SUMMARY
Antimicrobial peptides (AMPs) are among the repertoire of host innate immune defenses. In the oral cavity, several AMPs are present in saliva and have antimicrobial activities against oral bacteria, including Streptococcus mutans, a primary etiologic agent of dental caries. In this study, we hypothesized that unique S. mutans strains as determined by DNA fingerprinting from sixty 13 year-old subjects with or without caries experience would have different susceptibilities to α-defensins-1-3 (HNP-1-3), β-defensins-2-3 (HBD-2-3) and LL-37. The salivary levels of these peptides in subjects also were measured by enzyme-linked immunosorbent assays (ELISA). We found that S. mutans strains from caries-active subjects showed greater resistance to salivary HNP-1-2, HBD-2-3 and LL-37 at varying concentrations than those from caries-free subjects. In addition, combinations of these peptides increased their antimicrobial activity against S. mutans either additively or synergistically. The salivary levels of these peptides were highly variable among subjects with no correlation to host caries experience. However, the levels of a number of these peptides in saliva appeared to be positively correlated within an individual. Our findings suggest that the relative ability of S. mutans to resist host salivary AMPs may be considered a potential virulence factor for this species such that S. mutans strains that are more resistant to these peptides may have an ecological advantage to preferentially colonize within dental plaque and increase the risk of dental caries.
Keywords: Dental caries, defensins, S. mutans, innate immunity, saliva
INTRODUCTION
The oral cavity represents a unique environment in that the wide array of microbial species inhabiting the dental plaque biofilm interacts closely with host saliva. The protective attributes of saliva, such as its physical cleansing and buffering capacities, as well as a number of host innate and adaptive immune components, function collectively to maintain the balance between health and disease. The significance of saliva in the prevention of dental caries is well-recognized. The potential roles of a number of salivary proteins in the dental caries process have been a subject of interest in dental research, including a variety of salivary cationic antimicrobial peptides (AMPs), primarily the human neutrophil peptide α-defensins (HNP-1-3), human β-defensins (HBD-1-3), LL-37 and histatins. These peptides are expressed from various sources, including salivary gland acini and ducts, oral epithelium and neutrophils. These peptides possess broad spectrum antimicrobial activity against various microorganisms. In the oral cavity, the defensins and LL-37 are potent against a number of bacterial species, while histatins are predominantly antifungal (Joly et al., 2004, Sugimoto et al., 2006, Bartie et al., 2008, Nishimura et al., 2004a).
The roles of AMPs in a range of other bacterial infections have been intensely investigated. Several studies suggested that virulence potentials of several bacterial species were determined by their abilities to resist host AMPs (Nizet et al., 2001, Groisman et al., 1992). In addition, it was shown in murine models that AMPs were required for host resistance against certain infections and that differential expression of selected peptides could determine host susceptibility to diseases (Wu et al., 2009a, Wu et al., 2009b).
Evidence for a role of AMPs in dental caries has been reported. The level of HNP-1-3 was shown to be higher in saliva from caries-free children, compared to that from caries-active children, whereas no significant correlations between host caries experience and levels of HBD-3 or LL-37 were observed (Tao et al., 2005). These findings suggest a role for HNP-1-3 in dental caries protection and also raise the possibility of using salivary HNP-1-3 levels as a predictor of caries risk.
A handful of studies have investigated the susceptibility of S. mutans, primarily laboratory strains, to host AMPs in saliva (Maisetta et al., 2003, Ouhara et al., 2005, Joly et al., 2004, Nishimura et al., 2004b). The susceptibility patterns appeared to be strain-specific. However, the potential contribution and association of the susceptibility profiles of this cariogenic bacterium with host caries experience has not been defined.
In this study, we hypothesized that various strains of S. mutans possess different inherent susceptibility/resistance profiles to host salivary AMPs and that host-specific quantities of these peptides may influence plaque colonization by particular S. mutans strains. We showed that S. mutans strains from caries-free subjects were more susceptible to host AMPs than those from caries-active subjects. The differences in strain susceptibility to these peptides may influence the colonization of S. mutans strains in biofilms and potentially contribute to an individual’s relative risk for dental caries. Examination of salivary concentrations of these peptides showed that their levels were highly variable among individuals and not associated with caries experience in this subject group.
METHODS
Study sample and sample collection
Saliva and plaque samples were obtained from sixty children enrolled in the longitudinal Iowa Fluoride Study (Levy et al., 2001). For these analyses, equal numbers of subjects were randomly selected from among caries-free and caries-active groups. All children were 13 years old at the time of sampling. Subjects were designated high-caries when they had 3 or more carious or filled surfaces (D2-3F). Oral examination and sample collection were performed by trained and calibrated investigators using a standardized protocol (Warren et al., 2001). Subjects were constrained from eating, drinking (other than plain water), chewing gum or brushing teeth for 60 minutes before sampling. Plaque samples were taken from pits and fissures of occlusal surfaces of second molars using a sterile CytoSoft® Brush. Stimulated saliva was collected using Parafilm® wax. Samples were frozen at −80°C for later analysis. Investigators were blinded from demographic data and caries status of subjects while assays were performed.
S. mutans isolation and genotyping
Plaque samples were diluted in Trypticase Soy Broth supplemented with Yeast Extract (TSBYE) and plated on blood agar and Mitis-Salivarius-Kanamycin-Bacitracin (MSKB) agar for determination of total bacterial count and MS count, respectively. Ten colonies of presumed S. mutans per subject were selected and isolates were identified as S. mutans when they were positive for fermentation of mannitol, raffinose, salicin, sorbitol and negative for arginine hydrolysis and further confirmed by Gram stain and negative catalase tests. Ten S. mutans isolates per subject were frozen in TSBYE with 10% glycerol and kept at −80°C.
DNA extractions from S. mutans isolates were performed using the MasterPure™ Gram Positive DNA extraction kit (Epicentre Biotechnologies, Madison, WI). Genotypic characterization of S. mutans isolates were performed by arbitrarily primed polymerase chain reaction (AP-PCR) using 2 oligonucleotide primers, OPA-2 (5′-TGCCGAGCTG) and OPA-13 (5′-CAGCACCCAC). PCR reactions were performed in a volume of 25 μl containing 2.5 μl of 10X reaction buffer, 7 mM MgCl2, 1.25 units of Taq DNA Polymerase (Applied Biosystem, Foster City, CA), 200 mM of dNTP mix (Invitrogen, Calsbad, CA), 100 pmole of primer and 50 ng of template DNA. AP-PCR reactions were run for 45 cycles of 1 minute at 94°C, 1 minute at 36°C, and 2 minute at 72°C. Template DNA from S. mutans laboratory strain ATCC25175 was used as a control in all reactions. A negative control reaction in which template DNA was omitted was used to exclude the possibility of DNA contaminations. All PCR amplitypes were run on 1.5% agarose gel electrophoresis and subsequently stained with ethidium bromide.
AP-PCR fingerprinting patterns of S. mutans amplitypes from all subjects were analyzed using the GelCompar II program (Applied Maths Inc., Austin, TX). AP-PCR gel images were digitalized into the program database and normalized according to manufacturer’s instruction. Similarity coefficients were calculated by curve-based Pearson product-moment correlation. The unweighted pair group method using arithmetic averages (UPGMA) was used for clustering analysis of all strains. Dendrodrams were generated from the composite data set of both OPA2 and OPA13 fingerprinting profiles.
Antimicrobial susceptibility tests
Unique S. mutans strains from AP-PCR genotyping were subjected to antimicrobial susceptibility tests with a panel of AMPs, which included HNP-1-3, HBD-2-3 and LL-37. The selection of AMPs was based upon literature that reported their presence in saliva and their commercial availability. Additionally, the antimicrobial activity of three other peptides, HBD-1, lysozyme and histatin-5, against 16 S. mutans strains was also assessed in preliminary experiments. Data revealed no susceptibility of S. mutans strains to these peptides at concentrations 20 μg/ml or below; therefore, these three peptides were omitted from future experiments.
Antimicrobial susceptibility tests were performed using the alamarBlue® assay. In previous studies, as well as in our preliminary data, alamarBlue® assays and viable count assays for colony forming units (CFU) were comparable for evaluating bacterial viability (Vanitha & Paramasivan, 2004, Pettit et al., 2005, Montoro et al., 2005, Martin et al., 2005, Martin et al., 2003, Tenover et al., 1995, Yajko et al., 1995, DeForge et al., 2000). S. mutans strains were grown overnight in THBYE, centrifuged, and resuspended to 106 CFU/ml in Mueller-Hinton broth (MHB) for the HNP-1-3 tests or resuspended to 108 CFU/ml in 0.01 M sodium phosphate buffer (pH 7.4) for the HBD-2-3 and LL-37 tests. The choice to resuspend in MHB or sodium phosphate buffer and the selection of bacterial concentrations were based on conditions that optimized the activity of each particular peptide.
All peptides were tested at 3 different concentrations, 5 μg/ml, 1.5 μg/ml and 0.5 μg/ml, by incubating 90 μl of bacterial suspension with 10 μl of 10X stock of each peptide in BD Falcon™ black with clear bottom 96-well Microtest™ Optilux™ Plates. A growth control reaction was also set up using peptide diluent in place of each peptide. Following 2 hours incubation at 37°C, 10 μl of alamarBlue® reagent was added to each reaction and incubation continued for 4 hours. Fluorescent intensity was then measured at 560 nm and 590 nm using a fluorescent microplate reader (SpectraMax® M2e, Molecular Devices, Sunnyvale, CA). Assays were performed in duplicate. Percent viabilities of bacteria in each reaction were calculated using the following formula: Percent viability = (relative fluorescent units (RFU) of test peptide/relative fluorescent units of growth control) × 100.
AMP combination analysis
To examine the combined effect of these AMPs against S. mutans, the alamarBlue® assays were performed using serial dilutions of two peptides in a checkerboard assay format (Krogstad, 1986). In addition, the alamarBlue® assays were modified to permit the combination analysis of all peptide pairs. S. mutans clinical strains were grown as previously described, centrifuged and resuspended to 106 CFU/ml in 0.01 M sodium phosphate buffer (pH 7.4). The peptide concentrations used ranged from approximately twice the concentration that produced 50% killing of bacteria (EC50) to five serial two-fold dilutions below this concentration. The bacterial suspension was incubated with specified concentrations of peptides for 2 hrs at 37°C. Following incubation, 100 μl of MHB and 10 μl of alamarBlue® reagent were added to each reaction and incubation continued for 4 hrs. Plates were read using the fluorescent microplate reader as previously described.
Fractional inhibitory concentration (FIC) of each peptide was calculated as the concentration that gave 50% killing (EC50) when used in combination divided by the concentration that showed the same effect when used alone (Singh et al., 2000, Krogstad, 1986). The interaction of two peptides was also assessed using the FIC index, calculated by adding up their individual FIC values from the most effective combination. An FIC index approximately equal to 1 indicates an additive interaction. An FIC index of less than 1 reveals synergistic interaction, whereas an index of more than 1 is indicative of an antagonistic interaction. Peptide interactions were also evaluated by plotting an isobologram using the FIC values of different peptide combinations. If the peptide combination is additive, synergistic or antagonistic, the isobol will be straight, concave or convex, respectively. The degree of concavity or convexity also corresponds to the degree of synergy or antagonism (Tallarida, 2001, Tallarida, 2006).
Salivary AMP analysis
Saliva samples were thawed, cleared by centrifugation twice at 15,000 rpm for 10 minutes and used to measure the AMPs levels by enzyme-linked immunosorbent assays (ELISA). The assays for HNP-1-3 and LL-37 (Hycult Biotechnology, Uden, The Netherlands) were performed according to the manufacturer’s instructions, with the exception that, for the LL-37 assay, MgCl2 was added to the wash/dilution buffer to a final concentration of 300 nM. Sandwich ELISA assays for HBD-2 and HBD-3 were developed using recombinant peptide standards, capture and detection antibodies from Peprotech (Rocky Hill, NJ). Briefly, Nunc Maxisorp™ 96-well plates were coated with 100 μl of capture antibody overnight. The capture antibodies used were 2 μg/ml goat anti-HBD-2 and 3 μg/ml of rabbit anti-HBD-3 for HBD-2 and HBD-3 assays, respectively. Plates were blocked with 300 μl of 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 1 hr and incubated with recombinant peptide standards and saliva samples for 1 hr. Standards and saliva samples were diluted in the sample diluent (0.05% Tween-20, 0.1% BSA, 300 mM MgCl2 in PBS) and assayed in triplicate. Divalent cations were added to sample diluents to overcome the salivary masking effect of these peptides. MgCl2 (300 nM) improved peptide recovery to an optimal level and therefore was used in this study. Following sample incubation, plates were sequentially incubated with 100 μl of detection antibodies for 1 hr, avidin-horse radish peroxidase (HRP) conjugate for 30 minutes and 2,2′-Azinobis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) liquid substrate (Sigma-Aldrich, St. Louis, MO). Optical density (OD) readings were read at wavelengths of 405–650 nm. Detection antibodies used were biotinylated goat anti-HBD-2 and biotinylated rabbit anti-HBD-3 for HBD-2 and HBD-3 assays, respectively. Total protein concentrations in saliva samples were measured using the bicinchoninic acid (BCA) protein assay (Pierce Biotechnology, Rockford, IL) according to the manufacturer’s instructions.
Statistical analyses
Statistical analyses were performed using a two-sample t-test. A non-parametric Wilcoxon rank-sum test was used when the assumption of normally distributed data became invalid by the Shapiro-Wilks test. Correlation analyses were performed using a Pearson correlation test and Spearman rank correlation test. For all tests, the criterion for statistical significance was a p-value ≤0.05. SAS for Windows (v9.1, SAS Institute Inc, Cary, NC, USA) was used for the data analysis.
RESULTS
Subject demographics
Of the 60 subjects participating in this study, 38 subjects were females and 22 were males. The caries-active group consisted of 20 girls/10 boys and caries-free group consisted of 18 girls/12 boys. 58 subjects were Caucasian and one each were Hispanic and African-American. All subjects except two were in their permanent dentitions. Subjects were healthy with no current health conditions. One subject reported current antibiotics use. The majority of subjects (87%) brushed their teeth at least daily.
Mutans streptococci (MS) in dental plaque
To examine the relationship between the presence of MS in dental plaque and the caries experience of subjects, levels of MS and total cultivable bacteria from plaque samples were enumerated. Caries-active subjects showed significantly greater MS counts (p=0.04) and ratios of MS to total plaque bacteria (p=0.02) than caries-free subjects. The average level of MS recovered from caries-active subjects was 1.56×106 CFU/ml, whereas caries-free subjects harbored an average of 5.42×105 CFU/ml. Likewise, the average ratio of MS to total bacteria from dental plaque of caries-active subjects was 0.096, also significantly greater than the average value of 0.056 for caries-free subjects.
S. mutans genotyping
Ten S. mutans isolates were collected from plaque samples of each subject, giving a total of 600 isolates equally divided between caries-free and caries-active subjects. AP-PCR patterns generated from the DNA of these S. mutans isolates contained approximately 8–15 bands from each primer, representing fragments ranging from 0.3 to more than 1.5 kilobases in size. Overall, we found 74 distinct S. mutans amplitypes from 60 subjects (Table 1). A single distinct amplitype was observed in 47 subjects (78.33%). Twelve subjects (20%) harbored two distinct amplitypes and one subject (1.67%) harbored three amplitypes. Of the 13 subjects harboring more than one S. mutans amplitype, only 2 subjects (15.38%) were caries-free, whereas 11 subjects (84.62%) belonged to the caries-active group. Based on the nonparametric Wilcoxon rank-sum test, the number of S. mutans amplitypes was significantly greater in subjects with caries experience than in caries-free subjects (p<0.01). Overall, 42 genotypically distinct S. mutans strains were recovered from the caries-active group and 32 isolates were recovered from the caries-free group.
Table 1.
Numbers of subjects harboring 1-3 distinct S. mutans amplitypes.
| Subjects | Sex | Numbers of distinct S. mutans amplitypes per subjects |
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| All subjects | Girls (38) | 30 | 7 | 1 |
| Boys (22) | 17 | 5 | 0 | |
| All (60) | 47 | 12 | 1 | |
| Caries-active subjects | Girls (20) | 13 | 6 | 1 |
| Boys (10) | 6 | 4 | 0 | |
| All (30) | 19 | 10 | 1 | |
| Caries-free subjects | Girls (18) | 17 | 1 | 0 |
| Boys (12) | 11 | 1 | 0 | |
| All (30) | 28 | 2 | 0 | |
A dendrogram analysis demonstrated the genotypic variation among the 74 S. mutans strains obtained from all subjects (Fig. S1). The percentage similarities between each strain-pair based on the Pearson product-moment correlation ranged from 47.0% to 98.6%, indicating a large amount of genetic heterogeneity among the strains. Strains from 9 out of 13 subjects that harboured 2 or more amplitypes showed only a few band differences in the AP-PCR profiles compared to other genotypes from the same subject. The similarity values of these strains were more than 90% and tended to cluster closely together in the dendrogram (Fig. S2).
Susceptibility of S. mutans clinical strains to salivary AMPs and host caries experience
The 74 S. mutans clinical strains isolated were analyzed for their susceptibilities to HNP-1-3, HBD-2-3, and LL-37. Of all peptides tested, HBD-3 and LL-37 showed the most potent bactericidal activity against S. mutans strains. Among the α-defensins, S. mutans strains were most susceptible to HNP-1, followed by HNP-2 and HNP-3, respectively.
A two-sample t-test was used to compare the susceptibility profiles of S. mutans strains from caries-active and caries-free subjects. Since some subjects possessed more than one distinct strain and the proportion of each strain was variable in these subjects, we calculated the average susceptibilities of S. mutans from each clinical group using two methods: the first used the percentage viability of the predominant S. mutans strains from each subject to ensure equal weighting per subject, and the second method averaged the percentage viabilities of all distinct strains to account for all strains that were isolated. Results from analyses of predominant strains and all strains are shown in Fig. 1 and S3, respectively.
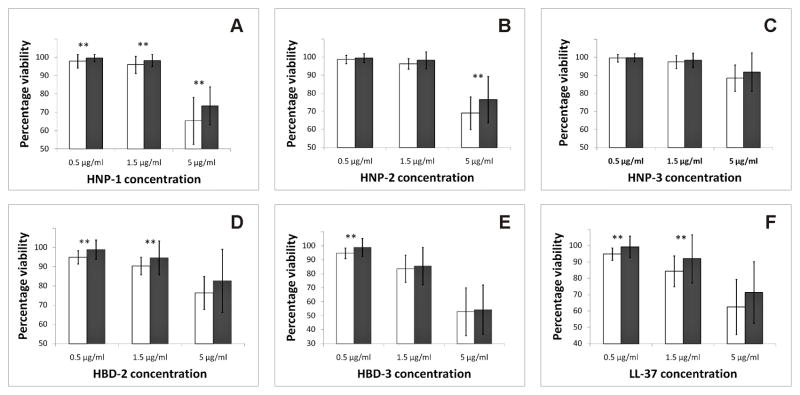
Fig. 1. Susceptibility of S. mutans from caries-active versus caries-free subjects: analysis of predominant strains from subjects.
(A) HNP-1; (B) HNP-2; (C) HNP-3; (D) HBD-2; (E) HBD-3; (F) LL-37. White bars represent caries-free subjects. Black bars represent caries-active subjects. Asterisks indicate statistically significant differences in viabilities between both groups (p<0.05).
Comparison of the susceptibilities to HNP-1 showed that strains from the two groups differed in their average percent viabilities for the predominant strains at all three HNP-1 concentrations. S. mutans strains from caries-free subjects showed significantly greater susceptibility to HNP-1 than those from children with caries experience at 5 μg/ml (p≤0.01), 1.5 μg/ml (p=0.04), and 0.5 μg/ml (p=0.03) (Fig. 1A). Analyzing the average percent viability of all strains resulted in significant differences at HNP-1 concentrations of 5 μg/ml (p=0.01) and 0.5 μg/ml (p=0.03) (Fig. S3A). A similar trend was also observed at 1.5 μg/ml HNP-1; however, this did not reach statistical significance (p=0.08).
S. mutans strains from caries-active and caries-free subjects also showed significant differences in their susceptibility to HNP-2. Analyses of both the predominant strains and all strains revealed that the average percentage viability of S. mutans exposed to 5 μg/ml HNP-2 was significantly greater when isolated from subjects with caries experience than when isolated from children with no caries experience (p=0.01 and 0.02, respectively) (Fig. 1B and S3B).
In contrast to results with HNP-1 and HNP-2, S. mutans isolates showed nearly 50% less susceptibility to HNP-3. At all three concentrations tested, no statistically significant differences in susceptibility to HNP-3 were detected between S. mutans strains from both groups (Fig. 1C and S3C).
Susceptibility tests with two β-defensins, HBD-2 and HBD-3, also showed significant differences between groups. S. mutans strains from caries-free subjects showed significantly greater susceptibility to HBD-2 at 1.5 μg/ml and 0.5 μg/ml and HBD-3 at 0.5 μg/ml, whether analyzing the predominant strains or all strains (p<0.05) (Fig. 1D–E and S3D–E).
Lastly, the tests of S. mutans strains against LL-37 also showed that S. mutans strains from caries-free subjects were more susceptible to LL-37 at 1.5 μg/ml and 0.5 μg/ml, compared to those from caries-active subjects. Though the difference did not reach statistical significance, the same trend was also noted at 5 μ/ml LL-37 for both the analyses of predominant strains (p=0.06) as well as all strains (p=0.06) (Fig. 1F and S3F).
Relationships between susceptibility profiles of S. mutans to AMPs
Our findings showed that all AMPs tested were active against S. mutans strains in a dose-dependent manner. These peptides share several common properties. They are small, cationic and are believed to exert their antimicrobial activities on the bacterial cytoplasmic membrane. It was of interest to examine whether S. mutans strains showed similar susceptibility patterns to these peptides. To determine this, percent viabilities of S. mutans exposed to different peptides at 5 μg/ml were correlated using Pearson correlation analysis. The data showed significant correlations between percentage viabilities to HNP-1 and HNP-2 (r=0.94, p<0.01), HNP-1 and HNP-3 (r=0.69, p<0.01), HNP-2 and HNP-3 (r=0.78, p<0.01), HBD-2 and HBD-3 (r=0.56, p<0.01), HBD-2 and LL-37 (r=0.60, p<0.01), and HBD-3 and LL-37 (r=0.53, p<0.01 (Table S1) Analyses between the pairs of three α-defensins and the β-defensins or LL-37 revealed no significant correlations.
Combined effects of salivary AMPs
The effects of AMPs in combination were examined. Pairwise combinations of these AMPs appear to enhance their antimicrobial activities either additively or synergistically. None of the combinations tested demonstrated antagonistic activity (Fig. S4).
Salivary AMP levels and host caries experience
Salivary analysis showed that AMP levels were highly variable among subjects despite normalization with total salivary protein concentrations. Mean total protein concentration in saliva was 935.2±273.4 μg/ml. Mean concentrations of HNP-1-3, LL-37, HBD-3 and HBD-2 were 1,913±1,157 ng/ml, 15.81±10.43 ng/ml, 2.233±2.183 ng/ml and 0.734±0.811 ng/ml, respectively (Table 2).
Table 2.
Levels of AMP in saliva.
| Total salivary protein (μg/ml) | Salivary concentration (ng/ml) | Salivary concentration relative to total protein (ng/mg) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HNP-1-3 | LL-37 | HBD-3 | HBD-2 | HNP-1-3 | LL-37 | HBD-3 | HBD-2 | ||
| Mean | 935.19 | 1913.56 | 15.80 | 2.23 | 0.73 | 2034.33 | 17.48 | 2.57 | 0.78 |
| SD | 273.36 | 1157.25 | 10.44 | 2.18 | 0.81 | 1069.12 | 11.37 | 3.07 | 0.81 |
| Median | 933.76 | 1443.36 | 14.20 | 1.67 | 0.39 | 1704.90 | 15.52 | 1.88 | 0.44 |
| Maximum | 1661.19 | 5213.06 | 71.02 | 11.56 | 3.89 | 5545.39 | 77.21 | 19.36 | 3.35 |
| Minimum | 385.57 | 548.63 | 3.93 | 0.15 | 0.08 | 631.38 | 4.02 | 0.11 | 0.08 |
To assess the relationships between the levels of AMPs in saliva and caries experience in children, two-sample t-tests were used to compare mean salivary levels of each peptide, with or without normalization to total salivary protein, for caries-free and caries-active subjects. For all peptides tested, we found no significant differences in salivary levels between the two groups: HNP1-3 (p=0.37), HNP1-3 relative to total protein (p=0.84); LL-37 (p=0.94), LL-37 relative to total protein (p=0.44); HBD-3 (p=0.81), HBD-3 relative to total protein (p=0.54); HBD-2 (p=0.23), and HBD-2 relative to total protein (p=0.54). Interestingly, our data showed that the mean total salivary protein level was significantly greater in children with caries experience than that observed in caries-free children (p=0.02). Mean total salivary protein concentrations were 1,018±306.3 μg/ml (range from 574.8 to 1,661 μg/ml) in caries-active subjects and 852.2±209.7 μg/ml (range from 385.6 to 1,241 μg/ml) in caries-free subjects (Table 3).
Table 3.
Mean salivary AMP levels between caries-active and caries-free subjects.
| Salivary concentrations | Caries-active | Caries-free | t-test p-values |
|---|---|---|---|
| HNP-1-3 (ng/ml) | 2047.44±1254.76 | 1779.68±1055.03 | p=0.37 |
| HNP-1-3 relative to total protein (ng/mg) | 2006.42±1125.17 | 2062.24±1028.52 | p=0.84 |
| LL-37 (ng/ml) | 15.92±7.50 | 15.70±12.86 | p=0.94 |
| LL-37 relative to total protein (ng/mg) | 16.33±8.29 | 18.64±13.84 | p=0.44 |
| HBD-3 (ng/ml) | 2.16±1.91 | 2.30±2.46 | p=0.80 |
| HBD-3 relative to total protein (ng/mg) | 2.33±2.39 | 2.82±3.65 | p=0.54 |
| HBD-2 (ng/ml) | 0.86±0.90 | 0.85±0.83 | p=0.23 |
| HBD-2 relative to total protein (ng/mg) | 0.61±0.71 | 0.72±0.81 | p=0.54 |
| Total protein (μg/ml) | 1,018.19±306.33 | 852.19±209.69 | p=0.02* |
Relationships between the salivary AMP levels
Since these AMPs to some extent play overlapping and/or collaborative roles in host defense against oral pathogens, we next evaluated the correlations between AMP levels in saliva within individuals. Results are shown in Figure S5. Pearson correlation analyses showed statistically significant correlations between the levels of LL-37 and HNP1-3 (r=0.66, p<0.01) and between the levels of LL-37 relative to total protein and HNP1-3 relative to total protein (r=0.65, p<0.01). In addition, statistically significant correlations were found between HBD-2 and HBD-3 levels (r=0.50, p<0.01) and between HBD-2 relative to total protein and HBD-3 relative to total protein (r=0.67, p<0.01). We also found significant correlations between HNP1-3 and HBD-2 concentrations in saliva (r=0.37, p<0.01) and between the levels of HBD-3 relative to total protein and LL-37 relative to total protein (r=0.26, p<0.05). No significant correlations were observed between other variables.
Relationships between the salivary AMP levels and MS or total bacteria in dental plaque
We further examined the relationships between the salivary levels of AMPs and MS levels within dental plaque. Analyses showed no significant correlations between the ratio of MS relative to total bacteria in dental plaque and salivary levels of any AMPs tested with or without normalization to total salivary protein. However, we found a significant positive relationship between the plaque MS count and HNP-1-3 concentration relative to total protein (r=0.28, p=0.03) (Fig. S6). The correlation analysis between MS count and the non-normalized salivary concentration of HNP-1-3 also revealed a similar trend but did not reach statistical significance (p=0.07). No significant correlations were found between MS counts and salivary concentrations of other peptides.
The relationships between salivary levels of AMPs and total cultivable bacteria in dental plaque were also examined. Spearman rank correlation tests showed no statistically significant correlations between salivary levels of each peptide, with or without normalization, and total plaque bacteria. However, it should be noted that there was a trend towards an increasing relationship between total plaque bacteria and salivary concentration of HBD-3 (p=0.06) and salivary concentration of HBD-3 relative to total protein (p=0.09), particularly when considering caries-active subjects only (data not shown).
Relationships between the salivary AMP levels and the susceptibility profiles of S. mutans
We next asked if there were relationships between salivary AMP levels and susceptibility to the corresponding peptide of S. mutans isolated from the same individual. It was expected that S. mutans strains would be most resistant to the AMPs that were represented at the highest levels in saliva. Pearson correlation analyses showed a significant positive relationship between the salivary level of HNP-1-3 relative to total protein and the percentage viability of S. mutans strains to 5 μg/ml HNP-1 (r=0.26, p=0.04) (Fig. S7).
DISCUSSION
In this study, several findings were demonstrated. For the first time, it was shown that S. mutans strains from individuals with caries experience showed greater resistance to salivary AMPs, compared to those obtained from caries-free individuals, suggesting that the ability of S. mutans to resist these peptides may be another factor that can help define the relative virulence of this bacterium. Significant differences were observed with HNP-1-2, HBD-2-3 and LL-37 at varying concentrations. We deliberately selected physiological concentrations of AMPs to assay. Although the susceptibility profiles of S. mutans strains to the three HNPs were highly correlated, significant differences between the susceptibilities to HNP-3 between both groups were not observed at the three concentrations tested, potentially due to the overall lower antimicrobial activity of this peptide. The susceptibility profiles of S. mutans strains to HBD-2, HBD-3 and LL-37 were also correlated, suggesting that these peptides may act by similar mechanisms, yet possess varying potentials for combating pathogens.
Based on our data, we speculate that S. mutans strains that are more resistant to host AMPs may have an ecological advantage over the more susceptible strains for surviving within dental plaque. It has been observed that S. mutans compete relatively poorly with other non-pathogenic microorganisms at neutral pH (Bradshaw et al., 1996). S. mutans appears to be more susceptible to host AMPs than oral streptococci that constitute the major proportion in normal non-pathogenic plaque, such as S. sanguinis, S. mitis and S. oralis (Bartie et al., 2008, Joly et al., 2004, Nishimura et al., 2004a, Ouhara et al., 2005). This suggests that salivary AMPs may play a role in maintaining the oral health by limiting the growth of potentially pathogenic organisms such as S. mutans and allowing the more resistant non-pathogenic bacteria to establish colonization and dominate. Therefore, differences in susceptibility profiles of S. mutans strains may be significant in providing the resistant strains with a competitive advantage over susceptible strains. In particular, under conditions favorable for growth, such as the increased presence of fermentable sugar and low pH, resistant S. mutans strains may better survive and populate, leading to greater cariogenic potential.
However, we did not find statistically significant differences between salivary levels of HNP-1-3, LL-37, HBD-3 and HBD-2 between caries-free and caries-active subjects. These results partially differed from those of a previous study (Tao et al., 2005), which showed significantly higher HNP-1-3 levels in saliva of caries-free subjects compared to that of caries-active subjects but no significant differences in LL-37 and HBD-3 levels between groups. This difference between studies may be the result of several factors. In this study, we used stimulated saliva as a source for AMP detection, whereas unstimulated whole saliva samples were used in the Tao et al. study. Differences between the concentrations of specific AMPs in stimulated versus unstimulated saliva have not been reported to date. However, it has been noted that salivary protein composition can differ, depending on whether saliva was collected under stimulated or unstimulated conditions (Oberg et al., 1982, Rantonen & Meurman, 2000). In addition, one notable difference is the subject ethnicity. While the overwhelming majority of our subjects were Caucasians, subjects in their study were mostly Hispanic. Several studies have observed not only differences in caries prevalence among people with diverse racial/ethnic backgrounds, but also variation in salivary protein composition (Everhart et al., 1973, Johnson, 2005, Sivakumar et al., 2009, Zakhary et al., 2007). However, no racial/ethnic differences in AMP expression or genetic polymorphisms have yet been confirmed. Future studies may potentially help clarify the effect, if any, of race/ethnicity on AMP expression.
Our results do not support the measurement of salivary AMP levels as a reliable tool to predict caries risk, at least not for stimulated saliva. As noted earlier, increased HNPs levels in saliva have been observed in patients with various oral inflammatory conditions (Mizukawa et al., 1998, Mizukawa et al., 1999a, Mizukawa et al., 1999b). The infiltration of neutrophils within these lesions is believed to be responsible for the increased defensin levels (Abiko & Saitoh, 2007). These findings support the role of salivary AMPs as a first line of host immune response in the oral cavity. However, their levels of expression appear to be associated with a wide range of inflammatory conditions in a rather non-specific manner. Any degree of simultaneous gingival or oral mucosal inflammation can contribute to the levels of these peptides in saliva irrespective of host caries activity.
It is of note that salivary HNP-1-3 levels detected in this study were well within the range of their antimicrobial activity, supporting a physiologically relevant role in oral immune defense. LL-37 and HBDs, however, were found within the ng/ml range in saliva in this subject group. These salivary concentrations are generally lower than their effective range in vitro, though the actual physiological concentrations of these peptides at different sites in the oral cavity are not known. The HBDs are expressed from oral keratinocytes as well as salivary duct cells, and their expression is particularly strong at regions close to mucosal surfaces such as at gingival margins around the tooth, allowing for close contact with supragingival plaque (Dale & Fredericks, 2005). Similarly, LL-37 is secreted from multiple sources, including salivary ducts, oral epithelium and neutrophils. The levels of HNP-1-3 and LL-37 are concentrated in gingival crevicular fluid, in close proximity to tooth surfaces. This could potentially increase the concentrations of these peptides substantially within the environment surrounding tooth structure and, thus, closely influence microbial ecology within dental plaque. These AMPs can work additively or synergistically to enhance their antimicrobial activity. Furthermore, the α-defensins, β-defensins, and LL-37, have other immunomodulatory roles, chemoattractant activity, and can stimulate the adaptive immune response, including enhancing the production of IgA or IgG (Yang et al., 2004). Therefore, the effect of saliva must be considered in its entirety to explain completely its contribution to dental health.
Unexpectedly, we found a significantly greater level of total salivary protein in subjects with caries experience than in caries-free subjects. Since protein concentrations in saliva are also dependent on salivary flow rate, it is possible that these caries-active subjects may have had lower salivary flow rates than caries-free subjects, which led to more concentrated protein in saliva and increased susceptibility to dental caries. Another explanation is that caries-active subjects may have higher concentrations of specific protein components in saliva that potentially facilitate dental caries formation. Several studies have suggested a role for specific salivary proteins in the adhesion of bacteria onto oral surfaces by forming adherent biofilms, or pellicles (Scannapieco, 1994, Bradway et al., 1989). A recent study also reported a positive correlation between the total protein and glycoprotein contents in saliva and saliva-promoted S. mutans adhesion to hydroxyapatite (Shimotoyodome et al., 2007). A number of salivary components, when adsorbed to oral surfaces, were described to mediate molecular interactions with oral bacteria, including mucins (Stinson et al., 1982), α-amylase (Scannapieco et al., 1995, Scannapieco et al., 1993), fibronectin (Dawson & Ellen, 1990) and PrPs (Russell & Mansson-Rahemtulla, 1989, Gibbons & Hay, 1989). Therefore, this pellicle-mediated bacterial adhesion could provide the basis for the robust formation of dental plaque populated with sufficient proportions of cariogens to increase risk of dental caries. Moreover, the role of salivary proteins as a source of nutrients to oral bacteria has also been suggested. During dental plaque formation, plaque bacterial colonizers may require specific salivary proteins that they can degrade to provide nutrients for their metabolism, allowing further growth, multiplication and aggregation to occur (Scannapieco, 1994, Bowden & Li, 1997).
Interestingly, we found that the salivary levels of several AMPs were correlated within an individual. The correlations between the salivary levels of HNP-1-3 and LL-37, and between HBD-2 and HBD-3, are consistent with the natural sources of these peptides, since the majority of HNP-1-3 and LL-37 are produced from neutrophils, and both HBD-2 and HBD-3 are secreted from oral epithelial cells and salivary duct cells. In addition, we also observed increasing relationships between salivary levels of HNP-1-3 and HBD-2 and between LL-37 and HBD-3. Collectively, it appears that these AMPs tend to be produced together in saliva, probably due to the similar types of inflammatory stimuli present as a part of the innate immune response. This may provide an appropriate setting for these peptides to work additively or synergistically to increase their antimicrobial activity against pathogens.
Also examined were the relationships between the salivary AMP levels and the MS levels in dental plaque. We found a significant positive correlation between the HNP-1-3 relative to total protein and the MS count. Likewise, the correlation analyses between total bacteria in dental plaque and salivary levels of HBD-3 or HBD-2 showed trends toward increasing relationships between these variables. These results are consistent with previous findings that the expression of HBD-3 and HBD-2 in oral epithelial cells is inducible upon bacterial contact or in response to pro-inflammatory stimuli, including IL-1β, TNF-α or IFN-γ (Chung & Dale, 2004, Joly et al., 2005, Krisanaprakornkit et al., 2000, Mathews et al., 1999, Taguchi & Imai, 2006). Their expression, however, varies with different bacterial species (Ji et al., 2007, Kimball et al., 2006, Krisanaprakornkit et al., 2000, Vankeerberghen et al., 2005). A recent study demonstrated that a S. mitis biofilm significantly upregulated HBD-2 gene expression in gingival epithelial cells, whereas S. mutans biofilm was a poor inducer (Eberhard et al., 2009). It is possible that the more resistant early colonizers in dental plaque may induce the expression of AMPs, which in turn limits the colonization and survival of more sensitive, potentially pathogenic microorganisms, such as S. mutans (Dale & Fredericks, 2005, Weinberg et al., 1998).
Lastly, we examined the relationship between levels of salivary AMPs and the susceptibility/resistance profiles of S. mutans isolated from the same individuals. A significant correlation was observed between the salivary level of HNP-1-3 relative to total protein and the percentage viability of S. mutans strains to 5 μg/ml HNP-1 (p=0.04), suggesting that individuals with higher levels of HNP-1-3 in saliva tend to harbor S. mutans strains that are more resistant to HNP-1. This finding is in the line with a previous study, which found that, in a series of selection experiments, microbial agents are able to evolve in vitro in response to a gradual increase of AMP concentrations such that, over several hundred generations, there is natural selection for organisms that are more resistant to the tested peptide (Perron et al., 2006). These data support our findings that increasing resistance to salivary AMPs is one mechanism that promotes S. mutans survival in the oral environment, potentially elevating the risk of dental caries.
In conclusion, our findings support the roles of host salivary AMPs in shaping S. mutans ecology by restricting the overall growth of this cariogenic bacterium. The presence of these peptides in saliva and dental plaque may be a factor in the selection of more resistant strains that better populate the plaque, thereby increasing the likelihood of initiating or propagating the dental caries process, particularly when the surrounding environment favors their growth. In other words, the relative ability of S. mutans to resist the antimicrobial activity of these peptides may constitute a virulence factor for this organism. However, the expression of AMPs in saliva is associated with the presence of local inflammatory stimuli, such as the dental plaque microflora, and is not specific to host dental caries experience.
Supplementary Material
Acknowledgments
This work was supported by The University of Iowa, College of Dentistry Seed grant, National Institutes of Health Grants R01-DE09551, R01-DE12101, R21-DE019475 and M01-MRR00059. E. Phattarataratip was supported by Royal Thai Government Scholarship.
References
- Abiko Y, Saitoh M. Salivary defensins and their importance in oral health and disease. Curr Pharm Des. 2007;13:3065–3072. doi: 10.2174/138161207782110417. [DOI] [PubMed] [Google Scholar]
- Bartie KL, Devine DA, Wilson MJ, Lewis MA. In vitro susceptibility of the Streptococcus milleri group to antimicrobial peptides. Int Endod J. 2008;41:586–592. doi: 10.1111/j.1365-2591.2008.01404.x. [DOI] [PubMed] [Google Scholar]
- Bowden GH, Li YH. Nutritional influences on biofilm development. Adv Dent Res. 1997;11:81–99. doi: 10.1177/08959374970110012101. [DOI] [PubMed] [Google Scholar]
- Bradshaw DJ, Marsh PD, Schilling KM, Cummins D. A modified chemostat system to study the ecology of oral biofilms. J Appl Bacteriol. 1996;80:124–130. doi: 10.1111/j.1365-2672.1996.tb03199.x. [DOI] [PubMed] [Google Scholar]
- Bradway SD, Bergey EJ, Jones PC, Levine MJ. Oral mucosal pellicle. Adsorption and transpeptidation of salivary components to buccal epithelial cells. Biochem J. 1989;261:887–896. doi: 10.1042/bj2610887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WO, Dale BA. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect Immun. 2004;72:352–358. doi: 10.1128/IAI.72.1.352-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale BA, Fredericks LP. Antimicrobial peptides in the oral environment: expression and function in health and disease. Curr Issues Mol Biol. 2005;7:119–133. doi: 10.1093/jac/dki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JR, Ellen RP. Tip-oriented adherence of Treponema denticola to fibronectin. Infect Immun. 1990;58:3924–3928. doi: 10.1128/iai.58.12.3924-3928.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForge LE, Billeci KL, Kramer SM. Effect of IFN-gamma on the killing of S. aureus in human whole blood. Assessment of bacterial viability by CFU determination and by a new method using alamarBlue. J Immunol Methods. 2000;245:79–89. doi: 10.1016/s0022-1759(00)00279-9. [DOI] [PubMed] [Google Scholar]
- Eberhard J, Pietschmann R, Falk W, Jepsen S, Dommisch H. The immune response of oral epithelial cells induced by single-species and complex naturally formed biofilms. Oral Microbiol Immunol. 2009;24:325–330. doi: 10.1111/j.1399-302X.2009.00518.x. [DOI] [PubMed] [Google Scholar]
- Everhart DL, Grigsby WR, Carter WH., Jr Human dental caries experience related to age, sex, race, and certein salivary properties. J Dent Res. 1973;52:242–247. doi: 10.1177/00220345730520021001. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Hay DI. Adsorbed salivary acidic proline-rich proteins contribute to the adhesion of Streptococcus mutans JBP to apatitic surfaces. J Dent Res. 1989;68:1303–1307. doi: 10.1177/00220345890680090201. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Parra-Lopez C, Salcedo M, Lipps CJ, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci U S A. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Kim Y, Min BM, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J Periodontal Res. 2007;42:503–510. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- Johnson RB. Racial differences in salivary sIgA concentrations in postmenopausal women. Spec Care Dentist. 2005;25:145–149. doi: 10.1111/j.1754-4505.2005.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Joly S, Maze C, McCray PB, Jr, Guthmiller JM. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–1029. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly S, Organ CC, Johnson GK, McCray PB, Jr, Guthmiller JM. Correlation between beta-defensin expression and induction profiles in gingival keratinocytes. Mol Immunol. 2005;42:1073–1084. doi: 10.1016/j.molimm.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Kimball JR, Nittayananta W, Klausner M, Chung WO, Dale BA. Antimicrobial barrier of an in vitro oral epithelial model. Arch Oral Biol. 2006;51:775–783. doi: 10.1016/j.archoralbio.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogstad D, Moellering RC. Antimicrobial combinations. In: Lorian V, editor. Antibiotics in Laboratory Medicine. Williams & Wilkins; Baltimore, M.D: 1986. [Google Scholar]
- Levy SM, Warren JJ, Davis CS, Kirchner HL, Kanellis MJ, Wefel JS. Patterns of fluoride intake from birth to 36 months. J Public Health Dent. 2001;61:70–77. doi: 10.1111/j.1752-7325.2001.tb03369.x. [DOI] [PubMed] [Google Scholar]
- Maisetta G, Batoni G, Esin S, Luperini F, Pardini M, Bottai D, Florio W, Giuca MR, Gabriele M, Campa M. Activity of human beta-defensin 3 alone or combined with other antimicrobial agents against oral bacteria. Antimicrob Agents Chemother. 2003;47:3349–3351. doi: 10.1128/AAC.47.10.3349-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Camacho M, Portaels F, Palomino JC. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother. 2003;47:3616–3619. doi: 10.1128/AAC.47.11.3616-3619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Morcillo N, Lemus D, Montoro E, Telles MA, Simboli N, Pontino M, Porras T, Leon C, Velasco M, Chacon L, Barrera L, Ritacco V, Portaels F, Palomino JC. Multicenter study of MTT and resazurin assays for testing susceptibility to first-line anti-tuberculosis drugs. Int J Tuberc Lung Dis. 2005;9:901–906. [PubMed] [Google Scholar]
- Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, Tack BF, McCray PB., Jr Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukawa N, Sugiyama K, Fukunaga J, Ueno T, Mishima K, Takagi S, Sugahara T. Defensin-1, a peptide detected in the saliva of oral squamous cell carcinoma patients. Anticancer Res. 1998;18:4645–4649. [PubMed] [Google Scholar]
- Mizukawa N, Sugiyama K, Ueno T, Mishima K, Takagi S, Sugahara T. Defensin-1, an antimicrobial peptide present in the saliva of patients with oral diseases. Oral Dis. 1999a;5:139–142. doi: 10.1111/j.1601-0825.1999.tb00078.x. [DOI] [PubMed] [Google Scholar]
- Mizukawa N, Sugiyama K, Ueno T, Mishima K, Takagi S, Sugahara T. Levels of human defensin-1, an antimicrobial peptide, in saliva of patients with oral inflammation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1999b;87:539–543. doi: 10.1016/s1079-2104(99)70130-7. [DOI] [PubMed] [Google Scholar]
- Montoro E, Lemus D, Echemendia M, Martin A, Portaels F, Palomino JC. Comparative evaluation of the nitrate reduction assay, the MTT test, and the resazurin microtitre assay for drug susceptibility testing of clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother. 2005;55:500–505. doi: 10.1093/jac/dki023. [DOI] [PubMed] [Google Scholar]
- Nishimura E, Eto A, Kato M, Hashizume S, Imai S, Nisizawa T, Hanada N. Oral streptococci exhibit diverse susceptibility to human beta-defensin-2: antimicrobial effects of hBD-2 on oral streptococci. Curr Microbiol. 2004a;48:85–87. doi: 10.1007/s00284-003-4108-3. [DOI] [PubMed] [Google Scholar]
- Nishimura E, Sakihama T, Setoguchi R, Tanaka K, Sakaguchi S. Induction of antigen-specific immunologic tolerance by in vivo and in vitro antigen-specific expansion of naturally arising Foxp3+CD25+CD4+ regulatory T cells. Int Immunol. 2004b;16:1189–1201. doi: 10.1093/intimm/dxh122. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Oberg SG, Izutsu KT, Truelove EL. Human parotid saliva protein composition: dependence on physiological factors. Am J Physiol. 1982;242:G231–236. doi: 10.1152/ajpgi.1982.242.3.G231. [DOI] [PubMed] [Google Scholar]
- Ouhara K, Komatsuzawa H, Yamada S, Shiba H, Fujiwara T, Ohara M, Sayama K, Hashimoto K, Kurihara H, Sugai M. Susceptibilities of periodontopathogenic and cariogenic bacteria to antibacterial peptides, {beta}-defensins and LL37, produced by human epithelial cells. J Antimicrob Chemother. 2005;55:888–896. doi: 10.1093/jac/dki103. [DOI] [PubMed] [Google Scholar]
- Perron GG, Zasloff M, Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc Biol Sci. 2006;273:251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit RK, Weber CA, Kean MJ, Hoffmann H, Pettit GR, Tan R, Franks KS, Horton ML. Microplate Alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother. 2005;49:2612–2617. doi: 10.1128/AAC.49.7.2612-2617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantonen PJ, Meurman JH. Correlations between total protein, lysozyme, immunoglobulins, amylase, and albumin in stimulated whole saliva during daytime. Acta Odontol Scand. 2000;58:160–165. doi: 10.1080/000163500429154. [DOI] [PubMed] [Google Scholar]
- Russell MW, Mansson-Rahemtulla B. Interaction between surface protein antigens of Streptococcus mutans and human salivary components. Oral Microbiol Immunol. 1989;4:106–111. doi: 10.1111/j.1399-302x.1989.tb00107.x. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Torres G, Levine MJ. Salivary alpha-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993;4:301–307. doi: 10.1177/10454411930040030701. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Torres GI, Levine MJ. Salivary amylase promotes adhesion of oral streptococci to hydroxyapatite. J Dent Res. 1995;74:1360–1366. doi: 10.1177/00220345950740070701. [DOI] [PubMed] [Google Scholar]
- Shimotoyodome A, Kobayashi H, Tokimitsu I, Hase T, Inoue T, Matsukubo T, Takaesu Y. Saliva-promoted adhesion of Streptococcus mutans MT8148 associates with dental plaque and caries experience. Caries Res. 2007;41:212–218. doi: 10.1159/000099321. [DOI] [PubMed] [Google Scholar]
- Singh PK, Tack BF, McCray PB, Jr, Welsh MJ. Synergistic and additive killing by antimicrobial factors found in human airway surface liquid. Am J Physiol Lung Cell Mol Physiol. 2000;279:L799–805. doi: 10.1152/ajplung.2000.279.5.L799. [DOI] [PubMed] [Google Scholar]
- Sivakumar T, Hand AR, Mednieks M. Secretory proteins in the saliva of children. J Oral Sci. 2009;51:573–580. doi: 10.2334/josnusd.51.573. [DOI] [PubMed] [Google Scholar]
- Stinson MW, Levine MJ, Cavese JM, Prakobphol A, Murray PA, Tabak LA, Reddy MS. Adherence of Streptococcus sanguis to salivary mucin bound to glass. J Dent Res. 1982;61:1390–1393. doi: 10.1177/00220345820610120101. [DOI] [PubMed] [Google Scholar]
- Sugimoto J, Kanehira T, Mizugai H, Chiba I, Morita M. Relationship between salivary histatin 5 levels and Candida CFU counts in healthy elderly. Gerodontology. 2006;23:164–169. doi: 10.1111/j.1741-2358.2006.00120.x. [DOI] [PubMed] [Google Scholar]
- Taguchi Y, Imai H. Expression of beta-defensin-2 in human gingival epithelial cells in response to challenge with Porphyromonas gingivalis in vitro. J Periodontal Res. 2006;41:334–339. doi: 10.1111/j.1600-0765.2006.00879.x. [DOI] [PubMed] [Google Scholar]
- Tallarida RJ. Drug synergism: its detection and applications. J Pharmacol Exp Ther. 2001;298:865–872. [PubMed] [Google Scholar]
- Tallarida RJ. An overview of drug combination analysis with isobolograms. J Pharmacol Exp Ther. 2006;319:1–7. doi: 10.1124/jpet.106.104117. [DOI] [PubMed] [Google Scholar]
- Tao R, Jurevic RJ, Coulton KK, Tsutsui MT, Roberts MC, Kimball JR, Wells N, Berndt J, Dale BA. Salivary antimicrobial peptide expression and dental caries experience in children. Antimicrob Agents Chemother. 2005;49:3883–3888. doi: 10.1128/AAC.49.9.3883-3888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Swenson JM, O’Hara CM, Stocker SA. Ability of commercial and reference antimicrobial susceptibility testing methods to detect vancomycin resistance in enterococci. J Clin Microbiol. 1995;33:1524–1527. doi: 10.1128/jcm.33.6.1524-1527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitha JD, Paramasivan CN. Evaluation of microplate Alamar blue assay for drug susceptibility testing of Mycobacterium avium complex isolates. Diagn Microbiol Infect Dis. 2004;49:179–182. doi: 10.1016/j.diagmicrobio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Vankeerberghen A, Nuytten H, Dierickx K, Quirynen M, Cassiman JJ, Cuppens H. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J Periodontol. 2005;76:1293–1303. doi: 10.1902/jop.2005.76.8.1293. [DOI] [PubMed] [Google Scholar]
- Warren JJ, Levy SM, Kanellis MJ. Prevalence of dental fluorosis in the primary dentition. J Public Health Dent. 2001;61:87–91. doi: 10.1111/j.1752-7325.2001.tb03371.x. [DOI] [PubMed] [Google Scholar]
- Weinberg A, Krisanaprakornkit S, Dale BA. Epithelial antimicrobial peptides: review and significance for oral applications. Crit Rev Oral Biol Med. 1998;9:399–414. doi: 10.1177/10454411980090040201. [DOI] [PubMed] [Google Scholar]
- Wu M, McClellan SA, Barrett RP, Hazlett LD. Beta-defensin-2 promotes resistance against infection with P. aeruginosa. J Immunol. 2009a;182:1609–1616. doi: 10.4049/jimmunol.182.3.1609. [DOI] [PubMed] [Google Scholar]
- Wu M, McClellan SA, Barrett RP, Zhang Y, Hazlett LD. Beta-defensins 2 and 3 together promote resistance to Pseudomonas aeruginosa keratitis. J Immunol. 2009b;183:8054–8060. doi: 10.4049/jimmunol.0902140. [DOI] [PubMed] [Google Scholar]
- Yajko DM, Madej JJ, Lancaster MV, Sanders CA, Cawthon VL, Gee B, Babst A, Hadley WK. Colorimetric method for determining MICs of antimicrobial agents for Mycobacterium tuberculosis. J Clin Microbiol. 1995;33:2324–2327. doi: 10.1128/jcm.33.9.2324-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- Zakhary GM, Clark RM, Bidichandani SI, Owen WL, Slayton RL, Levine M. Acidic proline-rich protein Db and caries in young children. J Dent Res. 2007;86:1176–1180. doi: 10.1177/154405910708601207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.