Abstract
The Microviridae comprises icosahedral lytic viruses with circular single-stranded DNA genomes. The family is divided into two distinct groups based on genome characteristics and virion structure. Viruses infecting enterobacteria belong to the genus Microvirus, whereas those infecting obligate parasitic bacteria, such as Chlamydia, Spiroplasma and Bdellovibrio, are classified into a subfamily, the Gokushovirinae. Recent metagenomic studies suggest that members of the Microviridae might also play an important role in marine environments. In this study we present the identification and characterization of Microviridae-related prophages integrated in the genomes of species of the Bacteroidetes, a phylum not previously known to be associated with microviruses. Searches against metagenomic databases revealed the presence of highly similar sequences in the human gut. This is the first report indicating that viruses of the Microviridae lysogenize their hosts. Absence of associated integrase-coding genes and apparent recombination with dif-like sequences suggests that Bacteroidetes-associated microviruses are likely to rely on the cellular chromosome dimer resolution machinery. Phylogenetic analysis of the putative major capsid proteins places the identified proviruses into a group separate from the previously characterized microviruses and gokushoviruses, suggesting that the genetic diversity and host range of bacteriophages in the family Microviridae is wider than currently appreciated.
Introduction
A number of ecological studies have revealed that microbial viruses predominate in the biosphere and outnumber their hosts by at least one order of magnitude [1], [2]. Due to their abundance and consequent influence on the composition and diversity of microbial communities, viruses can be rightfully considered to be the “major players in the global ecosystem” [3], [4]. Until recently, the majority of viruses in the environment were believed to possess double-stranded DNA genomes [2]. However, technological advances in single-stranded (ss) DNA amplification and sequencing from environmental samples revealed that viruses with ssDNA genomes are more prevalent in both soil and marine environments than previously recognized [5]–[8]. This realization precipitated an interest amongst environmental virologists in the diversity and distribution of ssDNA bacterial viruses in nature [7], [9]. Among ssDNA viruses that are most often identified in the environment using metagenomic approach are those belonging to the family Microviridae. However, the host organisms have yet to be determined.
The Microviridae comprises small isometric icosahedral viruses with circular single-stranded DNA genomes [10]. The members of this family are further divided into two subgroups based on structural and genomic differences. Viruses infecting enterobacteria belong to a genus Microvirus and are typified by microvirus phiX174. The other subgroup consists of viruses infecting obligate parasitic bacteria, such as Chlamydia, Bdellovibrio and Spiroplasma [11]. These viruses are grouped into subfamily Gokushovirinae (genera Chlamydiamicrovirus, Bdellomicrovirus and Spiromicrovirus) (http://www.ictvonline.org). Virions of phiX174-like microviruses are composed of four structural proteins (major capsid protein F, major spike protein G, DNA-binding protein J and DNA pilot protein H) [12]. In contrast, only two structural proteins, homologues of phiX174 proteins F and H, were identified in mature virions of gokushoviruses [13]. Furthermore, virion assembly in phiX174-like microviruses proceeds with the aid of two scaffolding proteins, internal scaffolding protein B and external scaffolding protein D [14]. The latter one does not have an equivalent in gokushoviruses. Consequently, the genomes of gokushoviruses are slightly smaller than those of microviruses (4.5 kb versus 5.3–6.2 kb). Viruses from both groups replicate their genomes via a rolling-circle (RCR) mechanism and encode dedicated RCR initiation proteins. All characterized members of the Microviridae are strictly lytic, unable to lysogenize their hosts [10]. However, the attempt to induce viruses from marine Synechococcus strains isolated from the Gulf of Mexico resulted in the production of icosahedral non-tailed virus-like particles that contained ssDNA [15], although detailed characterization of the virus-like particles was not performed. Furthermore, genomes of Chlamydophila caviae (formerly Chlamydia psittaci) and Chlamydia pneumoniae contain gene fragments showing sequence similarity to genes of Chlamydia-infecting gokushoviruses [16]. These observations suggest that the Microviridae might include not only lytic but also temperate members, as is the case for all other families of bacterial DNA viruses that possess circular genomes or replicate their genomes via a circular intermediate.
Unexplored diversity and abundance of the Microviridae viruses in the environment fuelled our interest in this virus group. In order to obtain more information about these viruses we analyzed the genomic sequences available in public databases for the presence of proviruses related to Microviridae. The rationale behind this approach is that a provirus, defective or not, represents a molecular record that a cell has been in contact with a particular virus [17]. In this study we identified seven proviruses that are related to members of the Microviridae. The proviruses are integrated in the genomes of different species of the order Bacteroidales (phylum Bacteroidetes). The identified proviruses are only distantly related to the previously characterized microviruses and gokushoviruses and may represent a new group or subfamily within the Microviridae. Searches against metagenomic databases suggest that these new viruses might be associated with the human gut microbiota. Our results presented here extend the knowledge on the evolution, diversity and host range of microviruses.
Results and Discussion
Identification of Microviridae-related proviruses
Bacterial and archaeal DNA viruses are often capable of integrating their genomes into the host chromosome thereby becoming proviruses. Even though proviruses related to Microviridae have not been previously reported, we set out to verify this possibility by performing searches against genomic sequences available in public databases. The ability to build a virion is the major feature distinguishing viruses from other mobile genetic elements, such as plasmids and transposons [18]. Therefore, to identify Microviridae-related proviruses the iterative BLAST searches were seeded with the major capsid protein (MCP) sequences of selected microviruses and gokushoviruses. Such targeted searches have previously yielded valuable information on the diversity and evolution of other bacterial and archaeal viruses [19], [20].
Iterative searches seeded with the MCP sequences of phiX174-like microviruses (protein F) did not return hits to proteins other than the orthologues encoded by microviruses and gokushoviruses. However, when the MCP sequences of gokushoviruses (protein VP1) were used as queries, significant hits to seven proteins encoded in the genomes of six different species of the phylum Bacteroidetes were obtained (Table 1). Notably, whereas protein sequences encoded by Bacteroidetes were obtained during the initial search (i.e., first iteration), the MCP orthologues encoded by microviruses were retrieved only after further iterations. This suggests that the MCPs of gokushoviruses are closer to the group of Bacteroidetes-encoded proteins than they are to the MCPs of microviruses.
Table 1. Coordinates of the putative BMV proviruses.
| Provirus | Host organism | Sourcea | Contig accession number | Coordinates | Size, bpb |
| BMV1 | Bacteroides sp. 2_2_4 | human; gastrointestinal tract | NZ_EQ973357 | 453860..460215 | 6356 |
| BMV2 | Bacteroides eggerthii DSM 20697 | human; feces | NZ_ABVO01000042 | 11368..17543 | ∼6176 |
| BMV3 | Bacteroides plebeius DSM 17135 | human; feces | NZ_ABQC02000012 | 89498..95972 | 6483 |
| BMV4 | Prevotella sp. oral taxon 317 str. F0108 | human; subgingival oral biofilm | NZ_GG740072 | 687371..687398 | 6733 |
| BMV5 | Prevotella buccalis ATCC 35310 | human; dental plaque | NZ_ADEG01000016 | 69569..75677 | 6109 |
| BMV6 | Prevotella bergensis DSM 17361 | human; soft tissue abscess | NZ_ACKS01000073+ NZ_ACKS01000072 | 1..700+78383..83169 | 700+4787 |
| BMV7 | Prevotella bergensis DSM 17361 | human; soft tissue abscess | NZ_ACKS01000036 | 22909..27491 | >4660 |
a – information regarding the source of species isolation was obtained either from original publications or from corresponding genome sequencing project homepages at NCBI.
b – distance between the first and the last nucleotides of two dif-like sites flanking the proviral region.
Analysis of the proviral regions
In order to test whether the identified putative MCP-coding genes are of viral origin we performed a genomic context analysis. The regions of Bacteroidetes genomes adjacent to the MCP-coding genes were analysed for the presence of other viral genes. In all cases, immediately upstream of the mcp gene we identified a gene for an initiator of the rolling-circle replication (RCR) (Fig. 1A, Table S1). All three motifs characteristic to RCR proteins were found to be conserved (Fig. S1). Notably, as is the case for all known members of the Microviridae, motif III of the identified Bacteroidetes RCR proteins contains two invariable catalytic tyrosine residues (Fig. S1), a signature of superfamily I RCR proteins [21].
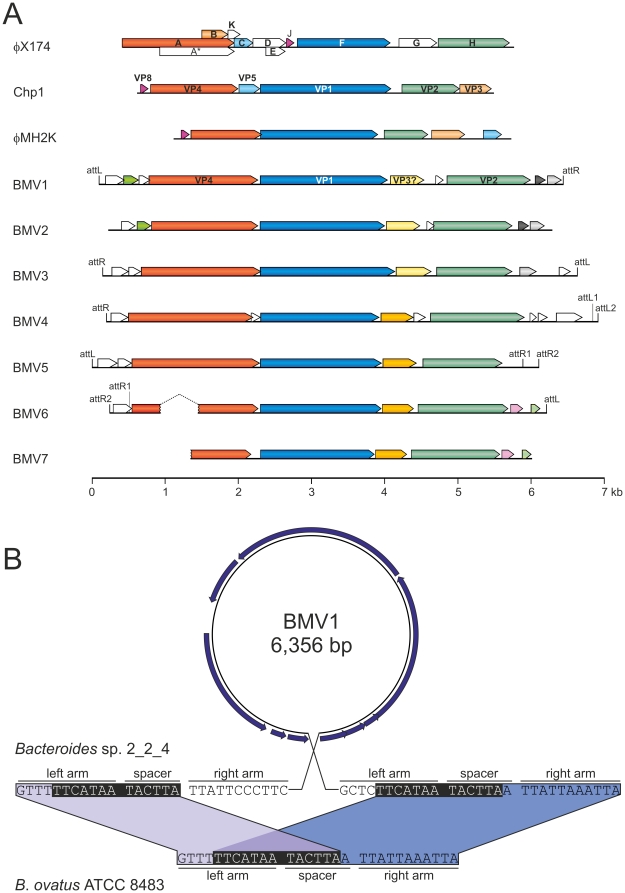
Figure 1. Bacteroidetes-associated, microvirus-related proviruses BMV1–7.
A. Genomic organization of the putative BMV proviruses residing in the genomes of different species of the phylum Bacteroidetes and two gokushoviruses (family Microviridae), Chlamydia phage 1 (Chp1) and Bdellovibrio-infecting virus φMH2K. Circular genome maps of Chp1 (GenBank accession number: D00624) and φMH2K (GenBank accession number: AF306496) are linearized for convenient alignment. Open reading frames (ORF; arrows) are labeled according to the gokushovirus and microvirus protein nomenclature. ORFs encoding homologous products are coloured similarly. attL and attR, left and right attachment sites, respectively. B. Comparison of the BMV1-containing genomic region of Bacteroides sp. 2_2_4 with the provirus-free genomic region of B. ovatus ATCC 8483 (GenBank accession number: NZ_AAXF02000049; nucleotide coordinates: 301224–301252). The putative attachment sites flanking BMV1 as direct repeats are highlighted in black background.
Transcriptionally downstream of the mcp genes we identified genes encoding homologues of the DNA pilot protein (protein H in microviruses or VP2 in gokushoviruses) (Fig. 1A, Table S1). The function of VP2/H-like proteins has been studied in the case of phiX174, but is yet to be fully understood [22]–[24]. Protein H is a multifunctional structural protein (12 copies per virion) required for piloting the viral DNA into the host cell interior during the entry process [10]. VP2 proteins of gokushoviruses share only limited primary structure similarity with H of microviruses [25]. However, VP2/H proteins from both groups of viruses share coiled-coil regions and predicted N-terminal transmembrane domains. Both these features are also characteristic to the VP2/H homologues (Table S1) encoded in the vicinity of the VP1/F-like mcp genes in the genomes of Bacteroidetes. Further sequence analysis did not reveal additional genes related to those of microviruses and/or gokushoviruses.
Mature virions of gokushoviruses are constructed of only two proteins, VP1 and VP2 [13]. Homologues of both proteins as well as the VP4-like RCR Rep protein are encoded as a block, within ∼6 kb region in the genomes of different species of Bacteroidetes (Fig. 1A). Furthermore, the organization of these genes is very similar to that found in the genomes of gokushoviruses (Fig. 1A). Consequently, these observations strongly suggest that this block of genes in Bacteroidetes genomes represents proviruses related to Microviridae. The seven proviral regions are refered to as BMV1–7, for Bacteroidetes-associated microviruses (Table 1).
Between the genes for VP1- and VP2-like proteins all BMVs contain open reading frames (ORFs) of approximately 150 codons (Fig. 1A, light and dark yellow arrows). Notably, despite being of similar size, ORFs from BMVs 1–3 (form Bacteroides species) share little sequence similarity (∼16 % identity at the protein level) with the corresponding ORFs from BMVs 4–7 (from Prevotella species) (Tables 1 and S1). The ORFs from either group have no homologues in public protein databases (except for those in BMVs). However, the conservation of the ORF within the two groups of BMVs suggested that it might encode an important function, possibly a scaffolding protein. To test this possibility, representative protein sequences from the two BMV groups were aligned with the sequence of VP3 scaffolding protein from φHM2K. The VP3 proteins of gokushoviruses are of ∼150 aa in length and share only limited sequence similarity with corresponding proteins from microviruses [25], [26]. Multiple sequence alignment revealed that proteins from the two BMV groups share a set of conserved residues not only with each other but also with the φHM2K VP3 (Fig. S2). We therefore predict that the conserved ORF following the one for the major capsid protein VP1 in all BMVs encodes a homologue of the internal scaffolding protein VP3.
Both microviruses and gokushoviruses encode very small (25–40 aa) DNA-binding proteins (protein J or VP8) that are rich in arginine and lysine residues [10]. The seven BMVs do not code for apparent homologues of J/VP8 proteins, nor were we able to identify homologues of any other proteins encoded by microviruses and gokushoviruses, including proteins C/VP5, D, G, or E.
BMVs most likely rely on the host chromosome dimer resolution system for integration
The vast majority of temperate bacterial and archaeal viruses lysogenize their hosts by site-specifically integrating into the cellular chromosome. The reaction is generally catalyzed by a virus-encoded recombinase [27]. Interestingly, none of the BMVs encodes a recognizable recombinase, suggesting that the mechanism of integration is different from that utilized by the majority of prokaryotic viruses. Filamentous ssDNA viruses (family Inoviridae) infecting Vibrio species are an exception to this rule [28]. Filamentous vibrioviruses do not encode a recombinase of their own, but rather highjack the chromosome dimer resolution system of their hosts. Cellular tyrosine recombinases XerC and XerD recognize the dif-like sequences within the viral genome [29] and promote the integration of either the single-stranded (e.g., CTXφ; [30]) or the replicative, double-stranded (e.g., VGJφ; [31]) form of the viral genomic DNA into the chromosome dimer resolution sites.
In order to define the precise integration sites of BMVs, we took advantage of the availability of the genomic sequence for Bacteroides ovatus ATCC 8483, a provirus-free species closely related to BMV1-harboring Bacteroides sp. 2_2_4. Comparison of the corresponding sequences from the two Bacteroides species revealed the exact attachment site on the bacterial chromosome (Fig. 1B). It appears that BMV1 was integrated in the intergenic region between the genes for DNA mismatch repair protein MutS (GI:237722019) and glycoside hydrolase (GI:237722028). Due to recombination, the attachment site (13 bp) has been duplicated to flank the integrated provirus as direct repeats (Fig. 1B). With the size of 6.3 kb (Table 1) BMV1 genome is larger than those of all currently described gokushoviruses (4.5 kb) and microviruses (5.3 – 6.2 kb).
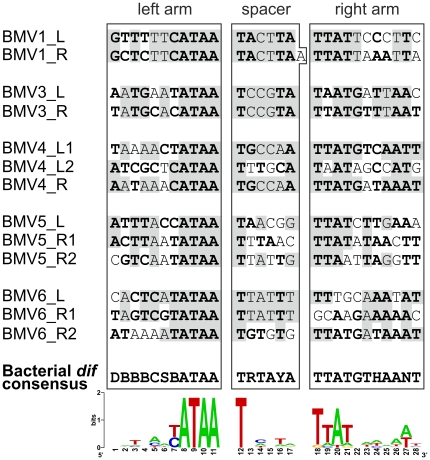
Bacterial and archaeal dif sites are typically 28 bp long, display palindromic structure and are situated in intergenic regions, close to the GC-skew shift-point and replication terminus [32]–[34]. XerC/D recombinases act at the dif site to resolve chromosome dimers following replication termination [35], [36]. Careful examination of BMV1 revealed that sequences flanking the provirus resemble bacterial dif sites (Fig. 1B, 2), suggesting that, like in the case of filamentous vibrioviruses, the integration of BMV1 might have been mediated by the cellular recombination machinery. It should be noted that, genome sequences for all BMV-harbouring species are available as WGS genomic libraries (Table 1), precluding a meaningful GC-skew analysis of these bacterial chromsomes.
Figure 2. Comparison of the predicted attachment sites of BMVs with the bacterial dif consensus sequence.
Left arm (XerC binding site), spacer and right arm (XerD binding site) regions of the dif sites are indicated. Bacterial dif consensus sequence [34] is indicated according to the IUPAC code. Nucleotide positions in the att sites of BMVs matching the dif consensus are shown in bold. Identical nucleotide positions in the left (L) and right (R) att sites, flanking the proviruses as imperfect direct repeats (see Figure 1), are shaded gray. BMV att consensus sequence is shown as sequence logo at the bottom of the figure.
For the remaining six BMVs (BMV2–7), the integration sites could not be unequivocally defined by direct comparison of provirus-containing and provirus-free strains, due to unavailability of the genomic sequences for the latter group. We therefore investigated the proviruses (along with the flanking sequences) for the presence of dif-like sequences, similar to those found in BMV1. Indeed, such sequences turned out not to be specific to BMV1, but could also be identified in BMV3–6 (Fig. 2), but not in BMV2 and BMV7. It should be noted, however, that BMV7 sequence is only partial, present on the extremity of a contig (NZ_ACKS01000036) and misses the 5′-distal region of the gene for the RCR Rep protein along with the upstream region, including the attachment site (Fig. 1A). Interestingly, for BMVs 4–6, an additional dif-like sequence was identified in each of these proviruses, close to one of the termini of the integrated viral genome (see Fig. 1A, 2). This is reminiscent of CTXφ-like vibrioviruses, where viral genomes have two different dif-like sequences in inverted orientations [29], allowing the single-stranded form of the genome to be recombined with the host chromosome [30].
Chromosome dimer resolution has not been studied in any member of the phylum Bacteroidetes. Therefore, to ascertain whether XerC/D system might potentially be involved in this process we searched for the homologues of the Escherichia coli genes xerC and xerD in the genomes of Bacteroidetes species for which genome sequences are available. Genes for the two proteins are present in both BMV-carrying (Table S2) and BMV-free Bacteroidetes species, suggesting that the two cellular recombinases might indeed be involved in the integration of viral genomes at the chromosomal dimer resolution sites.
BMVs are associated with human gut and oral microbiota
BMV-containing species fall into two different genera within the order Bacteroidales, Bacteroides (hosts for BMV1–3) and Prevotella (hosts for BMV4–7). Bacteroidales are gram-negative anaerobic bacteria that inhabit a variety of environments including the gastrointestinal tracts of mammals, the oral cavity of humans, soil and fresh water [37]–[39]. The six Bacteroidales species that contain BMV proviruses were isolated from humans (Table 1) and their genomes were sequenced as part of the Human Microbiome Project by NIH Human Microbiome Consortium [40]. In humans, Bacteroides constitute the dominant part of gut microbiota, whereas Prevotella are part of the normal flora of the human mouth and vagina [41]. Both Bacteroides and Prevotella are opportunistic human pathogens. Notably, Prevotella bergensis DSM 17361, the host for BMV6 and BMV7, was isolated from soft-tissue abscesses [42].
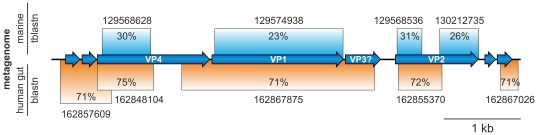
In order to get further insight into the distribution of BMV-like viruses in the environment, we searched the metagenomic databases at NCBI for the presence of sequences related to BMV proviruses. Searches seeded with the nucleotide sequence of BMV2 resulted in the most significant hits. These were to several contigs sequenced during the metagenomic analysis of the human gut microbiota [43]. The retrieved sequences were 71–75% identical (at the nucleotide level) to the BMV2 sequence and collectively covered ∼75% of the latter (Fig. 3). Notably, all five contigs matching the BMV2 provirus were generated by metagenomic sequencing of the faecal samples obtained from a single healthy male adult individual [43]. To identify more distantly related BMV-like sequences, further searches were performed against translated nucleotide sequences of the metagenomic databases using BMV2 protein sequences as queries. Sequences similar to the three conserved Microviridae proteins (VP1/F, VP2/H and VP4/A) of BMV2 were identified in the marine metagenome (Fig. 3), albeit the similarity was much lower (23–31% at the amino acid level) than to the human gut metagenome sequences. All marine samples containing BMV2-like sequences were collected during the Sorcerer II Global Ocean Sampling Expedition from surface marine waters along a voyage from Eastern North American coast to the Eastern Pacific Ocean [44]. It is not possible at the moment to tell whether the BMV-like sequences present in the metagenomic databases belong to free or integrated viruses. However, taking into account the information on the source of isolation of BMV-harbouring species and high sequence similarity to human gut-derived metagenomic sequences it is highly likely that BMV-like viruses are associated with human gut and oral microbiota.
Figure 3. BMV-related sequences in environmental databases.
Blastn (nucleotide query against nucleotide database) hits to the human gut metagenome and tblastn (protein query against translated nucleotide database) hits to marine GOS metagenome are depicted below and above the BMV2 genome map, respectively. Hit coverage and respective sequence identities (retrieved contigs are indicated by their GenBank identifiers) are also shown.
BMVs are closer to gokushoviruses but comprise a phylogenetically distinct group within Microviridae
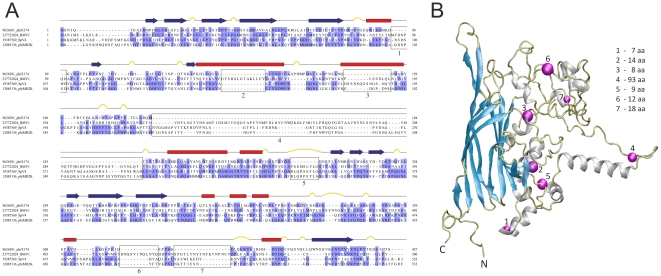
Virions of gokushoviruses possess 'mushroom-like' protrusions positioned at the three-fold axes of symmetry of their icosahedral capsids. These structures are formed by large insertion loops within the MCP of gokushoviruses and are absent in the microviral MCPs [45]. In order to find out whether equivalent loops are also characteristic to BMV MCPs, a multiple sequence alignment of MCP sequences from BMVs, gokushoviruses and microviruses was constructed (Fig. S3). BMV1 MCP was found to be more closely related to corresponding proteins from gokushoviruses, sharing with the latter proteins six insertions (larger than 5 aa), including the one responsible for formation of 'mushroom-like' structures (insertion 4 in Fig. 4). Notably, the latter insertion in BMVs is considerably longer (93 aa in BMV1; Fig. 4) than in gokushoviruses and is accountable for the larger size of the BMV capsid proteins. In addition, the BMV1 MCP displayed a specific insertion of 14 aa (insertion 2 in Fig. 4), not present in the gokushoviral MCPs. All insertions were located outside of the predicted eight-stranded antiparallel beta-barrel core structure (Fig. 4). Therefore, it appears that not only genomic organization of BMVs is closer to that of gokushoviruses (Fig. 1A), but also their capsid proteins are more closely related.
Figure 4. Analysis of the putative major capsid protein of BMV1.
A. Alignment of the BMV1 major capsid protein sequence to the corresponding protein sequences of φX174, SpV4 and φMH2K. The proteins are denoted by their GenBank identifier followed by the corresponding (pro)virus name. The alignment is coloured according to sequence conservation (BLOSUM62 matrix). The secondary structure determined from the X-ray structure of φX174 capsid protein F (PDB ID: 1CD3) is shown above the alignment with α helices, β strands, and turns represented by red rectangles, blue arrows, and yellow bulges, respectively. Insertions (>5 aa) relative to the capsid protein F of φX174 are boxed. B. Atomic structure of the major capsid protein F of microvirus φX174 (PDB ID: 1CD3). Magenta spheres highlight the equivalent positions of the putative capsid protein of BMV1, where insertions (larger than 5 aa; numbered 1–7) occur relative to the capsid protein F of φX174 (refer to panel A for the alignment). Size of each insertion is indicated on the right of the figure.
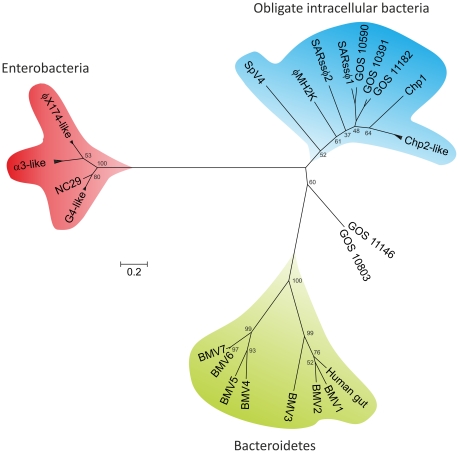
Previous phylogenetic analysis of the MCP proteins supported the division of Microviridae viruses into two distinct groups, microviruses on one side and gokushoviruses on the other [26]. To better understand the relationship of BMVs to other members of the Microviridae family we performed a phylogenetic analysis of their major capsid proteins (Fig. 5). Our maximum likelihood analysis supported the previous conclusion regarding the relationship of microviruses and gokushoviruses [26] and revealed that BMVs fall into a third group, separate from the other two (Fig. 5). Within the BMV cluster there is a separation between the BMV1-like (BMV1–3 and the MCP sequence obtained from the human gut metagenome; Fig. 3) and BMV4-like proviruses (BMV4–7). Notably, the division of BMVs into two groups based on the MCP phylogeny is consistent with the genomic content analysis (Fig. 1A, Table S1).
Figure 5. Unrooted phylogenetic tree of the φX174 F-like major capsid proteins.
The evolutionary history was inferred by using the Maximum Likelihood method based on the Whelan and Goldman model [58]. The bootstrap consensus tree was inferred from 1000 replicates. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The scale bar represents the number of substitutions per site. The analysis involved 35 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 356 positions in the final dataset. φX174-like virus group includes φX174 (GI:9626381), WA10 (GI:71843040), S13 (GI:11095662), NC11 (GI:71842956), ID1 (GI:71842872). α3-like virus group includes α3 (GI:9625363), φK (GI:2493329), st-1 (GI:242346750). G4-like virus group includes G4 (GI:9626346), WA6 (GI:71843160), ID12 (GI:71843172). CPAR39-like virus group includes. Chp2-like virus group includes Chp2 (GI:9634949), Chp3 (GI:47566141), Chp4 (GI:77020115), CPAR39 (GI:9791178), φCPG1 (GI:17402851). SARssφ1 (GI:313766927) and SARssφ2 (GI:313766923) are uncultured microviruses the genomes of which were assembled by Tucker et al [9]. GOS sequences are microvirus-like major capsid proteins obtained during the Sorcerer II Global Ocean Sampling (GOS) Expedition [44]: GOS_10590 (GI:142008996), GOS_10391 (GI:142009231), GOS_11182 (GI:142008205), GOS_11146 (GI:142008257), GOS_10803 (GI:142008696).
Whereas the BMV MCPs (VP1) are closer to corresponding proteins from gokushoviruses, the VP2 proteins of BMVs are more similar to their homologues in microviruses (protein H), as judged from the BLAST analysis (Table S1). We therefore investigated the possibility of horizontal gene transfer (HGT) between the two groups of BMVs. For that we performed a phylogenetic analysis of the three proteins conserved in all BMVs, i.e., homologues of VP1/F, VP2/H and VP4/A. No signs of recent inter- or intra-group gene transfer events could be detected for the three analysed proteins, suggesting that HGT might be rare not only in lytic members of the Microviridae [10], [46], but also among the temperate BMV group.
Concluding Remarks
Studies on various bacterial and archaeal proviruses have provided valuable information on the diversity, phylogenetic distribution and evolution of corresponding virus groups [19], [47]–[50]. Analysis of the putative proviruses described in the present study expands our knowledge on the viral family Microviridae. Not only is this the first time that members of the Microviridae are implicated in lysogenization of their hosts, but also the association of this virus group with Bacteroidetes has not been previously recognized. The host range of members of this virus family now covers four different bacterial phyla, namely Proteobacteria (microviruses and bdellomicrovirus), Tenericutes (spiromicrovirus), Chlamydiae (chlamydiamicroviruses), and Bacteroidetes (BMVs). Notably, BMVs are clearly distinct from the previously recognized microviruses and gokushoviruses. Consequently, if confirmed to produce genuine viruses, BMVs may represent a new group or subfamily within the Microviridae, which we propose to name Alpavirinae (Alpa: Sanskrit for 'small', 'minute'). BMVs appear to be associated with human gut and oral microbiota. In the future, it will be very interesting to explore the diversity of viruses infecting Bacteroides and Prevotella, to see what other new viral groups, in addition to BMVs, are associated with these bacteria. BMVs identified here are likely to integrate into the genomes of their hosts at the chromosome dimer resolution dif sites with the aid of cellular XerC/D recombination machinery, a route thought to be exclusively employed by filamentous vibrioviruses of the family Inoviridae. Studies on the integration of BMVs into the cellular chromosome therefore promise to provide further exciting insights on how bacterial viruses with small genomes highjack cellular machineries for their own needs.
Methods
Identification and analysis of proviral sequences
Putative microvirus-related prophages were identified by homology-based searches against the nonredundant protein database at NCBI. The major capsid protein (MCP) sequences of representative microviruses (phiX174, GI:9626381; α3, GI:9625363; G4, GI:9626346) and gokushoviruses (Chp1, GI:9629155; phiMH2K, GI:12085136; SpV4, GI:19387569) were used as queries in the PSI-BLAST searches [51] with the default parameters (BLOSUM62 matrix, 0.005 as an E-value cutoff). When MCP sequences of gokushoviruses were used as queries, in addition to homologues in other members of the Microviridae, significant hits were obtained (during the first or second iteration) to seven protein sequences encoded in the genomes of six different species belonging to the phylum Bacteroidales. The genomes of the six bacterial species (Table 1) were sequenced as part of the Human Microbiome Reference Genomes Project by NIH Human Microbiome Consortium [40]. Contigs encoding the MCP-like proteins were downloaded from NCBI and analysed for the presence of other viral proteins encoded in proximity of the mcp genes using CLC Genomics Workbench software package (CLC Bio, Inc.). Protein sequences of the identified proviruses are provided in Table S3.
Transmembrane domains were predicted using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) or TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). Coiled-coil regions were identified using COILS (http://www.ch.embnet.org/software/COILS_form.html) [52]. Sequence logo was created with WebLogo (http://weblogo.berkeley.edu/) [53].
Phylogenetic analysis
For phylogenetic analysis multiple sequence alignments were constructed using PROMALS3D [54] and MUSCLE [55], manually examined and edited. Sequence alignments were visualized using Jalview [56]. Maximum likelihood analysis was carried out using MEGA5 software [57] with a WAG amino acid substitution model [58]. The robustness of the trees was assessed by bootstrap analysis (1,000 replicates).
Supporting Information
Alignment of the three conserved motifs (I–III) of superfamily I rolling circle replication proteins with corresponding motifs from the putative replication proteins of the BMV proviruses. The protein sequences are denoted by their GenBank identifiers followed by the corresponding (pro)virus name. The limits of the depicted motifs are indicated by the residue positions on each side of the alignment, with the total length of the protein given in parenthesis. The numbers within the alignment indicate the distance between the motifs.
(PDF)
Multiple alignment of the putative internal scaffolding proteins from BMVs 1 and 6 with the VP3 protein from Bdellovibrio gokushovirus φMH2K (GI:12085142).
(PDF)
Phylogenetic analysis of proteins conserved in BMV proviruses. The evolutionary history of the VP1/F-like, VP2/H-like and VP4/A-like proteins encoded by BMV proviruses was inferred by using the Maximum Likelihood method based on the Whelan and Goldman amino acid substitution model. Numbers at the branch-points represent bootstrap values (1000 replicates). The outgroups were chosen based on the BLAST analysis.
(PDF)
Annotation of BMV1 and its comparison to BMV2–7.
(DOC)
XerC and XerD homologues in organisms of Bacteroidales containing microvirus-related proviruses.
(DOC)
BMV protein sequences.
(DOC)
Acknowledgments
We are grateful to Dr. Virgainija Cvirkaitč-Krupovič and three reviewers for their helpful suggestions and comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the European Molecular Biology Organization (Long-Term Fellowship ALTF 347-2010 to MK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, et al. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159:349–357. doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. Marine viruses - major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 5.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- 7.Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, et al. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol. 2008;74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, et al. High diversity of the viral community from an Antarctic lake. Science. 2009;326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- 9.Tucker KP, Parsons R, Symonds EM, Breitbart M. ISME J in press; 2010. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fane BA, Brentlinger KL, Burch AD, H afenstein SL, Moore E, et al. φX174 et al., the Microviridae. In: Calendar R, editor. The Bacteriophages. 2nd ed. New York: Oxford University Press; 2006. pp. 129–145. [Google Scholar]
- 11.Brentlinger KL, Hafenstein S, Novak CR, Fane BA, Borgon R, et al. Microviridae, a family divided: isolation, characterization, and genome sequence of phiMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J Bacteriol. 2002;184:1089–1094. doi: 10.1128/jb.184.4.1089-1094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna R, Xia D, Willingmann P, Ilag LL, Krishnaswamy S, et al. Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications. Nature. 1992;355:137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke IN, Cutcliffe LT, Everson JS, Garner SA, Lambden PR, et al. Chlamydiaphage Chp2, a skeleton in the phiX174 closet: scaffolding protein and procapsid identification. J Bacteriol. 2004;186:7571–7574. doi: 10.1128/JB.186.22.7571-7574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fane BA, Prevelige PE Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 15.McDaniel LD, delaRosa M, Paul JH. Temperate and lytic cyanophages from the Gulf of Mexico. J Mar Biol Ass UK. 2006;86:517–527. [Google Scholar]
- 16.Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krupovic M, Spang A, Gribaldo S, Forterre P, Schleper C. A thaumarchaeal provirus testifies for an ancient association of tailed viruses with archaea. Biochem Soc Trans. 2011;39:82–88. doi: 10.1042/BST0390082. [DOI] [PubMed] [Google Scholar]
- 18.Krupovic M, Bamford DH. Order to the viral universe. J Virol. 2010;84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupovic M, Bamford DH. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics. 2007;8:236. doi: 10.1186/1471-2164-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krupovic M, Bamford DH. Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology. 2008;375:292–300. doi: 10.1016/j.virol.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 21.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azuma J, Morita J, Komano T. Process of attachment of phi X174 parental DNA to the host cell membrane. J Biochem. 1980;88:525–532. doi: 10.1093/oxfordjournals.jbchem.a132999. [DOI] [PubMed] [Google Scholar]
- 23.Cherwa JE, Young LN, Fane BA. Uncoupling the functions of a multifunctional protein: The isolation of a DNA pilot protein mutant that affects particle morphogenesis. Virology. 2011;411:9–14. doi: 10.1016/j.virol.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Ruboyianes MV, Chen M, Dubrava MS, Cherwa JE, Fane BA. The expression of N-terminal deletion DNA pilot proteins inhibits the early stages of phiX174 replication. J Virol. 2009;83:9952–9956. doi: 10.1128/JVI.01077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu BL, Everson JS, Fane B, Giannikopoulou P, Vretou E, et al. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J Virol. 2000;74:3464–3469. doi: 10.1128/jvi.74.8.3464-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garner SA, Everson JS, Lambden PR, Fane BA, Clarke IN. Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila pecorum. Virus Genes. 2004;28:207–214. doi: 10.1023/B:VIRU.0000016860.53035.f3. [DOI] [PubMed] [Google Scholar]
- 27.Reiter WD, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 29.McLeod SM, Waldor MK. Characterization of XerC- and XerD-dependent CTX phage integration in Vibrio cholerae. Mol Microbiol. 2004;54:935–947. doi: 10.1111/j.1365-2958.2004.04309.x. [DOI] [PubMed] [Google Scholar]
- 30.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Das B, Bischerour J, Barre FX. VGJφ integration and excision mechanisms contribute to the genetic diversity of Vibrio cholerae epidemic strains. Proc Natl Acad Sci U S A. 2011;108:2516–2521. doi: 10.1073/pnas.1017061108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carnoy C, Roten CA. The dif/Xer recombination systems in proteobacteria. PLoS One. 2009;4:e6531. doi: 10.1371/journal.pone.0006531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortez D, Quevillon-Cheruel S, Gribaldo S, Desnoues N, Sezonov G, et al. Evidence for a Xer/dif system for chromosome resolution in archaea. PLoS Genet. 2010;6:e1001166. doi: 10.1371/journal.pgen.1001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendrickson H, Lawrence JG. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol Microbiol. 2007;64:42–56. doi: 10.1111/j.1365-2958.2007.05596.x. [DOI] [PubMed] [Google Scholar]
- 35.Ip SC, Bregu M, Barre FX, Sherratt DJ. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 2003;22:6399–6407. doi: 10.1093/emboj/cdg589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, et al. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duncan MJ. Genomics of oral bacteria. Crit Rev Oral Biol Med. 2003;14:175–187. doi: 10.1177/154411130301400303. [DOI] [PubMed] [Google Scholar]
- 38.Ohkuma M, Noda S, Hongoh Y, Kudo T. Diverse bacteria related to the Bacteroides subgroup of the CFB phylum within the gut symbiotic communities of various termites. Biosci Biotechnol Biochem. 2002;66:78–84. doi: 10.1271/bbb.66.78. [DOI] [PubMed] [Google Scholar]
- 39.O'Sullivan LA, Weightman AJ, Fry JC. New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon. Appl Environ Microbiol. 2002;68:201–210. doi: 10.1128/AEM.68.1.201-210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah HN, Gharbia SE, Duerden BI. Bacteroides, Prevotella and Porphyromonas. In: Collier L, Balows A, Sussman M, editors. Topley & Wilson's Microbiology and Microbial Infections. London: Arnold; 1998. pp. 1305–1330. [Google Scholar]
- 42.Downes J, Sutcliffe IC, Hofstad T, Wade WG. Prevotella bergensis sp. nov., isolated from human infections. Int J Syst Evol Microbiol. 2006;56:609–612. doi: 10.1099/ijs.0.63888-0. [DOI] [PubMed] [Google Scholar]
- 43.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chipman PR, Agbandje-McKenna M, Renaudin J, Baker TS, McKenna R. Structural analysis of the Spiroplasma virus, SpV4: implications for evolutionary variation to obtain host diversity among the Microviridae. Structure. 1998;6:135–145. doi: 10.1016/s0969-2126(98)00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rokyta DR, Burch CL, Caudle SB, Wichman HA. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol. 2006;188:1134–1142. doi: 10.1128/JB.188.3.1134-1142.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 48.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 49.Jalasvuori M, Pawlowski A, Bamford JK. A unique group of virus-related, genome-integrating elements found solely in the bacterial family Thermaceae and the archaeal family Halobacteriaceae. J Bacteriol. 2010;192:3231–3234. doi: 10.1128/JB.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krupovic M, Forterre P, Bamford DH. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol. 2010;397:144–160. doi: 10.1016/j.jmb.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 58.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the three conserved motifs (I–III) of superfamily I rolling circle replication proteins with corresponding motifs from the putative replication proteins of the BMV proviruses. The protein sequences are denoted by their GenBank identifiers followed by the corresponding (pro)virus name. The limits of the depicted motifs are indicated by the residue positions on each side of the alignment, with the total length of the protein given in parenthesis. The numbers within the alignment indicate the distance between the motifs.
(PDF)
Multiple alignment of the putative internal scaffolding proteins from BMVs 1 and 6 with the VP3 protein from Bdellovibrio gokushovirus φMH2K (GI:12085142).
(PDF)
Phylogenetic analysis of proteins conserved in BMV proviruses. The evolutionary history of the VP1/F-like, VP2/H-like and VP4/A-like proteins encoded by BMV proviruses was inferred by using the Maximum Likelihood method based on the Whelan and Goldman amino acid substitution model. Numbers at the branch-points represent bootstrap values (1000 replicates). The outgroups were chosen based on the BLAST analysis.
(PDF)
Annotation of BMV1 and its comparison to BMV2–7.
(DOC)
XerC and XerD homologues in organisms of Bacteroidales containing microvirus-related proviruses.
(DOC)
BMV protein sequences.
(DOC)