Abstract
The biochemical activity of a stunning diversity of cell types and organ systems is shaped by a 24-hour (circadian) clock. This rhythmic drive to a good deal of the transcriptome (up to 15% of all coding genes) imparts circadian modulation over a wide range of physiological and behavioral processes (from cell division to cognition). Further, dysregulation of the clock has been implicated in the pathogenesis of a large and diverse array of disorders, such as hypertension, cancer and depression. Indeed, the possibility of utilizing therapeutic approaches that target clock physiology (that is, chronotherapy) has gained broad interest. However, a deeper understanding of the underlying molecular mechanisms that modulate the clock, and give rise to organ-specific clock transcriptomes, will be required to fully realize the power of chronotherapies. Recently, microRNAs have emerged as significant players in circadian clock timing, thus raising the possibility that clock-controlled microRNAs could contribute to disorders of the human circadian timing system. Here, we highlight recent work revealing a key role for microRNAs in clock physiology, and discuss potential approaches to unlocking their utility as effectors of circadian physiology and pathophysiology.
Introduction
Deep within the brain ticks a 24-hour clock that imparts circadian control over virtually every aspect of human physiology. This clock is located in the suprachiasmatic nucleus (SCN), which is a tiny region of the ventral hypothalamus that contains approximately 20,000 neurons [1,2]. The inherent timing capacity of the SCN is derived from autonomous neuronal oscillators, which form a coherent and precisely phased pattern of rhythmic neuronal activity and hormonal output that serves as a phasing cue to ancillary oscillators found throughout the brain and peripheral organs [3,4]. The need for a circadian control center likely evolved as an adaptive response to the daily changes in the light-dark cycle resulting from the axial rotation of the earth. As such, the inherent clock timing process is tightly regulated by changes in the daily lighting cycle.
As discussed in detail below, dysregulation of the clock appears to play a central role in the genesis and progression of a broad range of disorders, and therapeutic approaches designed to target clock physiology (that is, chronotherapeutics) have been effective in treating a range of ailments, including seasonal affective disorder, asthma, hypertension and cancer [5-9]. Many of these treatment strategies are based on the integration of clock phase into the drug delivery schedule. However, if we are to develop novel chronotherapeutic approaches, then a deeper understanding of clock physiology (from cellular clock communication to mechanisms of molecular timing and organ-specific clock gene expression patterns) will be required. To this end, recent work by a number of groups has revealed a role for microRNAs in clock physiology. MicroRNAs are small (approximately 22 nucleotides), single-stranded, non-coding RNA species that act as potent gene silencers and are important to a diverse array of physiological and pathophysiological processes [10-13]. As we will discuss in detail below, microRNAs appear to play distinct roles in clock physiology, and dysregulation of microRNA expression can lead to marked alterations in circadian timing and output. Hence, microRNAs present novel therapeutic targets for disorders of the circadian clock. In this review, we will provide an overview of circadian biology, and highlight genome-scale and integrated approaches designed to reveal the role of microRNAs in clock physiology. Finally, we will provide some perspective on microRNAs as potential therapeutic targets for circadian disorders. By understanding the role of microRNAs in circadian biology on molecular, cellular and organ-systems levels, a cohesive and integrated approach to chronotherapeutics can be developed.
Circadian timing
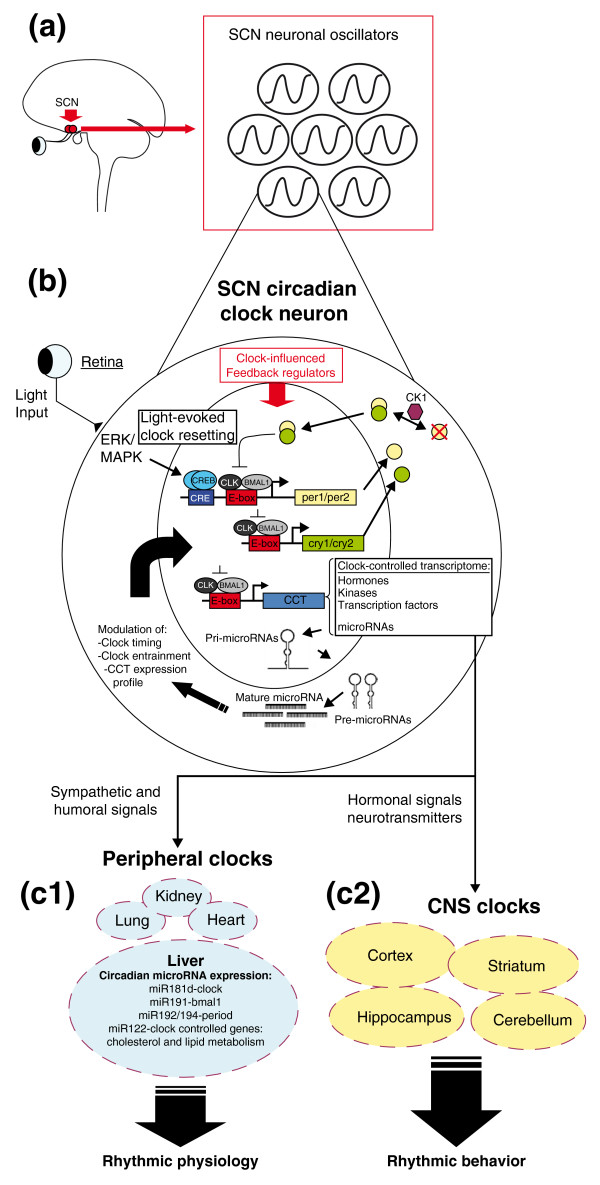
At a molecular level, the inherent 24-hour time-keeping capacity of the SCN stems from a well-characterized transcriptional/post-translation feedback loop (Figure 1). Central to this molecular clock is the rhythmic expression of the period (per1 and per2) and cryptochrome (cry1 and cry2) families of genes. Circadian transcription of per and cry genes is mediated by an E-box-binding basic helix-loop-helix transcription factor consisting of CLOCK (or NPAS2) and BMAL1. The gene products of per and cry accumulate in the cytoplasm, dimerize, enter the nucleus, and repress transactivation of the CLOCK-BMAL1 complex [14,15].
Figure 1.
Schematic overview of molecular circadian clock timing. (a) Suprachiasmatic nucleus (SCN; red circles) located within the ventral hypothalamus. The two nuclei serve as the master circadian clocks, orchestrating rhythmicity and phasing of ancillary clocks found in peripheral organ systems (c1) and throughout the brain (c2). Entrainment of the SCN clock to light is mediated by a direct projection from the retina. (b) The molecular clock feedback loop is centered on the rhythmic expression of the period (per1/2) and cryptochrome (cry1/2) gene families. PERIOD and CRY dimers translocate to the nucleus and negatively regulate their own production via suppression of CLOCK (CLK)- and BMAL1-mediated transcription. Casein kinase 1 (CK1)-mediated phosphorylation triggers degradation of PER proteins, thus relieving transcriptional repression and, in turn, allowing for robust 24-hour clock gene cycling. In addition to the core clock genes, a large fraction of the transcriptome is under the direct or indirect influence of the circadian clock. The clock-controlled transcriptome (CCT) within the SCN includes hormones, kinases, transcription factors and microRNAs. Many of these gene products serve as phasing cues to ancillary oscillators, or function as feedback regulators of the core molecular clock. (c1) Clock-regulated microRNA expression in the liver: denotation of several rhythmically expressed microRNAs that are either predicted (that is, miR181 d and miR191 [69]) or have been shown (that is, miR192/194 [71]) to target components of the core molecular clock. MiR122 has been shown to modulate clock-controlled gene expression involved in hepatic cholesterol and lipid metabolism [72,74]. In total, these data strongly indicate that microRNAs play a central role in sculpting the circadian gene expression profile and, in turn, regulate associated physiological and behavioral processes.
If this negative feedback loop were the entire story, one would anticipate only a single robust round of per/cry gene expression would occur, and then a steady, basal level of gene expression would persist. Rather, rhythmic relief of per/cry repression is achieved in part via progressive phosphorylation of PER by casein kinase 1 delta and epsilon, which targets the proteins for polyubiquitylation and degradation via the 26 S proteosomal pathway [16,17].
Here, it should also be pointed out that this core molecular feedback loop is influenced by multiple clock-influenced feedback regulators, including kinases such as protein kinase A and the extracellular signal-related kinase (ERK)/mitogen-activated protein kinase (MAPK) cascade. Likewise, oscillations in transcriptional pathways mediated by the nuclear hormone receptor Rev-erb, the bZIP transcription factor E4BP4 [18] and the basic helix-loop-helix transcription factors Dec1 and Dec2 [19] modulate the amplitude and phasing of the clock and also serve as output pathways that impart circadian control over an array of biochemical and physiological processes [20,21].
Entrainment of the biological clock to external time cues appears to be mediated in part by rapid changes in the expression of per1 and per2. Along these lines, photic input drives a rapid increase in clock gene expression via an ERK/MAPK-cAMP response-element-binding protein (CREB) signaling cassette in the SCN [22] (Figure 1). Conversely, non-photic entrainment cues have been shown to trigger a rapid degradation of per1 and per2 expression [23]. As state variables of the clock, changes in the expression of per1 and per2 would lead to resetting of the molecular oscillator, and in turn re-entrainment of the circadian rhythm.
A deeper understanding of the functional significance of the circadian timing process was realized through gene-array-based analysis, which showed that large and diverse functional classes of genes within the SCN are under the control of the core-clock timing processes [24,25]. Likewise, robust rhythmicity in the periphery has been demonstrated for tissues, including the liver and heart [26-28]. A truly remarkable finding was the organ-specific nature of circadian gene expression profile. Hence, beyond the canonical core clock genes, only small subsets of genes have been found to oscillate in multiple organ systems; along these lines, Panda et al. [25] reported that 28 genes (out of approximately 650) were found to oscillate in both the SCN and the liver. These findings reveal that clock-controlled gene expression is tailored to match the physiological role of the tissue.
Circadian modulation of disease
The finding that circadian timing mechanisms are distributed through the body raises important questions about the relevance of the clock to human health and disease. Some of the most interesting work on this subject has utilized animal models to assess the physiological effects that disruption of timing has on disease progression. Along these lines, work by Filipski et al. [29] showed that destruction of the SCN accelerated malignant tumor growth by up to threefold. Likewise, experimental models that lead to a marked disruption of clock synchrony, such as constant light, and repetitive shifting of the light-dark cycle, have been shown to accelerate tumor growth and, relatedly, increase mortality rates [30-32]. In a telling parallel to these studies, a number of epidemiological studies have found that shift workers are at an elevated risk for several types of cancer, including breast, colorectal and prostate cancer [33-35]. Given work showing that the circadian clock modulates hormone output, immune cell function and cell division, the postulation that disruption of the circadian clock occurs as part of the disease progress may not be surprising. However, many of these studies have also hinted at the possibility that disruption of the clock may be a key factor in the initiation of a disease state. In this regard, work in mouse models in which components of the core clock timing process are disrupted via genetic-based approaches have been particularly enlightening. Along these lines, Clock mutant mice are obese and hyperphagic, exhibiting hyperleptinemia, hepatic steatosis hyperglycemia and hypoinsulinemia [36]. In an interesting affirmation of the connection between the clock and metabolic disorders, SNP analysis of lean and obese humans has revealed an association between CLOCK polymorphisms and susceptibility to obesity [37]. In addition, haplotype analysis has identified CLOCK gene variants that may be protective against the development of obesity or that contribute to the development of metabolic syndrome, type 2 diabetes and cardiovascular disease [38]. It also appears that key components of the molecular clock are affected by alterations in metabolic activity. Along these lines, mice maintained on a high-fat diet, thus leading to obesity and metabolic syndromes, exhibited a marked alteration in core Clock gene expression within the liver and kidneys [39]. Hence, one could envision an intertwining of genetic and lifestyle factors that impact metabolic fitness via alterations in circadian clock function.
Disruption of clock timing has also been shown to contribute to an array of nervous system disorders. Along these lines, familial advanced sleep phase syndrome (FASPS) is an autosomal dominant disorder characterized by a 3- to 4-hour advance in sleep onset and awakening [40]. Hence, an individual with FASPS may fall asleep at 7 pm rather than 11 pm, and with arousal at 3 am instead of 7 am. Linkage analysis revealed that FASPS was localized to a point mutation in the PER2 gene [41]. At a mechanistic level, this mutation is thought to eliminate a phosphorylation motif for casein kinase 1 epsilon, thus leading to excessive accumulations of PER2 and, in turn, an altered/advanced circadian cycle, initiating premature drive for sleep onset. Interestingly, dysregulation of the clock also appears to contribute to disturbances of the sleep-wake cycles in individuals with Alzheimer's disease. These spontaneous night-time awakenings in patients with Alzheimer's disease are one of the most challenging issues for caregivers [42]. Likewise, individuals suffering from Huntington's disease exhibit clock-regulated disruption of neuroendocrine output, and of the sleep-wake cycles [43-45]. These examples are a mere shortlist of the profound functional roles that the clock plays in organismal physiology. The role of the clock in such diverse processes as cell division and xenobiotic responses, and its roles in regulation of behavior, cognition, arousal and aging are reviewed elsewhere [46-48].
Molecular control through microRNA
To further our understanding of the molecular underpinnings of the circadian clock, researchers have begun to explore the potential role of the non-coding class of small RNA species known as microRNAs. MicroRNAs are potent negative regulators of mRNA translation and, as such, play a central role in sculpting gene expression patterns. MicroRNAs are principally coded within introns and exons of non-coding RNA (that is, intergenic microRNA), and from introns of protein encoding genes (that is, introgenic microRNA) [49,50]. MicroRNAs are frequently coded close to other microRNAs and transcribed as long RNA that contains multiple microRNAs in a cluster (polycistronic microRNAs) [50,51]. The biogenesis of microRNAs occurs via a multistep process, which is described in detail elsewhere [52-54]. In short, microRNAs are transcribed by RNA polymerase II as long single-stranded RNA, forming stem-loop (hairpin) structures, referred to as primary microRNAs. Primary microRNAs are processed to precursor microRNAs (55 to 70 nucleotides) by a protein complex of RNAase III enzyme Drosha and the double-stranded RNA-binding domain protein DGCR8/Pasha in the nucleus [55-57]. Precursor microRNAs are then exported to the cytoplasm where they are further processed to approximately 22 bp microRNA/microRNA* double-stranded species by a protein complex of RNAase III enzyme, Dicer, and double-stranded RNA-binding domain proteins TRBP/PACT/loquacious [58,59]. Finally, microRNAs are loaded into the RNA-induced silencing complex, through which the gene silencing function of microRNAs occurs via a reduction in mRNA translation efficiency or mRNA stability [60-62].
MicroRNA and clock timing
Rhythmic control of microRNA expression appears to be conserved from plants to mammals, and has significant consequences for circadian timing. Along these lines, genome-wide tiling arrays have been used to profile the transcriptome of Arabidopsis thaliana as a function of the circadian cycle. Beyond the observation that approximately 25% of the coding genes exhibited circadian rhythms of expression, several rhythmically regulated microRNAs (that is, miR160B and miR167D) were identified [63]. This experiment was not designed to detect microRNA, and thus it is likely that the results underestimate the total number of oscillating microRNAs.
In addition to circadian control mechanisms, biologically relevant time cues, such as the daily light cycle, also drive rhythmic microRNA expression. Along these lines, Arabidopsis miR167, miR168, miR171 and miR398 have recently been shown to oscillate, with higher levels during the daytime than during the night [64] (Table 1). Interestingly, rhythmicity of these microRNAs was not governed by an internal circadian clock, suggesting photic control of some microRNAs. Within humans, it would not be surprising to observe a similar pattern of microRNA expression driven by the clock, exogenous time cues or both.
Table 1.
Light and clock-controlled microRNAs
| Model | Organ | MicroRNAs | Reference(s) |
|---|---|---|---|
| Cell culture | miR328, miR383 | [70] | |
| Arabidopsis | miR160, miR167, miR168, miR171, miR398 | [63,64] | |
| Drosophila | miR263 | [65] | |
| Chicken | Photoreceptors | miR26a | [78] |
| Rat | Intestine | miR16, miR20a, miR141 | [76] |
| Mouse | Liver | miR122, miR181, miR191 | [69,73-75] |
| Retina | miR96, miR103, miR106, miR124, miR182, miR422 | [77] | |
| Suprachiasmatic nucleus | miR219, miR132 | [12] | |
| Human | miR182 | [81] |
Circadian-regulated microRNA expression has also been described in Drosophila melanogaster. Array analysis of wild-type flies revealed that miR263a and miR263b showed robust circadian oscillations, which were absent in clock mutant flies [65] (Table 1). In a later study, Kadener et al. [66] found that abrogation of microRNA biogenesis led to both a marked rise in circadian-regulated gene expression and a disruption of circadian-regulated behavioral rhythms, thus indicating a role for microRNA in clock timing. AGO1 pull-down assays revealed that gene products involved in clock timing were targeted for microRNA-mediated degradation [64], indicating that core clock molecular feedback is not only regulated at a transcriptional and post-translational level, but also through post-transcriptional/microRNA-mediated control mechanisms.
In the mammalian SCN, microRNAs play a role in clock timing and entrainment. Cheng et al. [12] demonstrated that both miR132 and miR219 show oscillatory expression in wild-type mice, but not in circadian mutant mice. Both microRNAs have CRE-enhancer sequences allowing for CREB-dependent regulation [67], but only miR132 was shown to be inducible by light [12]. Importantly, miR219 appears to be under direct control of the molecular clock via the binding of CLOCK and BMAL1 to an E-box within its promoter region. In contrast, miR132 does not contain an E-box motif, and thus its rhythmicity is likely driven by an ancillary CREB-actuated transcriptional oscillator [12]. In vivo antisense silencing studies demonstrated that miR219 shortens the circadian period and that miR132 negatively regulates light-dependent resetting of the clock [12]. Both miR132 and miR219 affect per1 expression, and thus influence the core circadian transcriptional loop [12,68]. However, neither miR132 nor miR219 appears to directly target per1; thus, the precise mechanisms by which these microRNAs affect the clock are not known.
MicroRNA and peripheral circadian oscillators
In addition to rhythmic microRNA expression in the SCN clock, peripheral oscillators also exert circadian control over microRNA expression. A microarray-based study of mouse liver revealed that over 13% of the probed microRNAs (85 out of 640) exhibited circadian expression patterns [69]. Target prediction analysis indicated that a number of cycling microRNAs were capable of binding to components of the core clock timing process and, for a subset of these microRNAs, there was an inverse expression pattern with their predicted clock gene targets. For example, expression of miR181 d and its putative target CLOCK, and miR191 and its putative target BMAL1, were inversely correlated [69]. Conversely, miR328 and miR383 were positively correlated with the expression of their corresponding targets, per1 and cry. This unexpected relationship may be the result of a phase-lag in downregulation, or it may represent an example of positive microRNA regulation that has been uncovered in recent years [70].
In another study, a forward genetic screen was used to identify the miR192/194 gene cluster as inhibitors of the per (per1 to 3) gene family [71]. Significantly, the miR192/194 cluster is highly expressed in the liver and kidneys (two oscillating organs) [72] and overexpression studies found that miR192/194 shortens the length of the circadian rhythm via a decrease in per gene expression.
The expression of miR122, a known regulator of liver metabolism [73], is also subject to circadian control. Although the mature strand microRNA does not oscillate, both primary and precursor miR122 show robust daily changes in expression, suggesting that transcription is under circadian control [74]. Knockdown of miR122 revealed a marked alteration in the expression of clock-controlled genes, many of which are involved in cholesterol and lipid metabolism. In an additional study, Kojima et al. [75] found that the rhythmic expression of the mRNA deadenylase Nocturnin is modulated by miR122 in the liver, suggesting that miR122 is a key intermediate in circadian regulation of hepatic metabolic activity.
Intestine physiology is also governed by the circadian oscillator, which synchronizes enterocyte proliferation and, in turn, nutrient absorbance. Within this context, it is interesting that miR16, miR20a and miR141 all oscillate in the intestine, and that miR16 regulates the rate of proliferation in jejunal crypts by suppressing essential G1/S cell cycle regulators [76]. Hence, microRNA may serve as a link in the circadian control of intestinal physiology.
Finally, several microRNAs have been found to exhibit diurnal oscillations in the mouse retina, including miR96, miR124a, miR103, miR182, miR106b, miR422a and miR422b [77]. Of these, both miR96 and miR182 show potential targeting and downregulation of the clock-regulated gene adenylyl cyclase 6. In a separate study, miR26a, which is regulated by both CLOCK and CREB, was found to drive circadian rhythms in the production of the L-type voltage-gated calcium channel α1C subunit in cone photoreceptors [78]. This observation is of functional significance, given the central role that L-type calcium channels play in rhythmic melatonin synthesis and, in turn, circadian retinal physiology [79,80]. Finally, in humans, a polymorphism in pre-miR182 (which exhibits diurnal rhythmicity in the retina [77]) is significantly associated with major depression [81]. Relative to wild-type miR182, mutant miR182 was found to trigger a marked reduction in the expression of the circadian gene Clock. These data raise the possibility that mutant miR182 contributes to circadian clock ailments associated with major depression [82].
Chronotherapeutic approaches and future outlook
The finding that microRNA affects key clock timing processes (that is, the core molecular oscillator, clock entrainment and clock output) raises the prospect that chronotherapeutic approaches designed to affect microRNA physiology might prove efficacious.
Along these lines, some of the most tractable ailments associated with clock physiology may result from a damped SCN oscillator, which is observed as part of the aging process [83-85]. Hence, approaches to increase the robustness of the clock timing process may offset fragmentation of the sleep-wake cycle, which is often observed as a function of aging. This approach could also prove beneficial to individuals suffering from age-regulated neurodegenerative disorders, such as Alzheimer's disease. Therapeutic approaches could take the form of microRNA antisense or 'sponge' constructs [86], designed to target particular microRNAs, and in turn limit their ability to suppress components of the core clock timing process, or clock output genes. Of note, in mouse models, microRNA knockdown using cholesterol-conjugated 2-O-methyl RNA antisense (that is, antagomirs [87]) has already been used to assess the functional roles of miR132 and miR219 in SCN clock physiology [12]. Conversely, transgenic RNA interference-based approaches designed to harness the endogenous microRNA processing machinery could be used to selectively uncouple an aberrant clock-timing gene or clock-output gene from a specific physiological process. This approach could prove particularly useful as part of a cancer treatment strategy.
Obviously, there are numerous challenges that will need to be addressed for these types of approaches to be implemented. For example, the inability of RNA constructs (for example, antagomirs and sponges) to cross the blood-brain barrier is a significant impediment to minimally invasive therapeutic strategies. Further, issues related to the timing of antisense administration and organ-specific therapeutic targeting will need to be addressed. At the least, the practical application of these methods will require a deeper understanding of microRNA biology, the clock, and its role in the regulation of a myriad of gene networks that contribute to human physiology and pathology.
Abbreviations
CREB: cAMP response-element-binding protein; ERK: extracellular signal-related kinase; FASPS: familial advanced sleep phase syndrome; MAPK: mitogen-activated protein kinase; SCN: suprachiasmatic nucleus; SNP: single-nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
All three authors contributed to the writing of the manuscript.
Contributor Information
Katelin F Hansen, Email: hansen.293@buckeyemail.osu.edu.
Kensuke Sakamoto, Email: Sakamoto.20@osu.edu.
Karl Obrietan, Email: Obrietan.1@osu.edu.
Acknowledgements
This work was supported by grants MH62335, NS066345 and NS067409 from the National Institutes of Health.
References
- Moore RY, Speh JC, Leak RK. Suprachiasmatic nucleus organization. Cell Tissue Res. 2002;309:89–98. doi: 10.1007/s00441-002-0575-2. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ. A clinician's primer on the circadian clock: its localization, function, and resetting. Adv Intern Med. 1993;38:81–106. [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Winkler D, Pjrek E, Iwaki R, Kasper S. Treatment of seasonal affective disorder. Expert Rev Neurother. 2006;6:1039–1048. doi: 10.1586/14737175.6.7.1039. [DOI] [PubMed] [Google Scholar]
- Magnusson A, Boivin D. Seasonal affective disorder: an overview. Chronobiol Int. 2003;20:189–207. doi: 10.1081/CBI-120019310. [DOI] [PubMed] [Google Scholar]
- Innominato PF, Levi FA, Bjarnason GA. Chronotherapy and the molecular clock: clinical implications in oncology. Adv Drug Deliv Rev. 2010;62:979–1001. doi: 10.1016/j.addr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Portaluppi F, Smolensky MH. Perspectives on the chronotherapy of hypertension based on the results of the MAPEC study. Chronobiol Int. 2010;27:1652–1667. doi: 10.3109/07420528.2010.510788. [DOI] [PubMed] [Google Scholar]
- Burioka N, Fukuoka Y, Koyanagi S, Miyata M, Takata M, Chikumi H, Takane H, Watanabe M, Endo M, Sako T, Suyama H, Ohdo S, Shimizu E. Asthma: chronopharmacotherapy and the molecular clock. Adv Drug Deliv Rev. 2010;62:946–955. doi: 10.1016/j.addr.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. MicroRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss AW, Chen W. MicroRNAs in brain function and disease. Curr Neurol Neurosci Rep. 2008;8:190–197. doi: 10.1007/s11910-008-0031-0. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–2980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG. E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Bioessays. 2002;24:1023–1029. doi: 10.1002/bies.10176. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Mrosovsky N, Field MD, Hastings MH. Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc Natl Acad Sci USA. 1999;96:15211–15216. doi: 10.1073/pnas.96.26.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol. 2003;15:991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, Claustrat B, Hastings MH, Levi F. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- Filipski E, Delaunay F, King VM, Wu MW, Claustrat B, Grechez-Cassiau A, Guettier C, Hastings MH, Francis L. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 2004;64:7879–7885. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- Blask DE, Dauchy RT, Sauer LA, Krause JA, Brainard GC. Growth and fatty acid metabolism of human breast cancer (MCF-7) xenografts in nude rats: impact of constant light-induced nocturnal melatonin suppression. Breast Cancer Res Treat. 2003;79:313–320. doi: 10.1023/A:1024030518065. [DOI] [PubMed] [Google Scholar]
- Cos S, Mediavilla D, Martinez-Campa C, Gonzalez A, Alonso-Gonzalez C, Sanchez-Barcelo EJ. Exposure to light-at-night increases the growth of DMBA-induced mammary adenocarcinomas in rats. Cancer Lett. 2006;235:266–271. doi: 10.1016/j.canlet.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Wood PA, Yang X, Hrushesky WJ. Clock genes and cancer. Integr Cancer Ther. 2009;8:303–308. doi: 10.1177/1534735409355292. [DOI] [PubMed] [Google Scholar]
- Haus E, Smolensky M. Biological clocks and shift work: circadian dysregulation and potential long-term effects. Cancer Causes Control. 2006;17:489–500. doi: 10.1007/s10552-005-9015-4. [DOI] [PubMed] [Google Scholar]
- Stevens RG. Working against our endogenous circadian clock: breast cancer and electric lighting in the modern world. Mutat Res. 2009;680:106–108. doi: 10.1016/j.mrgentox.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky AS, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Gemma C, Gianotti TF, Burgueno A, Castano G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87:1606–1615. doi: 10.1093/ajcn/87.6.1606. [DOI] [PubMed] [Google Scholar]
- Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes. 2008;32:658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- Hsieh MC, Yang SC, Tseng HL, Hwang LL, Chen CT, Shieh KR. Abnormal expressions of circadian-clock and circadian clock-controlled genes in the livers and kidneys of long-term, high-fat-diet-treated mice. Int J Obes. 2010;34:227–239. doi: 10.1038/ijo.2009.228. [DOI] [PubMed] [Google Scholar]
- Jones CR, Campbell SS, Zone SE, Cooper F, DeSano A, Murphy PJ, Jones B, Czajkowski L, Ptacek LJ. Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Ownby RL, Saeed M, Wohlgemuth W, Capasso R, Acevedo A, Peruyera G, Sevush S. Caregiver reports of sleep problems in non-Hispanic white, Hispanic, and African American patients with Alzheimer dementia. J Clin Sleep Med. 2010;6:281–289. [PMC free article] [PubMed] [Google Scholar]
- Ferrari E, Nappi G, Vailati A, Martignoni E, Bossolo PA, Polleri A. Circadian periodicity of plasma prolactin in some neurological diseases. Int J Chronobiol. 1979;6:231–242. [PubMed] [Google Scholar]
- Petersen A, Hult S, Kirik D. Huntington's disease-new perspectives based on neuroendocrine changes in rodent models. Neurodegener Dis. 2009;6:154–164. doi: 10.1159/000225377. [DOI] [PubMed] [Google Scholar]
- Aziz NA, Anguelova GV, Marinus J, Lammers GJ, Roos RA. Sleep and circadian rhythm alterations correlate with depression and cognitive impairment in Huntington's disease. Parkinsonism Relat Disord. 2010;16:345–350. doi: 10.1016/j.parkreldis.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Benca RM, Duncan MJ, Frank E, McClung C, Nelson RJ, Vicentic A. Biological rhythms, higher brain function, and behavior: Gaps, opportunities, and challenges. Brain Res Rev. 2009;62:57–70. doi: 10.1016/j.brainresrev.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khapre RV, Samsa WE, Kondratov RV. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Ann Med. 2010;42:404–415. doi: 10.3109/07853890.2010.499134. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:271–276. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Liu N, Okamura K, Tyler DM, Phillips MD, Chung WJ, Lai EC. The evolution and functional diversification of animal microRNA genes. Cell Res. 2008;18:985–996. doi: 10.1038/cr.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Olive V, Jiang I, He L. miR17-92, a cluster of microRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:E236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by microRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO, Kay SA. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 2009;10:R17. doi: 10.1186/gb-2009-10-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire C, Moreno AB, Garcia-Chapa M, Lopez-Moya JJ, San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 2009;583:1039–1044. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. MiRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na YJ, Sung JH, Lee SC, Lee YJ, Choi YJ, Park WY, Shin HS, Kim JH. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp Mol Med. 2009;41:638–647. doi: 10.3858/emm.2009.41.9.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Clijsters L, Agami R. The microRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Champhekar A, Harris RB, Bittman EL. Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci USA. 2005;102:3111–3116. doi: 10.1073/pnas.0409734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. MiR122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, Schibler U. Integration of microRNA miR122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One. 2010;5:E11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan A, Stearns AT, Park PJ, Dreyfuss JM, Ashley SW, Rhoads DB, Tavakkolizadeh A. MicroRNA miR16 is anti-proliferative in enterocytes and exhibits diurnal rhythmicity in intestinal crypts. Exp Cell Res. 2010;316:3512–3521. doi: 10.1016/j.yexcr.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (microRNA) transcriptome of mouse retina and identification of a sensory organ-specific microRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltage-gated calcium channel alpha1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanova TN, Iuvone PM. Circadian rhythm and photic control of cAMP level in chick retinal cell cultures: a mechanism for coupling the circadian oscillator to the melatonin-synthesizing enzyme, arylalkylamine N-acetyltransferase, in photoreceptor cells. Brain Res. 2003;991:96–103. doi: 10.1016/j.brainres.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Ko ML, Liu Y, Dryer SE, Ko GY. The expression of L-type voltage-gated calcium channels in retinal photoreceptors is under circadian control. J Neurochem. 2007;103:784–792. doi: 10.1111/j.1471-4159.2007.04816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saus E, Soria V, Escaramís G, Vivarelli F, Crespo JM, Kagerbauer B, Menchón JM, Urretavizcaya M, Gratacòs M, Estivill X. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19:4017–4025. doi: 10.1093/hmg/ddq316. [DOI] [PubMed] [Google Scholar]
- Mendlewicz J. Disruption of the circadian timing systems: molecular mechanisms in mood disorders. CNS Drugs. 2009;23:15–26. doi: 10.2165/11318630-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Bentivoglio M, Deng XH, Nygard M, Sadki A, Kristensson K. The aging suprachiasmatic nucleus and cytokines: functional, molecular, and cellular changes in rodents. Chronobiol Int. 2006;23:437–449. doi: 10.1080/07420520500545797. [DOI] [PubMed] [Google Scholar]
- Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing Res Rev. 2006;5:33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Weinert D. Age-dependent changes of the circadian system. Chronobiol Int. 2000;17:261–283. doi: 10.1081/CBI-100101048. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]