Abstract
Reported is the structure and biosynthesis of ansalactam A, an ansamycin class polyketide produced by an unusual modification of the polyketide pathway. This new metabolite, produced by a marine sediment-derived bacterium of the genus Streptomyces, possesses a novel spiro γ-lactam moiety and a distinctive isobutyryl polyketide fragment observed for the first time in this class of natural products. The structure of ansalactam A was defined by spectroscopic methods including X-ray crystallographic analysis. Biosynthetic studies with stable isotopes further led to the discovery of a new, branched chain polyketide synthase extender unit derived from (E)-4-methyl-2-pentenoic acid for polyketide assembly observed for the first time in this class of natural products.
Introduction
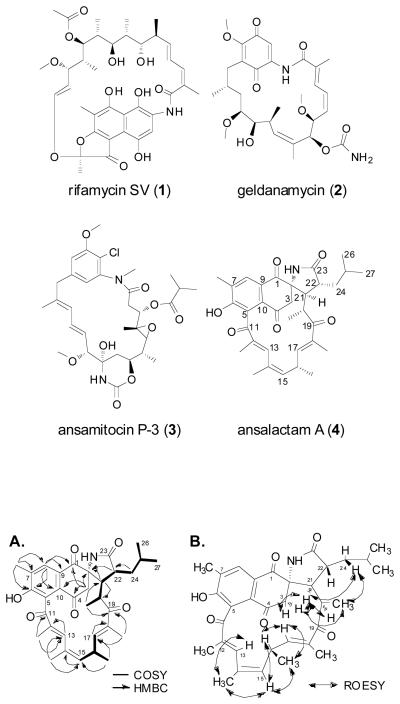
Ansamycin polyketides include potent antibiotic and anticancer agents such as rifamycin SV (1), geldanamycin (2), and ansamitocin P-3 (3).1 These natural product macrolactams are characterized by an mC7N structural unit biosynthetically derived from the aromatic acid 3-amino-5-hydroxybenzoate (AHBA), which is carboxy extended by multimodular polyketide synthases (PKSs) utilizing primarily acetate and propionate building blocks prior to macrolactam cyclization.1-3 Two ansamycin variations incorporating either benzyl or naphthyl ring systems have emerged based on the AHBA-derived structural unit. As part of our ongoing efforts to discover and biosynthetically characterize novel marine microbial natural products,4,5 we investigated Streptomyces sp. strain CNH-189 isolated from marine sediments retrieved off shore of Oceanside, California. Herein we report the discovery of the new ansamycin ansalactam A (4) from a marine sediment-derived streptomycete that uniquely contains a spiro γ-lactam functionality. Biosynthetic studies with stable isotopes further led to the discovery of a new, branched chain PKS extender unit for polyketide assembly observed for the first time in this class of natural products.
Results and Discussion
Structure determination of ansalactam A
LC-MS analysis of a whole culture crude extract of Streptomyces sp. strain CNH-189 revealed several medium molecular weight compounds with an unusual UV chromophore indicative of highly conjugated aromatic compounds. Fractionation of a 60-L culture extract of this strain by standard silica column chromatography, followed by reversed-phase HPLC separation, yielded ansalactam A (4, 70 mg), the full stereostructure of which was characterized by NMR methods and confirmed by X-ray analysis.
The molecular formula of 4 was deduced as C33H39NO6 based on analysis of high-resolution electrospray ionization mass spectrometry data and on interpretation of 13C NMR data (Table 1). Ansalactam A (4) showed strong UV absorptions at 225, 250, and 290 nm, which indicated a highly conjugated aromatic compound. The IR spectrum of 4 further revealed broad absorptions for hydroxy groups at 3354 cm−1 and a broad band at 1691 cm−1 indicating the presence of multiple conjugated carbonyl groups. The planar structure of 4 was established by thorough analysis of 13C and 2D NMR spectroscopic data. Two spin systems consisting of eight- and four-carbon units were constructed from the observation of COSY data (Figure 1A). Long-range HMBC correlations from the methyl doublet 20-Me and the singlet methyl 18-Me provided the connectivity from C-17 to C-20, thereby joining the eight- and four-carbon COSY-based fragments. The connection from C-11 to C-15 was further established by HMBC correlations from the methyl singlets 12-Me and 14-Me and the olefinic protons H-13 and H-15 to complete the remainder of the ansa side chain.
Table 1.
NMR Spectroscopic Data for Ansalactam A (4) (methanol-d4)a
| No. | δC, mult.b | δH (J in Hz) | COSY | HMBC |
|---|---|---|---|---|
| 1 | 194.7, C | |||
| 2 | 69.0, C | |||
| 3 | 51.6, CH2 | 3.38, d (14.0) 2.92, d, (14.0) | 1, 2, 4, 10, 21 | |
| 4 | 195.8, C | |||
| 5 | 129.8, C | |||
| 6 | 161.7, C | |||
| 7 | 134.9, C | |||
| 8 | 133.4, CH | 7.73, s | 1, 6, 7, 9, 10 | |
| 9 | 128.1, C | |||
| 10 | 134.1, C | |||
| 11 | 202.5, C | |||
| 12 | 138.7, C | |||
| 13 | 138.4, CH | 6.91, s | 11, 12-Me, 14-Me | |
| 14 | 135.4, C | |||
| 15 | 144.9, CH | 5.62, d (4.0) | 16 | 13, 14-Me, 16-Me, 17 |
| 16 | 33.4, CH | 3.17, m | 15, 17 | 16-Me, |
| 17 | 149.5, CH | 6.19, d (6.6) | 16 | 15, 16, 18-Me, 19 |
| 18 | 136.8, C | |||
| 19 | 203.0, C | |||
| 20 | 39.8, CH | 3.24, dq (6.8, 2.4) | 20-Me, 21 | 2, 19 |
| 21 | 58.4, CH | 2.43, dd (10.1, 2.4) | 20, 22 | 1, 2, 3, 4, 19 |
| 22 | 42.3, CH | 3.70, br s | 21, 24 | |
| 23 | 183.8, C | |||
| 24 | 41.3, CH2 | 1.52, m, 1.63, m | 22, 25 | 23, 25, 26, 27 |
| 25 | 27.2, CH | 1.97, m | 24, 26, 27 | 22, 24, 26, 27 |
| 26 | 23.7, CH3 | 0.95, d (6.8) | 25 | 24, 25, 27 |
| 27 | 22.5, CH3 | 0.92, d (6.8) | 25 | 24, 25, 26 |
| 7-Me | 16.9, CH3 | 2.31, s | 6, 7, 8 | |
| 12-Me | 12.8, CH3 | 2.27, s | 11, 13 | |
| 14-Me | 23.7, CH3 | 2.18, s | 13, 15 | |
| 16-Me | 20.3, CH3 | 1.05, d (6.8) | 16 | 15, 16, 17 |
| 18-Me | 12.4, CH3 | 1.19, s | 17, 19 | |
| 20-Me | 22.1, CH3 | 1.26, d (6.8) | 20 | 19, 20, 21 |
600 MHz for 1H NMR and 75 MHz for 13C NMR.
Numbers of attached protons were determined by analysis of 2D DEPT spectra.
Figure 1.
Structure determination of ansalactam A (4). (A) COSY and key HMBC correlations for structure elucidation of ansalactam A (4). (B) Selected key ROESY correlations for determination of geometries and the relative configurations for ansalactam A (4).
The naphthoquinone moiety in 4 was also established by analysis of HMBC spectroscopic data, in particular by analysis of correlations from the aromatic singlet proton H-8, the aromatic methyl singlet 7-Me, and the nonequivalent H-3 methylene protons (Figure 1A). These data and the chemical shift of C-6 (δC 161.7) suggested the presence of the naphthoquinone moiety with one hydroxy group substituent at C-6. The planar structure of 4 was completed by further analysis of HMBC data in which a two-bond correlation from H-24 to C-22 along with a three-bond correlation from H-24 to C-23 provided the C-22-C-23 attachment. The chemical shifts of C-2 (δC 69.0) and C-23 (δC 183.8) together with the molecular formula of 4 indicated their connection through nitrogen. The observation of key HMBC correlations established the connection of C-2 and C-21 with the spiro ring system as shown. Lastly, the gross structure of 4 was completed by the connection of C-11 to C-5.
The geometries of the three double bonds as 12E, 14Z, and 17E were determined by interpretation of ROESY correlation data. Strong ROESY correlations between H-21 and both H-3 methylene protons suggested a 21R* configuration (Figure 1B). The large vicinal coupling constant of 10.1 Hz between H-21 and H-22, and the ROESY correlation between H-21 and H-24 also indicated that H-21 and H-22 have a trans relationship.
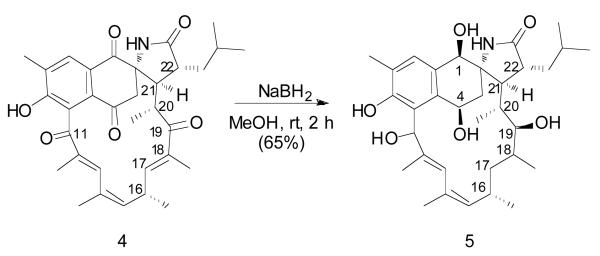
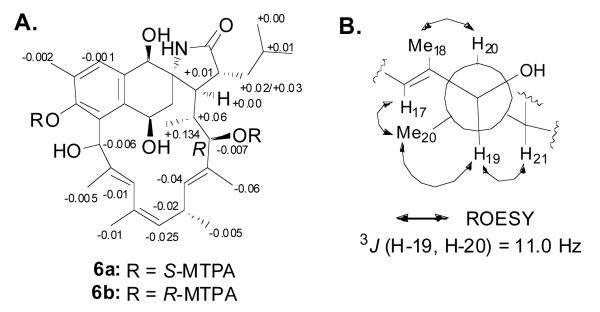
To determine the absolute configuration of C-20, we reduced the ketones in 4 (Scheme 1) and then applied the advanced Mosher's method6 on the major product, 1,4,11,19-octahydroansalactam A (5). Preparation of bis-S-MTPA ester 6a and bis-R-MTPA ester 6b (Figure 2A) provided positive ΔδS-R values for H-20, 20-Me, H-21, and H-22, and negative ΔδS-R values for H-16, 16-Me, H-17, 18-Me, thereby permitting the absolute configuration of C-19 to be assigned as R (Figure 2A). Next, the 11.0 Hz vicinal coupling between H-19 and H-20 indicated their anti-relationship. ROESY correlations between 20-Me and H-17, between 20-Me and H-19, between H-20 and Me-18, and between H-19 and H-21 revealed an R configuration for C-20 (Figure 2B).
Scheme 1.
Preparation of polyol 5 from ansalactam A (4).
Figure 2.
Mosher's ester analysis of the polyol 5. (A) ΔδS-R of 1H for S- and R-bis-MTPA esters of 5, and (B) rotation model for determination of relative configurations for C-19 and C-20
We undertook HETLOC7 and HSQMBC8,9 NMR experiments to establish the configurations of C-21 and C-22 but were unable to measure the coupling constants of 3J(H-20, C-22) and 3J(C-19, H-21) because neither signal was observed in either pyridine-d5 or methanol-d4. Thus, we carefully analyzed the ROESY NMR data to assign their configurations. There were only two possible stereoisomers, (20R, 21R) or (20R, 21S), for the alcohol product 5 (Figure S1, Supporting Information). The strong ROESY correlations between H-21 and both H-3 methylene protons supported the 21R configuration. These data allowed the absolute configurations at C-2, C-20, C-21, C-22 to be assigned as S, R, R, R, respectively.
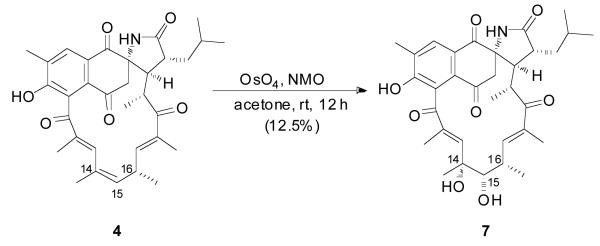
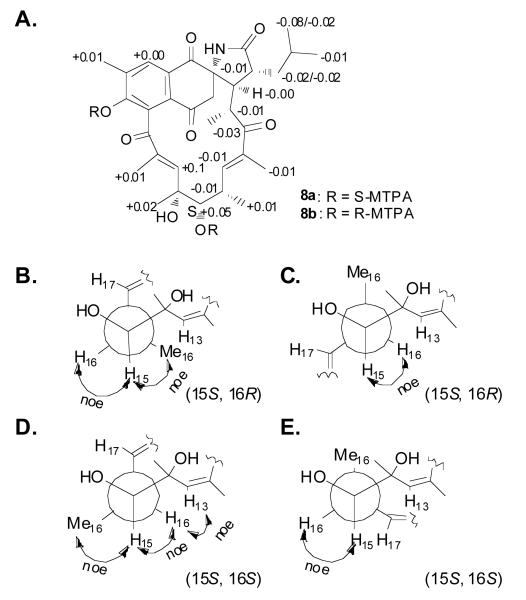
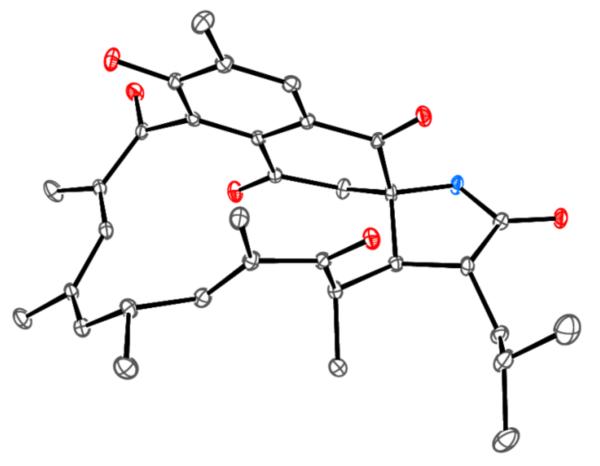
We performed synthetic modifications to determine the absolute configuration of the last remaining center at C-16. OsO4 treatment gave the corresponding 14,15-diol product, 7, (Scheme 2),10 which was then subjected to Mosher esterification to give the bis-S-MTPA ester 8a and bis-R-MTPA ester 8b (Figure 3). Calculation of ΔδS-R values, derived from NMR data from the MTPA esters, established the absolute configuration of C-15 as S. To establish the relative configurations of C-15 and C-16, we measured ROESY correlations of H-15 with H-16 and Me-16 and of H-16 with H-13, thereby permitting the assignment of C-16 as R. The absolute stereochemistry and the overall structure of 4 were confirmed by single-crystal X-ray analysis, which confirmed the absolute configuration of ansalactam A (4) as 2S,16R,20R,21R,22R (Figure 4).
Scheme 2.
Preparation of diol 7 from ansalactam A (5)
Figure 3.
Mosher's ester analysis of the diol 7. (A) ΔδS-R of 1H for S- and R-bis-MTPA esters of 7, and (B-E) rotation models for determination of relative configuration for C-16.
Figure 4.
ORTEP plot of the final X-ray structure of 5 depicting the absolute configuration.
Biosynthetic studies
There are several features of 4 that make it unique amongst previously reported ansamycins. Notably, the AHBA-derived amino group is involved in a γ-lactam residue with the aliphatic side chain. This lactam ring is spiro fused to the naphthalenic backbone, which is reduced in comparison to other ansamycins. A second unusual structural feature involves the C-22-C-27 aliphatic side chain of the lactam that suggests the possibility of a novel polyketide biosynthetic building block. To address whether 4 is indeed a new member of the ansamycin structure class and to explore the concept of a new PKS building block involved in 4 assembly, we explored its biosynthesis.
To determine whether Streptomyces sp. strain CNH-189 had the biosynthetic capacity to synthesize 4, we first employed a molecular approach and tested for the presence of a gene-encoded AHBA synthase, which is indicative of this structure class. Using degenerate PCR primers designed for the high throughput identification of AHBA synthases,11 we amplified a ~755 bp product from the genomic DNA of strain CNH-189. Phylogenetic analysis of the amplicon revealed that the CNH-189 AHBA synthase claded with AHBA synthases from characterized naphthalenic ansamycin gene clusters (Figure S2, Supplemental Information), which is consistent with the structure of 4.
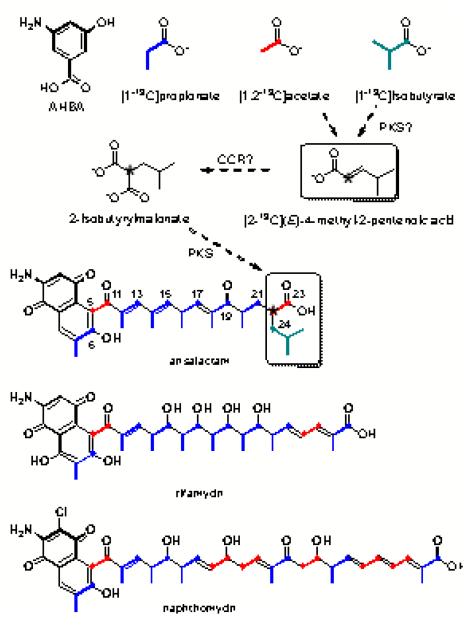
To explore the origin of the carbon backbone of 4, we next administered 13C-labeled precursors to strain CNH-189. Due to peak broadening of key carbon signals (C-20, 20-Me, C-21, and C-22) in the 13C NMR spectrum of 4, the 13C-enriched natural product was reduced with NaBH4 to 5 to provide sharp NMR signals for isotope analyses. Based on biosynthetic precedence in the rifamycin and naphthomycin series,3 we predicted that the polyketide backbone of 4 is assembled from malonate, six methylmalonates, and possibly 2-isobutyrylmalonate as a new PKS extender unit. As expected, sodium [1-13C]propionate enriched six carbons at ~17% corresponding to the carboxy position of each predicted propionate-derived extender unit (Figure 5). Similarly, sodium [1,2-13C]acetate was assimilated intact into the predicted C-5/C-11 unit (2J = 48 Hz). Additionally, we measured a second incorporated acetate unit at C-22/C-23 (2J = 46 Hz), suggesting acetate is incorporated into the C-1/C-2 position of the putative 2-isobutyrylmalonate extender unit (Figure 5).
Figure 5.
Biosynthetic origin of the carbons of the linearized PKS precursors of 4 compared to well characterized ansamycins and proposed biosynthesis of the C-22-C-27 branched extender unit.
Formation of the 2-isobutyrylmalonate polyketide extender unit
We first hypothesized that the non-traditional extender unit of 4 was derived from L-leucine, which would be consistent with incorporation of acetate at C-22/C-23. However, neither [1-13C]leucine or [1,2-13C]leucine could be incorporated. Consequently, we next postulated that the 2-isobutyrylmalonate unit may instead originate from the condensation of isobutyrate and acetate (via malonate) and that the resulting olefinic diketide would be utilized as the terminal PKS building block of 4. Support for this proposal was first provided by the specific incorporation of sodium [1-13C]isobutyrate at C-24 (4%) (Figure 5).
Recently, a new paradigm has surfaced for the assembly of 2-substituted malonyl-CoAs in polyketide biosynthesis in which α,β-unsaturated acyl-CoAs are reductively carboxylated with crotonyl-CoA reductase/carboxylase (CCR) homologs. Examples include ethylmalonate12,13 (ie, monensin A and tylosin), chloroethylmalonate14 (salinosporamide A), propylmalonate15 (salinosporamide E), allylmalonate16,17 (FK506), and putatively 2-(2-methylbutyryl)malonate18 (polyoxypeptin A) and 2-isovalerylmalonate19 (leupyrrin). In the case of the unique FK506 extender unit allylmalonyl-CoA, a discrete diketide synthase initiates its biosynthesis via the intermediate (E)-2-pentenyl-acyl carrier protein prior to CCR-mediated reductive carboxylation by TcsC. A similar pathway may also operate in 4 biosynthesis to give 2-isobutyrylmalonyl-CoA via (E)-4-methyl-2-pentenyl-CoA. To further explore this scenario, we synthesized [2-13C](E)-4-methyl-2-pentenoic acid (see Experimental Methods for details) and administered it to strain CNH-189. The resultant 4 was reduced and analyzed by 13C NMR to reveal a specific enrichment at C-22 (2%), thereby providing further support that the six-carbon C-22–C-27 fragment in 4 may be derived from 2-isobutyrylmalonyl-CoA via CCR-mediated biochemistry (Figure 5).
In conclusion, we isolated and structurally characterized a unique ansamycin natural product, ansalactam A (4), which incorporates a novel branched-chain PKS extender unit into its ansa side chain. We are presently exploring the bioactivity of 4 and a series of interesting structural analogs from this strain and will report those findings along with further biosynthetic and genomic studies in due course.
Experimental Methods
General experimental procedures
The optical rotations were measured using a Rudolph Research Autopol III polarimeter with a 10-cm cell. UV spectra were recorded in a Varian Cary UV-visible spectrophotometer with a path length of 1 cm and IR spectra were recorded on a Perkin-Elmer 1600 FTIR spectrometer. 1H and 2D NMR spectra data were recorded at 500 or 600 MHz in DMSO-d6, pyridine-d5, or methanol-d4 solution containing Me4Si as internal standard on Varian Inova spectrometers. 13C NMR spectra for structure determination were acquired at 75 MHz on a Varian Inova spectrometer. 13C NMR spectra for 13C-labeling studies were acquired at 125 MHz on a Varian VX-500 spectrophotometer equipped with an XSens cold probe. High resolution ESI-TOF mass spectra were provided by the Scripps Research Institute, La Jolla, CA or by the mass spectrometry facility at the UCSD Department of Chemistry. Low resolution LC/MS data were measured using a Hewlett-Packard series 1100 LC/MS system with a reversed-phase C18 column (Phenomenex Luna, 4.6 mm × 100 mm, 5 μm) at a flow rate of 0.7 mL/min.
Cultivation and extraction
The bacterium (strain CNH-189) was isolated from a near-shore marine sediment collected off Oceanside, California. It was identified as a Streptomyces sp. based on 16S rRNA gene sequence analysis (accession number HQ214120). It was cultured in sixty 2.8 L Fernbach flasks each containing 1 L of A1 production media (10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3•4H2O, 100 mg KBr) and shaken at 230 rpm at 27 °C. After seven days of cultivation, sterilized XAD-16 resin (20 g/L) was added to adsorb the organic products, and the culture and resin were shaken at 215 rpm for 2 hours. The resin was filtered through cheesecloth, washed with deionized water, and eluted with acetone. The acetone was removed under reduced pressure, and the resulting aqueous layer was extracted with EtOAc (3 × 500 mL). The EtOAc-soluble fraction was dried in vacuo to yield 4.5 g of crude extract.
Isolation of ansalactam A
The crude extract (4.5 g) was fractionated by open column chromatography on silica gel (25 g), eluted with a step gradient of CH2Cl2 and MeOH. The dichloromethane/methanol 50:1 fraction contained a mixture of ansalactams, which were purified by reversed-phase HPLC (Phenomenex Luna C-18 (2), 250 × 100 mm, 2.0 mL/min, 5 μm, 100 Å, UV = 210 nm) using an isocratic solvent system from 65% CH3CN to afford ansalactam A (4, 70.0 mg).
Ansalactam A (4)
yellow oil; [α]D21 −78 (c 0.5, MeOH); IR (KBr) νmax 3354, 2962, 1691, 1331, 1025, 746 cm−1; UV (MeOH) λmax (log ε) 225 (4.4), 250 (4.3), 290 (4.2), 340 (3.2) nm; 1H and 2D-NMR (500 MHz, methanol-d4), see Table 1; HRESIMS [M+H]+ m/z 546.2852 (C33H40NO6, calcd [M+H]+ 546.2850).
Reduction of ansalactam A (4) to yield polyol 5
Ansalactam A (4, 20 mg) was dissolved in 2 mL of dry MeOH. Twenty milligrams of NaBH4 was added to the solution, and the reaction mixture was stirred for 2 hours. The reaction was quenched by 2.5% aqueous NH4Cl solution and then the mixture was extracted with EtOAc. The solvent was removed in vacuo and the residual material was purified by reversed-phase HPLC (Phenomenex Luna 5μ C18 (2) 100 Å, 250 × 100 mm, 2.0 mL/min, UV detection at 210 nm) using an isocratic solvent system with 65% CH3CN in H2O. The polyol 5 (13.0 mg) was obtained with a retention time of 18 min.
1,4,11,19-octahydroansalactam A (5)
pale yellow oil; [α]D21 +10 (c 0.2, MeOH); 1H NMR (600 MHz, methanol-d4): δ 7.45 (1H, s, H-8 ), 5.78 (1H, s, H-11), 5.53 (1H, s, H-13), 5.44 (1H, d, J = 6.2 Hz, H-4), 5.36 (1H, d, J = 8.0 Hz, H-15), 4.94 (1H, d, J = 11.0 Hz, H-17), 4.48 (1H, s, H-1), 3.54 (1H, d, J = 6.2 Hz, H-19), 3.24 (1H, ddd, J = 14.0, 11.0, 6.6 Hz, H-22), 2.54 (1H, m, H-16), 2.40 (1H, dd, J = 14.0, 6.2 Hz, H2-3), 2.33 (1H, ddt, J = 11.0, 6.8, 1.0 Hz, H-20), 2.25 (1H, dd, J = 11.0, 1.0 Hz, H-21), 2.22 (3H, s, 7-Me), 2.22 (1H, m, H-25), 2.01 (3H, s, 12-Me), 2.00 (1H, d, J = 14.0 Hz, H2-3), 1.84 (3H, s, 14-Me), 1.55 (3H, s, 18-Me), 1.50 (1H, m, H2-21), 1.36 (1H, m, H2-21), 1.02 (3H, d, J = 6.8 Hz, 16-Me), 0.95 (3H, d, J = 6.8 Hz, H-26), 0.92 (3H, d, J = 6.8 Hz, H-27), 0.68 (3H, d, J = 6.8 Hz, 20-Me); 13C NMR (75 MHz, methanol-d4): δ 183.0 (C-23), 153.2 (C-6), 142.9 (C-18), 141.7 (C-12), 137.9 (C-15), 134.6 (C-10), 132.7 (C-14), 132.0 (C-9), 131.1 (C-17), 129.1 (C-5), 128.4 (C-13), 128.1 (C-8), 125.6 (C-7), 80.0 (C-19), 77.9 (C-1), 77.5 (C-11), 65.6 (C-4), 62.5 (C-2), 57.4 (C-21), 45.2 (C-3), 43.6 (C-24), 43.4 (C-22), 41.1 (C-20), 32.2 (C-16), 27.0 (C-25), 24.7 (C-27), 23.5 (20-Me), 23.0 (14-Me), 22.2 (C-26), 20.8 (18-Me), 19.4 (16-Me), 18.3 (12-Me), 17.0 (7-Me); ESI-MS m/z 554 [M+H]+, ESI-MS m/z 576 [M+Na]+.
Bis-MTPA esters of 5 (6a/6b)
Polyol 5 (5.0 mg) was dissolved in freshly distilled dry pyridine (2 ml) and several dry crystals of dimethylaminopyridine were added. The mixture was stirred for 15 min at RT. Treatment with R-(-)-MTPA-Cl yielded S-MTPA at 60 °C after 12 hrs. The reaction was quenched with 2 mL of MeOH. After removal of solvent under vacuum, the residue was purified by reversed-phase HPLC (Phenomenex Luna 5u C18 (2) 100 Å, 250 × 100 mm, 2.0 mL/min, UV detection at 210 nm) using an isocratic solvent system from 95% CH3CN in H2O. One of the major products, bis-S-MTPA ester was obtained at 27 min. Bis-R-MTPA ester was prepared with S-MTPA-Cl in the same manner. ΔδS-R values for the S- and R-MTPA esters of 6a/6b were recorded in ppm in methanol-d4.
Bis-S-MTPA ester of 5 (6a)
1H NMR (600 MHz, methanol-d4): δ 7.66 (2H, d, J = 7.8 Hz), 7.58 (1H, s), 7.50-7.46 (4H, m), 7.47-7.36 (4H, m), 6.26 (1H, s), 5.35 (1H, d, J = 4.0 Hz), 5.33 (1H, d, J = 12.0 Hz ), 5.28 (1H, d, J = 5.0 Hz), 4.74 (1H, dd, J = 9.0, 6.0 Hz), 4.58 (1H, s), 4.57 (1H, s), 3.68 (3H, s), 3.46 (3H, s), 2.81 (1H, m), 2.77 (1H, m), 2.58 (1H, dd, J = 11.0, 10.0 Hz), 2.42 (1H, m), 2.22 (1H, m), 2.15 (3H, s), 1.91 (1H, m), 1.89 (1H, dd, J = 11.0, 4.4 Hz), 1.81 (3H, s), 1.70 (3H, s), 1.68 (1H, m), 1.34 (1H, m), 0.99 (3H, d, J = 6.8 Hz), 0.97 (3H, d, J = 6.8 Hz), 0.95 (3H, d, J = 6.8 Hz), 0.92 (3H, d, J = 6.8 Hz), 0.86 (3H, s); ESI-MS m/z 968 [M-H2O]+, ESI-MS m/z 990 [M+Na-H2O]+.
Bis-R-MTPA ester of 5 (6b)
1H NMR (600 MHz, methanol-d4): δ 7.71 (2H, d, J = 7.8 Hz), 7.53-7.49 (4H, m), 7.50 (1H, s), 7.48-7.35 (4H, m), 6.27 (1H, br s), 5.37 (1H, d, J = 12.0 Hz), 5.35 (1H, d, J = 4.0 Hz ), 5.27 (1H, d, J = 6.0 Hz), 4.76 (1H, dd, J = 9.0, 6.0 Hz), 4.58 (1H, br s), 4.55 (1H, s), 3.66 (3H, br s), 3.45 (3H, s), 2.831 (1H, m), 2.77 (1H, m), 2.59 (1H, dd, J = 11.0, 10.0 Hz), 2.32 (1H, m), 2.17 (1H, m), 2.15 (3H, s), 1.92 (1H, m), 1.85 (1H, dd, J = 11.0, 4.4 Hz), 1.81 (3H, s), 1.65 (1H, m), 1.33 (1H, m), 1.28 (3H, s), 0.97 (3H, d, J = 6.8 Hz), 0.96 (3H, d, J = 6.8 Hz), 0.93 (3H, d, J = 6.8 Hz), 0.92 (3H, s), 0.86 (3H, d, J = 6.8 Hz); ESI-MS m/z 968 [M-H2O]+, ESI-MS m/z 990 [M+Na-H2O]+.
Oxidation of Ansalactam A
To a solution of ansalactam A (4, 20.0 mg) in acetone (2 ml) were added 4-methylmorpholine N-oxide (NMO, 4.8 mg) and a solution of OsO4 in H2O (10 μL), and the mixture was stirred for 10 hr at RT. The reaction was quenched by the removal of acetone, and the residue was chromatographed using silica gel eluting with mixtures of EtOAc and MeOH (1:1). After removal of solvent under vacuum, the residue was purified by reversed-phase HPLC (Phenomenex Luna 5μ C18 (2) 100 Å, 250 × 100 mm, 2.0 ml/min, UV detection at 210 nm) using an isocratic solvent system 75% aqueous CH3CN (0.01% TFA). 7 (2.5 mg) was obtained at 18 min.
Vicinal Diol 7
pale yellow oil; [α]D21 5 (c 0.2, MeOH; 1H NMR (600 MHz, methanol-d4): δ 7.74 (1H, s, H-8), 6.80 (1H, d, J = 9.3 Hz, H-17), 6.11 (1H, s, H-13), 3.41 (1H, d, J = 1.0 Hz, H-15), 3.40 (1H, br s, H2-3), 3.34 (1H, m, H-20), 3.19 (1H, m, H-22), 2.95 (1H, d, J = 14.0, H2-3), 2.67 (1H, m, H-16), 2.31 (3H, s, 7-Me), 2.55 (1H, br s, H-21), 2.27 (3H, s, 12-Me), 2.00 (1H, m, H-25), 1.65 (1H, m, H2-24), 1.54 (1H, m, H2-24), 1.28 (3H, s, 14-Me), 1.24 (3H, br s, 20-Me), 1.22 (3H, s, 18-Me), 1.05 (3H, d, J = 6.8 Hz, 16-Me), 0.94 (3H, d, J = 6.8 Hz, H-26), 0.91 (3H, d, J = 6.8 Hz, H-27); ESI-MS m/z 580 [M+H]+, ESI-MS m/z 602 [M+Na]+.
Bis-MTPA esters of 7 (8a/8b)
Vicinal diol 7 (1.0 mg) was dissolved in freshly distilled dry pyridine (1 ml) and several dry crystals of dimethylaminopyridine were added. The mixture was stirred for 15 min at RT. Treatment with R-(-)-MTPA-Cl for 12 hrs at 60 °C yielded the S-MTPA ester. The reaction was quenched with 1 mL of MeOH. After removal of solvent under vacuum, the residue was purified by reversed-phase HPLC (Phenomenex Luna 5u C18 (2) 100 Å, 250 × 100 mm, 2.0 ml/min, UV detection at 210 nm) using an isocratic solvent system from 95% CH3CN in H2O (0.01% TFA). Bis-S-MTPA ester was eluted at 35 min. Bis-R-MTPA ester was prepared with S-MTPA-Cl in the same manner. ΔδS-R values for the S- and R-MTPA esters of 8a/8b were recorded in ppm in DMSO-d6.
Bis-S-MTPA ester of 7 (8a)
1H NMR (600 MHz, DMSO-d6) δ 7.99 (1H,s), 7.81 (1H, s), 7.59-7.44 (8H, m), 7.35 (3H, m), 6.43 (1H, d, J = 8.0 Hz), 5.89 (1H, s), 4.80 (1H, s), 3.51 (3H, s), 3.53 (1H, m), 3.47 (3H, s), 3.16 (1H, m), 3.06 (1H, m), 3.05 (1H, d, J = 14.6 Hz), 2.96 (1H, br s), 2.54 (3H, s), 2.18 (3H, s), 2.09 (3H, s), 1.99 (1H, m), 1.91 (1H, m), 1.51 (1H, m), 1.32 (1H, m), 1.25 (3H, s), 1.21 (3H, d, J = 6.8 Hz), 1.09 (3H, d, J = 6.8 Hz), 0.84 (3H, d, J = 6.8 Hz), 0.80 (3H, d, J = 6.8 Hz); ESI-MS m/z 1108 [M-H2O+CF3COOH+H]+, ESI-MS m/z 1130 [M-H2O+CF3COOH+Na]+.
Bis-R-MTPA ester of 7 (8b)
1H NMR (600 MHz, DMSO-d6) δ 8.02 (1H,s), 7.81 (1H, s), 7.59-7.46 (10H, m), 7.32 (1H, br s), 6.45 (1H, d, J = 8.0 Hz), 5.86 (1H, s), 4.75 (1H, s), 3.49 (3H, s), 3.53 (1H, m), 3.46 (3H, s), 3.17 (1H, m), 3.06 (1H, m), 3.05 (1H, d, J = 14.6 Hz), 2.96 (1H, br s), 2.54 (3H, s), 2.21 (3H, m), 1.99 (1H, m), 1.96 (3H, s), 1.91 (1H, m), 1.52 (1H, m), 1.35 (1H, m), 1.25 (3H, s), 1.23 (3H, d, J = 6.8 Hz), 1.08 (3H, d, J = 6.8 Hz), 0.85 (3H, d, J = 6.8 Hz), 0.81 (3H, d, J = 6.8 Hz); ESI-MS m/z 1108 [M-H2O+CF3COOH+H]+, ESI-MS m/z 1130 [M-H2O+CF3COOH+Na]+.
Identification of Streptomyces sp. strain CNH-189 AHBA synthase
Genomic DNA from Streptomyces sp. strain CNH-189, S. arenicola strain CNS-205, and S. tropica strain CNB-440 was extracted and purified using standard protocols.20 The 3-amino-5-hydroxybenzoic acid (AHBA) synthase gene (accession number HQ219709) was amplified from gDNA from each strain using primers and PCR conditions described by Huitu et al.11 The ~755 bp PCR-product from strain CNH-189 was recovered by gel electrophoresis and purified using a DNA fragment purification kit (Qiagen). The resulting DNA fragment was sequenced at both ends by Seqxcel (San Diego, CA). A neighbor joining distance tree was constructed in MEGA421 using aligned partial sequences (695 bp) from the NCBI database comprising AHBA synthase genes from characterized ansamycin biosynthetic gene clusters and from genome sequencing projects with no confirmed metabolite.
Stable isotope incorporation studies
For each of the stable isotope enrichment experiments, 20 mL of CNH-189 starter culture was inoculated into 2 X 1 L of A1 production media in 2.8 L Fernbach flasks and allowed to grow for 24 hrs before the addition of labeled substrate – sodium [1-13C]propionate, 50 mg/L; sodium [1,2-13C]acetate, 100 mg/L; sodium [1,2-13C]leucine, 100 mg/L; sodium [1-13C]leucine, 50 mg/L; [1-13C]isobutyrate, 30 mg/L; [2-13C]4-Me-2-pentenoic acid, 50 mg/L – dissolved in H2O. Cultures were grown for an additional 96 hrs before 4 was extracted, purified, and reduced to 5 for 13C NMR analysis as described above. 13C-enrichment was calculated as described by Bringmann et al.22
Synthesis of [2-13C]4-methyl-2-pentenoic acid
[2-13C]4-Methyl-2-pentenoic acid was synthesized from [2-13C]malonic acid and isobutyraldehyde by Doebner condensation according to reference 23. The spectroscopic data obtained was in good agreement with the literature: δH (CDCl3, 500 MHz) 7.06 (1H, dd, J = 15.7, 6.5 Hz, CHCOOH), 5.63 (1H, d, J = 15.7 Hz, CHCH(CH3)2), 2.50 (1H, m, CH(CH3)2), 1.09 (6H, d, J = 6.7 Hz, CH(CH3)2).23
Supplementary Material
ACKNOWLEDGMENT
Financial support was generously provided by the NIH (CA127622 to B.S.M. and CA044848 to W.F.), the California Sea Grant Program (R/NMP-100 to B.S.M. and P.R.J.) and a postdoctoral fellowship from the German Academic Exchange Service (DAAD) to T.A.M.G. We thank Drs. A. L. Rheingold and C. Moore for crystallography data and Drs. C. Hughes and X. Alvarez-Mico for their illuminating advice in the determination of stereochemistry.
Footnotes
Supporting Information Available: AHBA synthase phylogenetic analysis, 1H, 13C, and 2D NMR spectra for 4 and its synthesized derivatives, 13C NMR spectra for 13C-labeled 5, and complete ref 17. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Floss HG. J. Nat. Prod. 2006;69:158. doi: 10.1021/np058108l. [DOI] [PubMed] [Google Scholar]
- 2.August PR, Tang L, Yoon YJ, Ning S, Muller R, Yu TW, Taylor M, Hoffmann D, Kim CG, Zhang X, Hutchinson CR, Floss HG. Chem. Biol. 1998;5:69. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 3.Floss HG, Yu TW. Chem. Rev. 2005;105:621. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 4.Gulder TAM, Moore BS. Curr. Opin. Microbiol. 2009;12:252. doi: 10.1016/j.mib.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 6.Uhrin D, Barlow PN. J. Magn. Reson. 1997;126:248. [Google Scholar]
- 7.Williamson RT, Marquez BL, Gerwick WH, Kover KE. Magn. Reson. Chem. 2000;38:265. [Google Scholar]
- 8.Ohtani I, Kusumi T, Kashman Y, Kakisawa H. J. Am. Chem. Soc. 1991;113:4092. [Google Scholar]
- 9.Seco JM, Quinoa E, Riguera R. Chem. Rev. 2004;104:12. doi: 10.1021/cr2003344. [DOI] [PubMed] [Google Scholar]
- 10.VanReen V, Kelly RC, Cha DY. Tetrahedron Lett. 1976;17:1973. [Google Scholar]
- 11.Huitu Z, Linzhuan W, Aiming L, Guizhi S, Feng H, Qiuping L, Yuzhen W, Huanzhang X, Qunjie G, Yiguang W. J. Appl. Microbiol. 2009;106:755. doi: 10.1111/j.1365-2672.2008.04010.x. [DOI] [PubMed] [Google Scholar]
- 12.Erb TJ, Berg IA, Brecht V, Muller M, Fuchs G, Alber BE. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10631. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erb TJ, Brecht V, Fuchs G, Muller M, Alber BE. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8871. doi: 10.1073/pnas.0903939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12295. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Hazzard C, Eustaquio AS, Reynolds KA, Moore BS. J. Am. Chem. Soc. 2009;131:10376. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goranovic D, Kosec G, Mrak P, Fujs S, Horvat J, Kuscer E, Kopitar G, Petkovic H. J. Biol. Chem. 2010;285:14292. doi: 10.1074/jbc.M109.059600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mo S, et al. J. Am. Chem. Soc. accepted. [Google Scholar]
- 18.Umezawa K, Ikeda Y, Kawase O, Naganawa H, Kondo S. J. Chem. Soc., Perkin Trans. 1. 2001;2001:1550. [Google Scholar]
- 19.Bode HB, Wenzel SC, Irschik H, Hofle G, Muller R. Angew. Chem. Int. Ed. Engl. 2004;43:4163. doi: 10.1002/anie.200454240. [DOI] [PubMed] [Google Scholar]
- 20.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich: 2000. [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. Mol. Biol. Evol. 2007;24:1596. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Bringmann G, Haagen Y, Gulder TA, Gulder T, Heide L. J. Org. Chem. 2007;72:4198. doi: 10.1021/jo0703404. [DOI] [PubMed] [Google Scholar]
- 23.Fadnavis NW, Sharfuddin M, Vadivel SK, Bhalerao UT. J. Chem. Soc., Perkin Trans. 1. 1997:3577. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.