequences. All ICK/KRP proteins studied so far can interact with D-type cyclins (Wang et al., 1998; Fountain et al., 1999; Lui et al., 2000; De Veylder et al., 2001; Zhou et al., 2002, 2003a; Jasinski et al., 2002a, b, 2003; Schnittger et al., 2003; Pettko-Szandtner et al., 2006; Bisbis et al., 2006; Jakoby et al., 2006). Motif 2 resides in the cyclin-binding domain of ICK1 (Wang et al., 1998), and the role of this motif in the interaction with a D-type cyclin has been confirmed with a different CDK inhibitor, rice EL2 (Peres et al., 2007). Furthermore, reports have shown that ICK/KRPs can also interact with CDK/cyclin complexes other than the expected CDKA/CYCD complexes, including CDKB/CYCD2 (Nakai et al., 2006), alfalfa CDKB2;1 complex (Pettko-Szandtner et al., 2006) and maize CYCA complexes (Coelho et al., 2005). A clear phylogenetic tree is helpful to the analysis of differences among members in their interactions with CDKs and cyclins.
Motif 7 in arabidopsis ICK/KRPs confers nuclear localization of the fusion protein with GFP, as well as a punctate sub-nuclear pattern (Jakoby et al., 2006; Zhou et al., 2006; Bird et al., 2007). No experimental information is available on the functions of other motifs. The observation that motifs 3 and 4 are only present in A-class proteins while motifs 5 and 6 are not present in B-class proteins (Fig. 3) is one indication that there is probably functional differentiation among the three classes of ICK/KRP proteins.
In addition to the conserved sequence motifs, other putative functional motifs are also identified. Analysis shows that the majority of ICK/KRP proteins contain either a monopartite or a bipartitie NLS (Fig. 4A). Studies have shown that all plant ICK/KRP proteins analysed experimentally so far are localized in the nucleus (Jasinski et al., 2002a, 2003; Zhou et al., 2003b; Bird et al., 2007; Nafati et al., 2010). For Arath;ICK1/KRP1, three separate sequences in the N-terminal, central and C-terminal regions could confer nuclear localization of GFP fusion proteins (Jakoby et al., 2006; Zhou et al., 2006), even though only one NLS was predicted through bioinformatics analysis. As the almost exclusive nuclear localization of ICK/KRP proteins is unlikely due entirely to ‘piggybacking’ on another protein, it appears that not all nuclear localization sequences have been detected in ICK/KRP proteins by the bioinformatics analysis.
Timely removal of CDK inhibitors is important for normal cell cycle progression (Schnittger et al., 2003). It is known that Arath;ICK1/KRP1 and Arath;ICK2/KRP2 are unstable proteins (Zhou et al., 2003b; Verkest et al., 2005). The removal of the N-terminal region increased the stability of truncated ICK1/KRP1 protein (Zhou et al., 2003b). Recently it was shown that a RING-figure E3 ligase RKP probably targets arabidopsis ICK1/KRP1 and ICK2/KRP2 for degradation (Ren et al., 2008; Lai et al., 2009), while another RING-figure E3 RHF1a/2a was shown to interact and degrade, via proteasome, the arabidopsis ICK4/KRP6 during male gametogenesis (Liu et al., 2008). In addition, an F-Box-like 17 (FBL17), which forms an E3 ubiquitin ligase complex (SCFFBL17) consisting of ASK1, Cullin1 and FBL17, interacts and targets the arabidopsis ICK4/KRP6 and ICK5/KRP7 for degradation (Kim et al., 2008; Gusti et al., 2009). The degradation of arabidopsis ICK4/KRP6 and ICK5/KRP7 appears to be essential for male gametogenesis (Kim et al., 2008; Liu et al., 2008). The tobacco ICK/KRP protein NtKIS1a was found to interact with a homolog of the subunit 5 of COP9 signalosome, a conserved protein complex that functions in the ubiquitin–proteasome pathway (Le Foll et al., 2008). Motif 2 in tomato SlKRP1 has been found to be critical for this interaction, suggesting that motif 2 may also be involved in proteasome-mediated degradation of ICK/KRP proteins (Nafati et al., 2010).
PEST and D-box sequences have been shown to be involved in degrading cyclins (Rogers et al., 1986; Renaudin et al., 1998). In this regard, searching for putative PEST and D-box sequences would help to identify some of the sequences involved in the degradation of ICK/KRP proteins. Putative PEST sequences were detected in half of the ICK/KRP protein sequences tested, and putative D-box sequence only in Cheru;CDKI1 (data not shown). The possible roles of these putative PEST and D-box sequences are subject to experimental analysis.
Reversible protein phosphorylation/dephosphorylation events are essential during cell cycle progression (Nurse, 1994). It has been shown that Arath;ICK2/KRP2 can be phosphorylated by both CDKA;1 and CDKB1;1 and in a mutant with decreased CDKB activity, the level of Arath;ICK2/KRP2 increased, suggesting that phosphorylation of Arath;ICK2/KRP2 by CDKB decreased its stability (Verkest et al., 2005). An ICK/KRP CDK inhibitor KRPMt could be phosphorylated by a recombinant calmodulin-like domain protein kinase in vitro and this phosphorylation enhanced the CDK inhibition activity of KRPMt (Pettko-Szandtner et al., 2006). However, no specific phosphorylation sites have been experimentally identified in any of the ICK/KRP proteins. The present analysis shows that most of the 20 ICK/KRP proteins analysed carry putative phosphorylation sites, implying that many of them may be phosphorylated (Fig. 4A). Future experiments will determine whether some of them are actually phosphorylated in these proteins.
Analysis of gene expression patterns
Expression patterns of ICK/KRPs were analysed to find association of their expression with certain tissues and processes. The expression of arabidopsis ICK/KRP genes has been described in previous studies (Wang et al., 1998; Lui et al., 2000; De Veylder et al., 2001; Ormenese et al., 2004; Menges et al., 2005), but only for limited tissues and conditions. We analysed the expression patterns using a microarray dataset either to extend existing observations or to discover new patterns. Several observations are of interest. First, Arath;ICK1/KRP1 and Arath;ICK7/KRP4 are more strongly expressed in old and particularly senescing leaves. It is now becoming clear that some plant CKIs may function to inhibit cell proliferation during plant development. Arabidopsis Arath;ICK1/KRP1 and Arath;ICK2/KRP2 appear to function in arresting the cell cycle, and thus may promote cell differentiation. This suggestion is supported by the observations from several studies that showed increased expression when the cell cycle is inhibited (Menges et al., 2005), increased expression of Arath;ICK1/KRP1 in old leaves, and induction by abscisic acid and salt (Wang et al., 1998; Ruggiero et al., 2004), and promotion of endoreduplication when weakly overexpressed (Verkest et al., 2005; Weinl et al., 2005). The strong expression of Arath;ICK1/KRP1 in senescing leaves further suggests that Arath;ICK1/KRP1 expression may promote leaf senescence, consistent with the observation that tissue-specific expression of Arath;ICK1/KRP1 in trichomes leads to cell death (Schnittger et al., 2003). Second, both microarray and RT-PCR data show that in arabidopsis stem tissue, Arath;ICK2/KRP2 has a much higher level than other arabidopsis ICK/KRPs (Fig. 5). Lui et al. (2000) observed that the expression of Arath;ICK2/KRP2 is highest in stems among several different tissue types tested. The consistency of data from three different approaches (northern blots, microarray and RT-PCR) suggests that Arath;ICK2/KRP2 probably plays a role in the stem. While the level of Arath;ICK1/KRP1 is relatively high in differentiated leaves, Arath;ICK6/KRP3 is strongly expressed in the shoot apex, suggesting different roles for different members of the family.
In addition to extending previous observations, new patterns of expression for arabidopsis ICK/KRPs are also described from this study, including Arath;ICK2/KRP2, Arath;ICK7/KRP4 and Arath;ICK5/KRP7 in petals and Arath;ICK7/KRP4 in seeds in the early stage of germination. These observations are supported by both microarray and RT-PCR, adding confidence regarding the preferential expression in these tissues. Despite significant progress made in understanding plant CKIs, less is known about their specific functions and physiological effects in plants. The information on preferential expression in specific tissues will be useful to focus our attention to tissues where one or more ICK/KRPs are prominently expressed. Among rice ICK/KRPs, strong expression has been observed for Orysa;KRP5 and Orysa;KRP6 during panicle development and relatively strong expression for Orysa;KRP1 in mature leaves (Fig. 6). However, more data will be required to obtain a clear understanding of the relative levels of rice ICK/KRPs in various tissues under different conditions.
Conclusions
Our understanding of the fundamental aspects of the molecular and biochemical functions and regulation of the ICK/KRP plant CDK inhibitors has increased, particularly on a few prominent members such as Arath;ICK1/KRP1 and Arath;ICK2/KRP2 (Wang et al., 2006). An important challenge is to understand the differences among different members of this gene family and their specific roles in plant tissues and various processes. The in planta functional analysis of ICK/KRP genes has been hampered in the presence of multiple members in this gene family and likely functional redundancy. In addition, unlike other core cell cycle regulators such as CDKs and cyclins that have been classified into subfamilies, there had been no clear phylogenetic classification for the ICK/KRP CKIs. The classification of ICK/KRP genes into three classes as well as the expression patterns identified in this study can guide future efforts to determine the differences among the three sub-classes of these CKIs. Clarification of the relationship among ICK/KRP sequences from different species also allows us to propose a model for further tests regarding how these genes might have evolved. At the molecular level, conserved domains and motifs shared by ICK/KRP protein sequences from diverse plant species suggest that they probably have specific functions in the plant. Although functions for several motifs have been identified, it will be interesting to identify roles for the remaining motifs. Furthermore, some of the expression patterns identified would encourage functional analysis of individual ICK/KRPs in specific tissues or at specific stages of plant development.
SUPPLEMENTARY DATA
ACKNOWLEDGMENTS
We thank Rui Wen for total RNA samples of arabidopsis tissues. L.C.F. and H.W. gratefully acknowledge financial support from the Natural Sciences and Engineering Research Council of Canada.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barroco RM, Peres A, Droual AM, et al. The cyclin-dependent kinase inhibitor Orysa KRP1 plays an important role in seed development of Oryza sativa. Plant Physiology. 2006;142:1053–1064. doi: 10.1104/pp.106.087056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R, Brutlag D, Karp P, Lathrop R, Searls D, editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bemis SM, Torii KU. Autonomy of cell proliferation and developmental programs during Arabidopsis aboveground organ morphogenesis. Developmental Biology. 2007;304:367–381. doi: 10.1016/j.ydbio.2006.12.049. [DOI] [PubMed] [Google Scholar]
- Bird DA, Buruiana MM, Zhou Y, Fowke LC, Wang H. Arabidopsis cyclin-dependent kinase inhibitors are nuclear-localized and show different localization patterns within the nucleoplasm. Plant Cell Reports. 2007;26:861–872. doi: 10.1007/s00299-006-0294-3. [DOI] [PubMed] [Google Scholar]
- Bisbis B, Delmas F, Joubes J, et al. Cyclin-dependent kinase inhibitors regulate the CDK/cyclin complex activities in endoreduplicating cells of developing tomato fruit. Journal of Biological Chemistry. 2006;281:7374–7383. doi: 10.1074/jbc.M506587200. [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Research. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowe LM, Coat G, dePamphilis CW. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales' closest relatives are conifers. Proceedings of the National Academy of Sciences. 2000;97:4092–4097. doi: 10.1073/pnas.97.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. Journal of Molecular Evolution. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AL, Fowke LC, Wang H, John PCL. The effect of ICK1, a plant cyclin-dependent kinase inhibitor, on mitosis in living plant cells. Plant Cell Reports. 2002;20:814–820. [Google Scholar]
- Coelho CM, Dande RA, Sabelli PA, et al. Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiology. 2005;138:2323–2336. doi: 10.1104/pp.105.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1667. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Murray JA. The plant cell cycle. Annual Review of Plant Biology. 2003;54:235–264. doi: 10.1146/annurev.arplant.54.031902.134836. [DOI] [PubMed] [Google Scholar]
- Fountain MD, Renz A, Beck E. Isolation of a cDNA encoding a G1-cyclin-dependent kinase inhibitor (ICDK) from suspension cultured photoautotrophic Chenopodium rubrum L. cells. Plant Physiology. 1999;120:339. [Google Scholar]
- Guo J, Song J, Wang F, Zhang XS. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Molecular Biology. 2007;64:349–360. doi: 10.1007/s11103-007-9154-y. [DOI] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, et al. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One. 2009;4:e4780. doi: 10.1371/journal.pone.0004780. doi:10.1371/journal.pone.0004780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot R, Keller B. Ancestral genome duplication in rice. Genome. 2004;47:610–614. doi: 10.1139/g04-016. [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby MJ, Weinl C, Pusch S, et al. Analysis of the subcellular localization, function and proteolytic control of the Arabidopsis CDK inhibitor ICK1/KRP1. Plant Physiology. 2006;141:1293–1305. doi: 10.1104/pp.106.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Riou-Khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N. The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. Journal of Cell Science. 2002a;115:973–982. doi: 10.1242/jcs.115.5.973. [DOI] [PubMed] [Google Scholar]
- Jasinski S, Perennes C, Bergounioux C, Glab N. Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiology. 2002b;130:1871–1882. doi: 10.1104/pp.008573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Saraiva L, Perennes C, et al. NtKIS2, a novel tobacco cyclin-dependent kinase inhibitor is differentially expressed during the cell cycle and plant development. Plant Physiology and Biochemistry. 2003;41:503–676. [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Molecular Biology. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kang J, Mizukami Y, Wang H, Fowke L, Dengler NG. Modification of cell proliferation patterns alters leaf vein architecture in Arabidopsis thaliana. Planta. 2007;226:1207–1218. doi: 10.1007/s00425-007-0567-2. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, et al. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature. 2008;455:1134–1137. doi: 10.1038/nature07289. [DOI] [PubMed] [Google Scholar]
- Kreegipuu A, Blom N, Brunak S. PhosphoBase, a database of phosphorylation sites: release 2·0. Nucleic Acids Research. 1999;27:237–239. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lai J, Chen H, Teng K, et al. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant Journal. 2009;57:905–917. doi: 10.1111/j.1365-313X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
- Le Foll M, Blanchet S, Millan L, Mathieu C, Bergounioux C, Glab N. The plant CDK inhibitor NtKIS1a interferes with dedifferentiation, is specifically down regulated during development and interacts with a JAB1 homolog. Plant Science. 2008;175:513–523. [Google Scholar]
- Liu J, Zhang Y, Qin G, et al. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell. 2008;20:1538–1554. doi: 10.1105/tpc.108.059741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Wang H, DeLong C, Fowke LC, Crosby WL, Fobert PR. The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant Journal. 2000;21:379–385. doi: 10.1046/j.1365-313x.2000.00688.x. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends in Biochemical Sciences. 1996;21:375–382. [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mendenhall MD. Cyclin-dependent kinase inhibitors of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Current Topics in Microbiology and Immunology. 1998;227:1–24. doi: 10.1007/978-3-642-71941-7_1. [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant Journal. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annual Review of Cell and Developmental Biology. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nafati M, Frangne N, Hernould M, Chevalier C, Gévaudant F. Functional characterization of the tomato cyclin-dependent kinase inhibitor SlKRP1 domains involved in protein–protein interactions. New Phytologist. 2010;188:136–149. doi: 10.1111/j.1469-8137.2010.03364.x. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends in Biochemical Sciences. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nakai T, Kato K, Shinmyo A, Sekine M. Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2-associated kinases, including plant-specific B-type cyclin-dependent kinase. FEBS Letters. 2006;580:336–340. doi: 10.1016/j.febslet.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- Ormenese S, de Almeida Engler J, de Groot R, de Veylder L, Inzé D, Jacqmard A. Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Annals of Botany. 2004;93:575–580. doi: 10.1093/aob/mch077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres A, Churchman ML, Hariharan S, et al. Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. Journal of Biological Chemistry. 2007;282:25588–25596. doi: 10.1074/jbc.M703326200. [DOI] [PubMed] [Google Scholar]
- Pettko-Szandtner A, Meszaros T, Horvath GV, et al. Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant Journal. 2006;46:111–123. doi: 10.1111/j.1365-313X.2006.02677.x. [DOI] [PubMed] [Google Scholar]
- Pines J. Cyclins and cyclin-dependent kinases: a biochemical view. Biochemical Journal. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Carvajal GA, Morse AM, Davis JM. Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytologist. 2008;177:77–89. doi: 10.1111/j.1469-8137.2007.02240.x. [DOI] [PubMed] [Google Scholar]
- Ren H, Santner A, del Pozo JC, Murray JA, Estelle M. Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant Journal. 2008;53:705–716. doi: 10.1111/j.1365-313X.2007.03370.x. [DOI] [PubMed] [Google Scholar]
- Renaudin JP, Savour A, Philippe H, Van Montagu M, Inze D, Rouz P. Characterization and classification of plant cyclin sequences related to A- and B-type cyclins. In: Francis D, Dudits D, Inz D, editors. Plant cell division. Colchester: Portland Press; 1998. pp. 67–98. [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino Acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Ruggiero B, Koiwa H, Manabe Y, et al. Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiology. 2004;136:3134–3147. doi: 10.1104/pp.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Schnittger A, Weinl C, Bouyer D, Schöbinger U, Hülskamp M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell. 2003;15:303–315. doi: 10.1105/tpc.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Research. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes and Development. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Smith RF, Wiese BA, Wojzynski MK, Davison DB, Worley KC. BCM search launcher–an integrated interface to molecular biology data base search and analysis services available on the world wide web. Genome Research. 1996;6:454–462. doi: 10.1101/gr.6.5.454. [DOI] [PubMed] [Google Scholar]
- Sternberg MJE, Islam SA. Protein sequences – Homologies and motifs. Trends in Biotechnology. 1991;9:300–302. doi: 10.1016/0167-7799(91)90099-4. [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant Journal. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFzio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Verkest A, de O, Manes C-L, Maes S, et al. The cyclin-dependent kinase inhibitor KRP2 controls the mitosis-to-endocycle transition during Arabidopsis leaf development through a specific inhibition of the mitotic CDKA;1 kinase complexes. Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Delseny M. Isolation of Total RNA from Arabidopsis thaliana seeds. Analytical Biochemistry. 1999;268:412–413. doi: 10.1006/abio.1998.3045. [DOI] [PubMed] [Google Scholar]
- Vigneault F, Lachance D, Cloutier M, Pelletier G, Levasseur C, Séguin A. Members of the plant NIMA-related kinases are involved in organ development and vascularization in poplar, Arabidopsis and rice. Plant Journal. 2007;51:575–588. doi: 10.1111/j.1365-313x.2007.03161.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Fowke LC, Crosby WL. A plant cyclin-dependent kinase inhibitor gene. Nature. 1997;386:451–452. doi: 10.1038/386451a0. [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant Journal. 1998;15:501–510. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC. Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant Journal. 2000;24:613–623. doi: 10.1046/j.1365-313x.2000.00899.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Fowke LC. The emerging importance of cyclin-dependent kinase inhibitors in the regulation of the plant cell cycle and related processes. Canadian Journal of Botany. 2006;84:640–650. [Google Scholar]
- Wang H, Zhou Y, Torres-Acosta L, Fowke LC. CDK inhibitors. In: Inze D, editor. Cell cycle control and plant development. Oxford: Blackwell Publishing; 2007. pp. 62–86. [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJH, et al. Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell. 2005;17:1704–1722. doi: 10.1105/tpc.104.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R, Torres-Acosta JA, Pastushok L, et al. Arabidopsis UEV1D promotes lysine-63 linked polyubiquitination and is involved in DNA damage response. Plant Cell. 2008;20:213–217. doi: 10.1105/tpc.107.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society: Biological Sciences. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Wang J, He X, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Research. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Fowke LC, Wang H. Plant CDK inhibitors: studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Reports. 2002;20:967–975. [Google Scholar]
- Zhou Y, Wang H, Gilmer S, Whitwill S, Fowke LC. Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta. 2003a;216:604–613. doi: 10.1007/s00425-002-0935-x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Li G, Brandizzi F, Fowke LC, Wang H. The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant Journal. 2003b;35:476–489. doi: 10.1046/j.1365-313x.2003.01821.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Niu H, Brandizzi F, Fowke L, Wang H. Molecular control of nuclear and subnuclear targeting of the plant CDK inhibitor ICK1 and ICK1-mediated nuclear transport of CDKA. Plant Molecular Biology. 2006;62:261–278. doi: 10.1007/s11103-006-9019-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.