Abstract
Sequencing a single molecule of deoxyribonucleic acid (DNA) using a nanopore is a revolutionary concept because it combines the potential for long read lengths (>5 kbp) with high speed (1 bp/10 ns), while obviating the need for costly amplification procedures due to the exquisite single molecule sensitivity. The prospects for implementing this concept seem bright. The cost savings from the removal of required reagents, coupled with the speed of nanopore sequencing places the $1000 genome within grasp. However, challenges remain: high fidelity reads demand stringent control over both the molecular configuration in the pore and the translocation kinetics. The molecular configuration determines how the ions passing through the pore come into contact with the nucleotides, while the translocation kinetics affect the time interval in which the same nucleotides are held in the constriction as the data is acquired. Proteins like α-hemolysin and its mutants offer exquisitely precise self-assembled nanopores and have demonstrated the facility for discriminating individual nucleotides, but it is currently difficult to design protein structure ab initio, which frustrates tailoring a pore for sequencing genomic DNA. Nanopores in solid-state membranes have been proposed as an alternative because of the flexibility in fabrication and ease of integration into a sequencing platform. Preliminary results have shown that with careful control of the dimensions of the pore and the shape of the electric field, control of DNA translocation through the pore is possible. Furthermore, discrimination between different base pairs of DNA may be feasible. Thus, a nanopore promises inexpensive, reliable, high-throughput sequencing, which could thrust genomic science into personal medicine.
Index Terms: Deoxyribonucleic acid (DNA), nanopore, protein, sequencing, solid state
I. Introduction
Deoxyribonucleic acid (DNA), the code of life, is composed of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T), which are paired together in a complementary fashion (A to T and C to G) and ordered in a species-specific sequence. The aim of genomic science is to predict biological behavior using the information stored in the DNA within each cell. But, when the first draft sequence of the human genome emerged in early 2001 [1], despite its enormous value for genetics, it quickly became apparent that our understanding of the relationship between the genetic code and cellular function was deficient. For example, only 5% of the human genome is estimated to be functional and, of that, only 30% lies within the exons of known protein-encoding genes [2]. The rest lies in the so-called “dark matter” of the human genome—leading to efforts, such as the Encyclopedia of DNA Elements (ENCODE) [3], that strive to identify regulatory components. Identifying genes and controlling regions, such as promoter, insulator, and enhancer sites, turns out to be a major undertaking in itself.
To glean more information about how genetics informs cellular function and its affect on development and disease, it is essential to sequence rapidly and economically using only minute amounts of material. There are two major categories of sequencing tasks: de novo, or initial sequencing of unknown genomes, and resequencing of genomes with a known base sequence. Though much of the de novo sequencing is already complete for the human genome, along with most of the popular model organisms, there are still many species with unknown genomes. For example, the human microbiome, the genome sequences of the many different species of bacteria living in or on humans, still remains a mystery. The microbiome of the flora of our gut alone is estimated to contain ∼300 billion base pairs (Gbp), or ∼100 times the human genome [4]. However, the vast majority of the work ahead involves resequencing genomes with an already known base sequence. The first, obvious example is mutation sequencing, where recent work has shown that the majority of human cancers do not always have mutations in the same locations, or even the same genes [5]. Moreover, the mutations and genotype of the individual has been shown to be important for chemotherapeutic effectiveness; i.e., genomics can determine the effectiveness of a drug for an individual [6].
Sequencing can also provide clues to health and development beyond the actual genomic sequence itself. The proteins expressed by genes represent the machinery of the cell—they make things work. But an individual organism can express the same genes differently depending on the epigenetic profile. Sequencing can help determine this profile, e.g., it can give information on DNA-binding protein interactions using sequencing of immunoprecipitated DNA fragments (ChIP-seq) to find occupied binding sites [7]. Conceivably, with inexpensive high-throughput sequencing, we will be able to determine the difference between these binding sites in different tissue and under different conditions. Sequencing of bisuflite-treated DNA can be used to determine DNA methylation patterns, a reversible modification of cytosines (in mammals), which alters protein binding. Subsequent sequencing and alignment may be used to distinguish between methylated and unmethylated cytosines, allowing for delineation of the methylation pattern, the methylome [8]. It may also be advantageous for gene expression studies to sequence the transcriptome; i.e., the sequence of the RNA. This can give detailed information about the levels of expression, the splicing variation, and even allow for the identification of new noncoding RNAs, potentially involved in regulation, all of which are part of the “dark matter” of the genome [9].
However, sequencing all of these “-omes” is facilitated by technology that inexpensively and quickly determines sequence information from genetic samples.
II. Sequencing Methods
Since its development in 1977, the Sanger method of DNA sequencing has transformed biology [10]. It is the standard to which all other methods of sequencing are compared. The Sanger sequencing reaction is similar to the polymerase chain reaction (PCR), but using only one primer in combination with dideoxy nucleotide triphosphates (ddNTPs) to prematurely terminate the elongation reaction. By mixing fluorescently labeled dideoxynucleotides with deoxynucleotides, the polymerase reaction generates fragmentary single stranded copies of a DNA template with the last base labeled with a different fluorescent moiety depending on the base. Separating these fragments by size through electrophoresis, the sequence can be determined from the color of fluorescence produced at a given fragment length.
Though functional, this procedure is problematic for several reasons. The length of template that can be read using this method is limited to ∼1000 bp [11], [12]. As a result, either chromosome walking or shotgun sequencing must be used, both of which are time consuming and require reassembly of the completed sequence. The chain termination reaction is also time consuming, as is the electrophoretic separation, leading to the development of many different techniques for massively parallel methods for sequencing [13]. But the overwhelming problems with the Sanger sequencing method are the relatively large amounts of DNA required—amplification leads to errors—and the expense of reagents for labeling and separation.
There are emerging technologies under development that have the potential to supersede conventional Sanger sequencing and, in some cases, sequence the human genome for $1000 or less. Following Shendure et al. [14], the emerging technologies can be loosely categorized as: bioMEMS, an extension of conventional electrophoretic methods through miniaturization and integration [15], [16]; sequencing-by-hybridization, which uses the differential hybridization of oligonucleotide probes to decode the DNA sequence [17]; massively parallel signature sequencing (MPSS), which is not based on polymerase extension, but on cycles of restriction digestion and ligation [18]; cyclic-array sequencing, which can detect which base is added to a growing sequence using either fluorescence or luminescence [19], [20]; and finally, nonenzymatic, real-time single-molecule sequencing [21], [22]. BioMEMS has the advantage that it relies on the same tested principles as electrophoretic sequencing, which has already been used to sequence 1011 nucleotides. Using variations of the Sanger process, in conjunction with capillary array electrophoresis to separate DNA fragments, about 100 bp can be sequenced per minute at a cost of <$1 (or about $100 million per genome) with an accuracy of about 99.99%. This is considered to be the gold standard, but it seems unlikely that a factor of 100 000× cost reduction will be achieved through scaling and integration alone. Sequencing-by-hybridization has the advantage that the data collection method, i.e., scanning the florescence emitted by labeled DNA that has been hybridized to an array of probe sequences, is compatible with high-throughput methodology. However, probes have to be designed that avoid cross-hybridization to the wrong target, which renders 50% of the chromosome inaccessible. All methods that rely on some method of isolated clonal amplification, e.g., cyclic-array sequencing on amplified molecules and MPSS, are costly and often problematic. For example, these methods have a low frequency of nucleotide misincorporation or nonincorporation, which manifests itself in signal decay through “dephasing.” While single molecule cyclic-array sequencing, as compared to sequence-by-synthesis and MPSS, eliminates the costly PCR-amplification step, and hence requires less starting material with less risk of dephasing, achieving the signal-to-noise ratio (SNR) required for single molecule detection is still a challenge. For example, the Helicos HeliScope has recently been able to sequence a human genome from gDNA extracted from 2 mL of blood, with no additional amplification [23]. These techniques are all limited in read length, however, necessitating larger computing requirements for sequence assembly.
Right now, the Roche GS-FLX (454) sequencer uses cyclic-array sequencing coupled to emulsion PCR to produce 100 Mb of data in 7 h with a 250 bp read length (per bead) at a cost of $8439 ($84.40 per Mb) [24]. The Helicos single-molecule cyclic array sequencer offers a much higher data rate at a cost of ∼$60 000 to sequence a human genome [25], but with a higher error rate(∼95% accuracy) [26]—mostly due to dark reads (personal communication). In contrast, a run in an Applied Biosystems SOLiD (Sequencing by Oligo Ligation) sequencer requires 5 days and produces 3–4 Gb of sequence data with an average read length of 25–35 bp, costing $5.81 per Mb. Applied Biosystems estimates that their SOLiD system sequencer will be able to sequence an entire human genome for only $10 000 in just 2 weeks. According to Shendure et al. [14] for resequencing, the error rate of the sequence has to be less than the expected variation in the sequence for variation to be accurately detected. Since human chromosomes differ at approximately 1 in every 1000 bp, an error rate of 1/100 kbp would be needed to ensure confidence. If the accuracy of a raw read is 99.7% (current state-of-the-art), then 3× coverage of each base will yield a 1/100 kbp error rate. To ensure a minimum 3× coverage of >95% of the diploid human genome, 6.5× coverage is required; about 40 billion raw bases at a cost per base of <$10 000, or 4 million bases per $1 [14]. If an improvement over SOLiD performance is derived simply from an increase in the acquisition rate per device, we would therefore need to sequence at a rate of 330 000 bp/s to reach a $1000 genome. Resequencing a genome does not require complex alignment procedures; a read only needs to be long enough to allow it to be mapped to a unique location in an assembled reference genome. In the mammalian genome, only ∼73% of 20-bp genomic reads can be assigned to a single unique location; achieving >95% uniqueness will require reads >60 bp. Thus, a resequencing instrument that can deliver a $1000 human genome with reasonable coverage and accuracy will need to achieve >60-bp reads with 99.7% raw-base accuracy. A faster instrument with longer reads will be cheaper still.
Single molecule DNA sequencing represents the logical end-of-the-line for development of sequencing technology in which we extract the maximum amount of information from a minimum of material and pre-processing. If this were paired with a high-throughput and low cost instrument, it would change the genomic flow of data from a trickle to a deluge. Specifically, the low material requirement and quick results would allow for easy sequencing of precious primary samples from human patients. Fast, inexpensive, low material sequencing would thrust genomics within the grasp of personalized medicine. Moreover, it would represent a leap forward in determining the epigenome, the heritable nongenetic changes that affect gene expression.
III. Nanopore
Within the categories of emerging technologies, sequencing a single molecule of DNA with a nanopore is the most revolutionary. It is revolutionary because it combines the potential for long read lengths (>5 kbp) with high speed (1 bp/10 ns), while obviating the need for costly procedures like PCR amplification due to the exquisite single molecule sensitivity. The nanopore sequencing concept uses a radically new approach to detection that is reminiscent of Coulter’s original idea of using objects within a constricted current path to alter the electrical resistance [27]. As first articulated by Branton and Deamer and independently by Church, nanopore sequencing relies on the electric signal that develops when DNA translocates through a pore in a membrane [21]. By applying an electric field to a nanometer-diameter pore in a thin membrane, we can force individual polyanionic DNA molecules to move through the pore in a single-file sequential order, as if threading a needle. If each base has a characteristic electrical signature, then ostensibly a pore could by used to analyze the sequence by reporting all of the signatures in a single read without resorting to multiple DNA copies.
Electrical detection of DNA using a nanopore could have significant advantages over cyclic arrays or fluorescent microscopy. Usually single molecule sequencing relies on enzymatic incorporation of a fluorescently labeled mononucleotide through a polymerase and applying techniques that suppress the ambient radiation so that one molecule can be identified. The nanoporesequencing concept uses a radically new approach that does not require fluorescent labeling or any chemical treatment, but instead relies on an electrical signal that develops when DNA translocates through a pore in a membrane. To sequence DNA using a nanopore, one must first find a robust, nanoporous structure of an appropriate size. The primary equilibrium form of DNA, B-form double-stranded DNA (dsDNA), is a stiff, highly charged polymer with a solvated, helical structure about 2.6–2.9 nm in diameter, according to neutron scattering, that depends on the sequence and the number of strongly bound water molecules included in the primary hydration shell [28]. Single-stranded DNA (ssDNA) is about half this size—it can fit through a 1-nm pore [29]—and it is more flexible. Owing to the limited flexibility of the DNA, the direction of the polymer segment persists over a distance denoted as the persistence length. ss-DNA has a persistence length of 0.75–4 nm, depending on the salt concentration compared to ∼50 nm for dsDNA [30]. The prospects for low cost, high-throughput nanopore sequencing are currently being explored using as prototypes either α-hemolysin (α-HL) [22], [31], [32] and its mutants, or nanopores in solid-state membranes [33]–[35] as illustrated in Fig. 1.
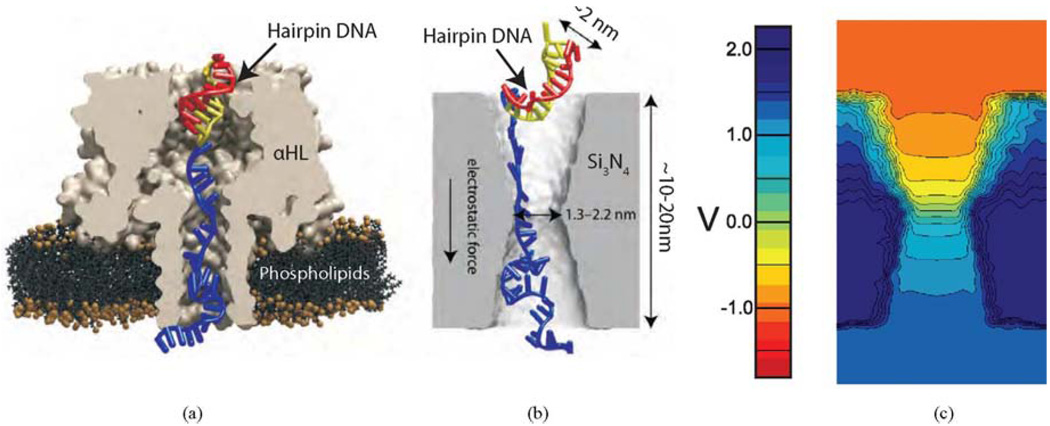
Fig. 1.
Nanopores in thin membranes used for sequencing DNA. (a) Cross section through an α-HL pore taken from a simulation showing a trapped DNA hairpin. The pore diameter (1.3 nm) is smaller than the stem of the hairpin. (b) Cross section through a solid-state pore in a silicon nitride membrane showing a hairpin trapped in the pore. The diameter of the solid-state pore can be any size. (c) Distribution of the electrostatic force and potential in a nanopore. Electrostatic potential contours inside a 1.0-nm-radius pore at 2.6 V without DNA present in the pore.
It has been speculated that it should be possible to distinguish between different bases or base pairs as they translocate across the membrane by measuring the ionic current through the pore. Since the translocation velocity through the pore can be very high, i.e., >1 nucleotide/10 µs for α-HL and >1 nucleotide/10 ns in a sold-state nanopore, it should, therefore, be possible to sequence a single DNA molecule quickly and inexpensively provided that the bases can be discriminated electrically, but single base resolution has not been demonstrated yet [36]–[39]. When applying a voltage to a nanopore spanning a membrane, the applied voltage is essentially dropped across the pore as evident in Fig. 1(c). This means that DNA has to first diffuse within range of the pore to be driven through by the electric field. The rate of DNA capture (and translocation) is roughly given by R = 2πCDr, where R is the capture rate, C is the concentration of DNA molecules, D is the diffusion constant of DNA in free solution, and r is the radius of probable capture by the pore, which depends on the voltage applied [40].
While a nanopore may be the ultimate analytical tool with single molecule sensitivity—this feature recommends it for third generation sequencing—there is a shortcoming in using it to sequence single molecules that is related to the diffusion equivalent capacitance [41], [42]. The diffusion capacitance governs the time required to capture a molecule, which is about 1 s for 109 molecules/µL concentration, and leads to a tradeoff between response time and the detectable concentration.
Once the DNA molecule is inside the pore, there are three main forces that affect it. The first and strongest force is the electric field, acting primarily on the negatively charged phosphate backbone of DNA. The electric field causes electrophoretic motion of the DNA molecule, driving it forward while the positively charged ion cloud surrounding it is driven back [43]. There is an electrostatic interaction with the pore walls, and/or a non-polar (van der Waals) interaction. And finally, there is a drag force associated with the movement of the polymer in solution, essentially a frictional force.
To determine the net force exerted on DNA in a nanopore at a given transmembrane bias, Luan and Aksementiev [44] used molecular dynamics (MD) to simulate the system illustrated in Fig. 2, which includes a 20-bp fragment of (dsDNA), 0.1 M KCl electrolyte, and a pore through a solid-state membrane fabricated from silicon nitride. MD simulations revealed three regimes for the dependence of the net force F on the applied electric field E, which are categorized according to the pore diameter. For a pore diameter larger than 5 nm, the interactions with the pore itself are negligible due to the small electrostatic screen length, or Debye length (1 nm), and the weak interaction of the van der Waals (r−6 dependence). When the pore diameter is between 3.6 and 5 nm, the electrolyte still behaves as it does in bulk solution, but direct interaction between the DNA molecule and the pore surface becomes important. Finally, when d < 3.6 nm, the viscosity of water in a thin film between DNA and a nanopore surface is larger than in the bulk and depends on the shearing velocity of the moving DNA [45]. In this regime, the interactions between DNA and the pore can be much stronger and the microscopic details of the pore surface strongly affect the friction. A nonlinear dependence of the force on the applied electric field is expected, which is optimal for sequencing, as it allows the force and velocity of DNA translocation to be easily affected. Moreover, the small diameter pore forces DNA molecules to move into and through the pore single file, as more than one double helix cannot fit in the pore at the same time. In a small pore, the DNA occludes much of the electrolytic current through the pore, maximizing the signal.
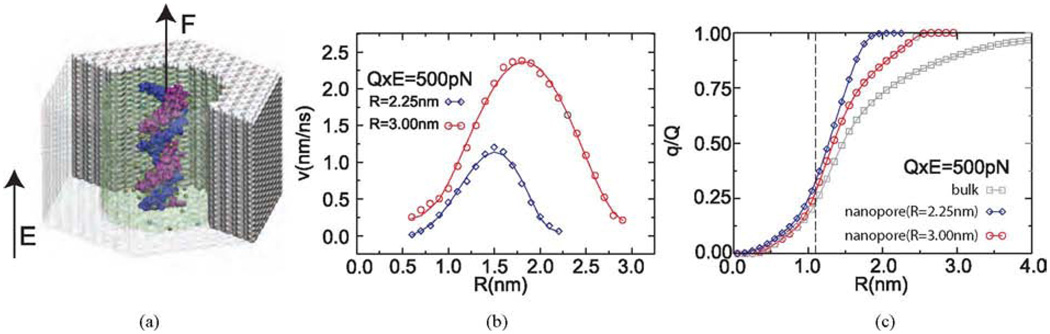
Fig. 2.
Analyzing the forces on DNA in a nanopore. (a) All-atom model of DNA solvated in 100 mM KCl electrolyte in a nanopore in a nitride membrane. DNA is simulated under simultaneous actions of force F, which is a harmonic spring used to measure the net force on DNA and an external electric field E. (b) Pattern of the electro-osmotic flow between DNA and the nanopore surface. The diameter of DNA is ∼2.4 nm. (c) Net charge of the electrolyte within the distance R from the central axis of the DNA, i.e., q(R) = Σ qion (r < R). q is the charge of the bare DNA. The dashed line indicates the position of the DNA surface. Figure adapted from Ref. [44].
The MD simulations have been thoroughly tested, by performing experiments to examine DNA interactions under the effects of different shapes of proteinaceous or solid-state pores. With large pores, it was found that dsDNA could translocate, even folded on itself or multiple molecules at a time, which is not advantageous for acquiring sequencing data [34], [35]. However, smaller pores have been created that demonstrate the ability to translocate dsDNA one base pair at a time through the pore, by appropriate sizing of the pore relative to the diameter of the DNA helix [33]. In fact, ssDNA can even be sifted from dsDNA using such a pore [29].
Once the DNA diffuses within range of the pore and after the initial acceleration by the electric field, the frictional forces acting on the DNA due to pore interaction and hydrodynamic resistance cause the molecule to reach a terminal velocity. The complex interactions of DNA with the flowing electrolyte can be decomposed into two independent motions: one motion is that of DNA dragged at a constant velocity by a nonelectric force F = ηυ; and the other is that of DNA drifting in an electric field E at constant velocity υ′ = µE, where η and μ are the friction coefficient and electrophoretic mobility, respectively. This description takes into account the hydrodynamic drag of the electro-osmotic flow that develops around DNA under the action of the applied field [see Fig. 2(b)], characterized by the electrophoretic mobility μ. The product η × μ is commonly misunderstood to be the effective charge on the DNA due to the counterion condensation qeff. As illustrated in Fig. 2(c), the distribution of counterions around DNA depends on the pore diameter; hence, the effective charge should also be different, but the measured net force on DNA due to the electric field of the same magnitude is almost the same. The average velocity of the electro-osmotic flow, Fig. 2(b), is found to be proportional to the effective screening force on DNA. By measuring directly the friction coefficient η and electrophoretic mobility μ of DNA from independent MD simulations, Luan and Aksimentiev found that the effective driving force in a nanopore obeys F = ηµE, which simplifies to F = qephE by introducing the electrophoretic charge qeph = ηµ. Note that the physical meaning of qeph is different from qeff, as the latter does not include the effect of the electroosmotic flow [43].
It seems, therefore, feasible to control the force in a nanopore by changing either the viscosity of the electrolyte or the interaction that the pore surface has with the DNA. For example, when the solution is doped with glycerol, the translocation time increases linearly with solution viscosity, increasing as much as five times [46]. However, the accompanying drop in ion mobility reduces the current as well, reducing the SNR, which adversely affects detection. Along the same lines, there have been some preliminary attempts to adjust the interaction between the pore and the DNA polymer [47]–[49] to slow DNA motion or increase the SNR. However, slowing the DNA adversely affects throughput.
If a continuous strand of DNA is driven electrophoretically past a detector at a fixed location, the motion of a DNA base relative to the detector will introduce “translocation noise” [50], which is succinctly captured in a 1-D transport model by the ratio of the drift (or migration) to diffusion velocities, i.e., υdrift/υdiff = µE/(D/Lm) = V/(kT/q) according to the Einstein relation, where D and μ are the diffusivity and the mobility of DNA in a nanopore, respectively, and the electric field is given by E = V/Lm, where V is the voltage applied across a membrane of thickness Lm. From this relation, it is apparent that large voltages compared to kT/q, the thermal voltage, are desirable to offset diffusive fluctuations associated with the motion of DNA in the pore. But large voltages can adversely membrane reliability (due to breakdown) and increase the translocation velocity, which forces high frequency operation to electrically read/sample each base pair. To obviate the need for continuous high voltage operation, while still suppressing positional noise, the base should ideally be trapped in the pore, read, and then impelled at high velocity to the next base in the sequence.
Thus, high fidelity reads demand stringent control over both the molecular configuration in the pore and the translocation kinetics. The molecular configuration determines how the ions passing through the pore come into contact with the nucleotides, while the translocation kinetics affect the time interval in which the same nucleotides are held in the constriction as the data is acquired. Until recently, no nanopore prototype proffered for sequencing has shown any prospect of satisfying both of these specifications at the same time.
There are two major sources of current passing through an unblocked nanopore. First, and most obvious, is the ionic current passing through the central cavity of the pore, the bulk pore current, which is screened from electrostatic interaction with the pore walls. This current consists of cations and anions from the electrolyte being driven through the pore. The second source of current is from the electrical double layer associated with the pore surface, the interfacial pore current. If there are surface charges on the inner walls of the pore, counterions will accumulate at the surface to generate a screening layer. These mobile ions will then move under the influence of the electric field, contributing to the overall detected current [51].
When a DNA molecule enters the pore, the ionic current through the pore alters drastically. The effective cross-sectional area of the pore changes, due simply to volume exclusion— as the DNA molecule occludes space that ions could previously travel through. This results in a reduction in the bulk pore current. Fig. 3(a)–(c) show such current traces observed when 48.502 kbp λ-DNA interacts with a 2.5-nm pore in a silicon nitride membrane [see Fig. 3(a). A DNA translocation is detected by a sharp decrease in the current as seen in Fig. 3(b). A histogram of the different current values in the trace indicates that the value of blockage currents form a Gaussian distribution, which is expected considering the different fluctuations and alterations of the blockage current discussed earlier. The same DNA and same pore can generate a variety of different transients, some of which are illustrated in Fig. 3(c). We speculate that the disparity in shape and duration of the current transients reflects the molecular configuration in the neighborhood of the pore [52] and the time required to disentangle the DNA into the single file of base-pairs required to permeate the pore. If the duration of the blockade corresponds with the interval that DNA occupies the pore, then the average transient width signifies that double stranded λ-DNA translocates through the pore in about 0.45 ± 0.2 ms, corresponding to a velocity of 48.5 kbp/0.5 ms = 1 bp/10 ns, which is in line with MD estimates [53], and about 1000× faster than ssDNA in α-HL.
Fig. 3.
Electrolytic current through a 2.5-nm pore in a silicon nitride membrane. The diameter is chosen to be comparable to the size of the double helix to maximize the current signal. (a) TEM micrograph of a 2.5× 2.0 ± 0.2 nm cross-section nanopore in a silicon nitride membrane about 15-nm thick. (b) (left) Electrolytic current measured in 100 mM KCl at 800 mV (blue) through the pore shown in (a) as a function of time. The open pore current at this voltage is about 2.85 nA. These current blockades are associated with λ-DNA translocating through the pore. Current blockades are presumably associated with the translocation of λ-DNA across the membrane through the pore. (Right) Frequency of blockades observed at 800 mV with a particular change in current normalized to the open pore current in the same pore. (c) Three examples of current blockades observed in the 2.5-nm pore shown in (a) as a function of time at V = 800 mV under the same conditions. Electrolytic current through a 2-nm pore in a membrane formed from an MOS capacitor, interacting with hairpin DNA. Voltage of 200 mV is applied across the membrane, which is smaller than the presumed threshold for stretching the hairpin. (d) High resolution transmission electron micrograph of the nanopore through a composite membrane approximately 45-nm thick. The shot noise in the center of the image is associated with the pore. (e) (left) Long duration (>1 s) current transients are observed both above and below the open pore current near 0.2 nA. For example, near 1394 s, a transient occurs that blocks about 92% of the open pore current (0.014 nA) and persists until 2812 s. Also notice that the current transients with current >0.35 nA, nearly double the open pore value. (Right) Histogram showing the values of the current observed over a 5000 s interval: 1) is identified with the current blockade; 2) and 3) are intermediate and recurring blockades; and 4) is a current enhancement above the open pore value. (f) Magnified view of the interval starting at 1226 s showing long duration current enhancement above the open pore value. The inset is a magnified view of the interval highlighted by the dashed box illustrating the well-defined current levels.
However, as previously mentioned, the bulk pore current is not the entire story. The charge on the DNA also has an effect on both the interfacial current [51], and brings along with it a cloud of counterions of its own [52], [54]. The interfacial current may be directly affected by the charge on the DNA, and if it is a cationic current, increased [51]. The counterions directly associated with DNA will also contribute to the current, as previously mentioned, moving in the opposite direction to DNA. The magnitude of this contribution will depend on both the concentration of the counterion, given by the ionic concentration used in the electrolyte, and the counterion’s association with the DNA, dictated by the type of ion. In fact, the contributions from these current sources may be so significant that they nullify the blockade of the bulk current, depending on the DNA concentration.
Thus, current transients may not be an unequivocal indication of a translocation. To establish correspondence, it is necessary to perform quantitative PCR (qPCR) on the cathode sample to determine the number of translocated DNA molecules. While both qPCR and the number of current transients indicate a threshold for permeation and count similar numbers of DNA molecules, the correspondence is imperfect. Nanopores may show a blockade in the pore even if the molecule is not translocating through it [53]. Furthermore, current enhancements above the open pore current value [53] are observed, if the increased concentration of ions in the pore (from screening charges around DNA) gives a larger contribution to the current than the bulk current occluded by the presence of DNA in the pore [51], [52], [55]. On the other hand, we have recently shown that it is possible to sort out the relationship between the current transients and the electrolyte concentration, pore charge, and molecular configuration by using molecular dynamics (MD) [52].
Fig. 3(d) shows a 2-nm diameter pore in a composite membrane fabricated from an ultrathin MOS capacitor approximately 50-nm thick. The membrane is formed on a silicon-on-insulator (SOI) substrate using conventional silicon processing technology. The electrodes of the capacitor are fabricated from heavily doped layers of silicon, appropriately thinned using a combination of oxidation and chemical-mechanical polishing (CMP). The capacitor dielectric is formed by growing an oxide on crystalline silicon using rapid thermal oxidation. According to ellip-sometry, the polysilicon, silicon, and SiO2 layers are 33 ± 2 nm, 12 ± 2 nm, and 1.6 nm thick, respectively. Thus, the membrane is about 47-nm thick.
Comer et al. studied the interactions between this pore and hairpin DNA (hpDNA) [52]. hpDNA is a single strand of DNA partially doubled over on itself and base-paired to its complement and stabilized by hydrogen bonds. Functionally, this means that the molecule consists of a single stranded tail or overhanging coil portion and a duplex head (or stem) region—it is an exceptionally attractive system for the purposes of DNA sequencing. Using a pore of the appropriate minimum diameter (1.0 < d < 2.5 nm), the hairpin can become trapped in the pore with the tail threaded through the constriction. The probability of translocation can, therefore, be controlled by varying the probability of the double helix’s rupture, through adjustment of the transmembrane voltage. The hpDNA used in this experiment consisted of an overhanging coil of 50 adenine nucleotides and a double helix of 12 pairs with an intervening 76-nucleotide loop.
Fig. 3(e) shows current transients superimposed on the open pore electrolytic current associated with an hpDNA molecule or molecules interacting with the pore observed in 1 M KCl for a 0.2-V transmembrane bias, which is below the translocation threshold. There are also transients >0.35 nA, nearly double the open pore value. On the right-hand side of Fig. 3(e) is a histogram that tallies the values of the current observed over the 5000-s interval shown to the left. The salient features are: a current blockade that peaks near 10 pA which is identified by 1) in the figure; along with intermediate and recurring blockades at 0.10 and 0.14 nA associated with 2) and 3), respectively; and a current enhancement above the open pore value centered at 0.35 nA denoted by 4). The histogram reflects that fact that the current through the pore assumes well-defined levels for extended periods of time and some of these values recur over time. For example, Fig. 3(f) shows a magnified view of transient current enhancements to a value centered near 0.35 nA and intermediate blockades observed in the time frame near 1230 s and the corresponding histogram of the current levels.
The largest current reductions likely imply that the loop at the apex of the double helix of hpDNA is in the pore’s constriction and the largest current enhancements (with currents about twice those in the absence of DNA) correspond to portions of the hpDNA threading through the constriction and occupying the anodic side of the pore. Hence, the same DNA can cause ionic currents < 0.1I0 and > 2I0 depending on the conformation of the DNA/nanopore system. Individual polynucleotides should have distinct electrical signatures that reflect their mobility through the pore and their composition due to the different size and chemical interaction that the bases have with pore and the ions co-occupying it. For example, Ashkenasy et al. were able to hold a DNA hairpin statically in a nanopore smaller in diameter than the stem while the ssDNA coil, which is smaller, threaded inside the pore. With the DNA held in place, they could measure the blockade current for a long enough time (2 s) to avoid SNR and bandwidth issues, granting a measurable difference dependent on the composition of the ssDNA strand—dC was distinguishable from dA [56]. An example of this is shown in Fig. 3(e), where some of the transients persist for a long time (>1 s) with multiple well defined levels that are in some cases much larger than the open pore values. There are multiple long duration current transients both above and below the open pore value (about 0.2 nA). We found an extraordinary example near 1894 s of a transient blockade, which persisted for about 1400 s in which only 8% of the open pore current (0.014 nA) flows. Stoddart et al. have expanded slightly on this method, using a modified form of α-HL with improved resolution of nucleotide differences, and a protein (streptavidin/biotin) bound to one end of the molecule to hold it in place [57].
It is not just the amplitude of the blockade current that can be affected by the sequence of the DNA strand; the speed of translocation may be affected as well. This both complicates the interpretation of current traces, and simplifies the prospect of gathering data from DNA translocation through a nanopore. There isn’t just amplitude modulation, but also frequency modulation. Meller et al. [32] have shown that the composition of DNA homopolymers affects the speed at which they translocate through α-HL pore; the interaction of purines versus pyrimidines is particularly clear, suggesting base stacking may be a factor [32], [58]. However, by doping a poly dC strand with either dA or dT at well-spaced intervals, base-stacking can be invalidated as the primary cause of altered translocation time; moreover, the difference in translocation time with dT doping demonstrates that it is not simply a purine/pyrimidine difference [59].
IV. Protein Pore
Kasianowicz et al. [22], [31] were among the first to adopt proteinaceous nanopores to detect and sort single DNA molecules. They selected Staphylococcus aureus α-HL, a mushroom shaped heptamer that assembles across a phospholipid membrane [see Fig. 1(a)]. It is composed of seven identical sub-units arranged around a central axis; the transmembrane portion is a β-barrel about 5nm long with a minimum diameter of 1.4 nm [60], [61]. By placing this protein within a lipid membrane, an electric field induced flow of ions through the protein can be measured. If ssDNA or RNA is added to the anodic side, the translocation through the α-HL pore and resulting current blockade can be detected, with the length of the current blockade proportional to the length of the molecule [22], [31]. The correspondence between current blockades and translocation of DNA between compartments was demonstrated by quantifying the DNA in the cathodic compartment using competitive PCR [22].
However, there are limitations to using α-HL for sequencing. First are the obvious structural limitations—the protein structure is difficult to change in a predictable way. Though it is possible to introduce subtle mutations into the protein, gross structural changes are inordinately difficult. Chief among these structural limitations in α-HL are the length of the nanopore, and hence, the thickness of the membrane, and the diameter of the pore. For example, the α-HL channel limiting aperture is only 1.5 nm in diameter—it will not admit dsDNA. The shape and length of the nanopore also means that it is functionally impossible to measure only one base at a time, making the sequence nontrivial to interpret, as multiple nucleotides are contributing to the signal. Finally, the lipid bilayer presents another limitation. The lipid bilayer membrane is typically 25–100 µm in diameter and only 5-nm thick. It ruptures after a few hours of use or after cycling the electrolyte a few times and the large size of the membrane produces a capacitance that adversely affects the frequency and noise performance.
An alternative approach using protein nanopores is to engineer α-HL in such a way to improve the signal to noise ratio, i.e., to hold a nucleotide in place for a longer period of time to perform more averaging. By modifying the α-HL such that a cyclodextrin is placed in the β-barrel, the time that the pore is occluded by a single nucleotide can be extended. This allows more accurate measurements (>90%) of what nucleotide is in the pore based on blockade current [62]. This method has been used recently to determine the composition of ssDNA using an exonuclease to cleave off individual nucleotides, then measure them as they are captured by the α-HL nanopore [48]. This could potentially be used on raw, double-stranded, genomic DNA, and is even sensitive to base modifications such as cytosine methylation. However, it suffers a problem with logistics, i.e., how to transport the cleaved nucleotides from the exonuclease to the pore, ensuring that they arrive in the same sequence as found in the original DNA strand, that none escape (missing a base), and that the exonuclease does not outpace the pore. Tethering the exonuclease to the nanopore has been proffered as a solution, but this scheme is a nontrivial extension to the original nanopore sequencing concept.
V. Solid-State Nanopore
In contrast to α-HL, in a solid-state membrane the pore geometry, the thickness and composition of the membrane can all be controlled with subnanometer precision using semiconductor nanofabrication practices, allowing for nanopore diameters and lengths to be tailored to a specific purpose. This precision translates directly into control of the distribution of the electric field, which has already led to the development of the most sensitive device for charge measurement: the single electron transistor [63]. Solid-state pores also offer vastly improved stability allowing for much harsher chemical and thermal environments useful for denaturing the DNA, as well as allowing for easier integration with other electrical or microfluidic components. Finally, through microfabrication techniques, the solid-state membrane can be reduced to submicrometer scales, in principle, mitigating parasitic capacitance effects and improving electrical performance. There are several different methods available to create nanopores in thin membranes, such as, ion-beam milling [64], ion-track etching [65], silicon dioxide reflow [66], or electron-beam ablation/sputtering [33]. These techniques may be used on a variety of different membrane materials allowing for different chemical properties, e.g., surface charge density and electrical properties, e.g., capacitance.
Even with the higher stability and greater degree of structural and electrical flexibility conferred by using solid-state nanopores, there are still the central issues of SNR and bandwidth to overcome. Some have attempted to increase the SNR and remove the bandwidth limitations by altering the method of measurement. Rather than trying to detect a small alteration in the ionic current through the nanopore, some groups are attempting to detect the difference between bases/base pairs through measurement of transverse tunneling or capacitive currents as the DNA passes through the nanopore [67], [68].
Thus far, the majority of experimental attention has been focused on measuring current that flows and is blocked by the DNA molecule in the pore. This is fraught with difficulty in teasing out the DNA sequence from the structural and electrostatic effects that the molecule has on current. What if, instead, we actually try to directly probe each base or base pair as it goes by, using electrodes, which are placed extremely close to the DNA molecule, orthogonal to the DNA backbone? Theoretical studies have shown that with such electrodes, the electron tunneling current should be significantly different between the different nucleotides [see Fig. 4(a)] [69], [70]. There are advantages to this approach—e.g., directly probing the nucleotide in question, rather than the blockage of ionic current. Lagerqvist et al. suggests that 107 current measurements per second should be sufficient to identify individual bases—or 10 MHz—which should be possible with proper electrode design [69]. However, differences in tunneling current have not yet been experimentally shown to differentiate individual bases, or base pairs, from each other in a single DNA strand using typical STM probes although prospects are brighter for chemically modified STM [71], [72]. Tunneling current is exquisitely sensitive to bases and position, which is a blessing and a curse, especially considering the thermal fluctuations predicted to occur. Construction of transverse electrodes with atomic precision is a nontrivial task, considering the strict nanofabrication rules in place for the construction of the nanopore. Different groups are attempting this in different ways: trying to place the electrodes within the pore or on the surface of the pore; or using carbon nanotubes placed over the pore aperture [73].
Fig. 4.
Sequencing DNA using a nanopore. Schematic representations of three different schemes for sequencing DNA using a nanopore. (a) Electrodes, produced from carbon nanotubes, oriented transverse to the DNA axis probe the chemoelectronic structure of DNA via tunneling or electrochemistry as it translocates through a nanopore. (b) Alternating external bias Vex drives a single stranded DNA strand back and forth through a nanometer-diameter pore in a synthetic membrane submerged in electrolyte solution. Insulating SiO2 (Shown in gray) that separates two conducting plates made of highly doped silicon (shown in yellow). SiO2 is also present at the pore’s surface. The silicon layers are used as electrodes to measure the electrostatic potentials induced by DNA motion in the pore. (c) Snapshot of dsDNA trapped in a 2.0-nm-diameter pore. The DNA preserved its canonical B-form structure outside the pore constriction, whereas in the constriction it is stretched. Changes in the ionic current can be used to identify the base pair sequence.
Another method of potentially detecting the nucleotide present in the nanopore is through capacitive detection [see Fig. 4(b)]. By placing electrodes in the nanopore, the electrostatic potential in the pore can be measured. If DNA is cycled back and forth through the pore (with an amplitude of oscillation of ∼10 Å), an effect due in part to the dipole moment of the base present in the pore constriction is measured. The dipole moment should be characteristic of the base in the pore, allowing for identification of the DNA sequence [68].
Each of these implementations have similar issues of SNR and bandwidth. Many different strategies have been proposed to attempt to control the velocity of DNA translocation, from the breakneck speed of 1 bp/10 ns. Controlling DNA translocation speed would allow for multiple measurements per base/basepair, improving SNR and removing the bandwidth issue. Fine control of DNA motion within the nanopore would even allow for a back-and-forth motion via an ac driving voltage—potentially allowing frequency filtering of the base/basepair characteristic signal.
VI. Progress Towards Sequencing
Using DNA hairpins can result in interesting behavior when interacting with a nanopore of appropriate size. For pore diameters below 1.5 nm, the hairpin unzips breaking the hydrogen bonds making up the basepairs in the duplex. However, if the nanopore is between 1.6 and 2.3 nm, respectively, the dsDNA head of the hairpin will actually stretch, transitioning from B-form to the S-form or stretched form of DNA due to the strong electric field present in the pore [52], [74]. S-form DNA is around 1.7 times longer and 30% smaller in diameter compared to B-form DNA [75]. Unzipping actually requires less energy than stretching the duplex, and correspondingly will occur at a lower electric field strength [74].
The fact that a nanopore can be used to stretch DNA has interesting implications. Using both DNA hairpins [74], and blunt dsDNA duplexes [29], [76], [77], we have shown that above a certain electric field strength, a DNA molecule can permeate pores <3 nm in diameter if it stretches. When forced into a pore smaller than the double helix, the leading edge of the dsDNA can penetrate the membrane via a nanopore a ∼2.5 nm in diameter. If the differential force acting on the leading nucleotides is insufficient to stretch the helix, the translocation stalls there. However, applying a voltage bias above threshold provides a differential force exceeding that required to stretch dsDNA (∼60 pN), and the molecule is pulled towards the center of and eventually through the membrane [29]. The DNA entering the pore is thus altered by a forced structural transition, similar to Akeson et al.’s original attempts with RNA molecules [31]. The two strands comprising the double helix, however, do not pass through pores with diameters 1.6 < d < 2.5 nm in the same way as they do through larger pores. The confinement of the smaller pores causes the base pairs to tilt. As this change is energetically unfavorable, we believe that there is activation energy required to begin this stretching transition on the leading edge of the DNA, and below a certain applied voltage, no DNA will be able to translocate. Moreover, we have found that the threshold has a dependence on the pH, the composition of the strand and the methylation profile. In particular, methylation profile seems to alter the E-field threshold dramatically [77].
An example of this stretching trap is shown in Fig. 5. Using the 2.3 × 2.0 nm nanopore pictured in Fig. 5(a), we first determined the voltage required for the stretching transition (300 mV) by measuring the frequency of current blockades at different voltages, as pictured in Fig. 5(b). We then applied 600 mV to initially force a DNA molecule into the pore, then 200 µs after a current blockade was detected, the voltage was rapidly reduced to 100 mV, and the resultant current trace is pictured in Fig. 5(c). The current blockade in this case lasted for ∼20 s for a λ-DNA molecule. Comparing histograms of the current measured during the blockade [see Fig. 5(d)] and the open pore current measured after the blockade [see Fig. 5(e)] reveals a difference in the width of the distribution. A close examination of the blockade current histogram reveals finer structure; a two Gaussian fit seems to fit the distribution better than a single Gaussian.
Fig. 5.
(a) TEM micrograph of a 2.3 × 2.0 nm nanopore in a silicon nitride membrane 15-nm thick. (b) Frequency of blockades observed with the same pore as a function of membrane voltage illustrating a frequency drop as voltage decreases below 0.3 V, which we attribute to the threshold for stretching. The dotted line represents a fit to the data. (c) Approximately 20 s current blockade. Triggered by the onset of a blockade in a 2.3-nm pore, the membrane voltage is switched from 600 mV (above the stretching threshold) to 100 mV (below threshold). As a result, the molecule is trapped in the pore till t= 23.4 s. (Inset) Snapshot taken from a molecular dynamics simulation showing dsDNA trapped in the nanopore. (d) Histogram showing the distribution of the current during the blockade in an interval t= 0.5s when λ-DNA is trapped and (e) open pore after the molecule exits the trap. The distribution for the trapped molecule must be fit to at least two Gaussians: one (solid blue) offset from the median (ΔI = 0) by +1.8 pA with a width of 14.4 pA; and another (solid red line) offset by −2.1 pA with a width of 12.9 pA. The black line represents the sum. In contrast, the data in (e) representing the open pore can be fit by a single Guassian with a width 8.6 pA.
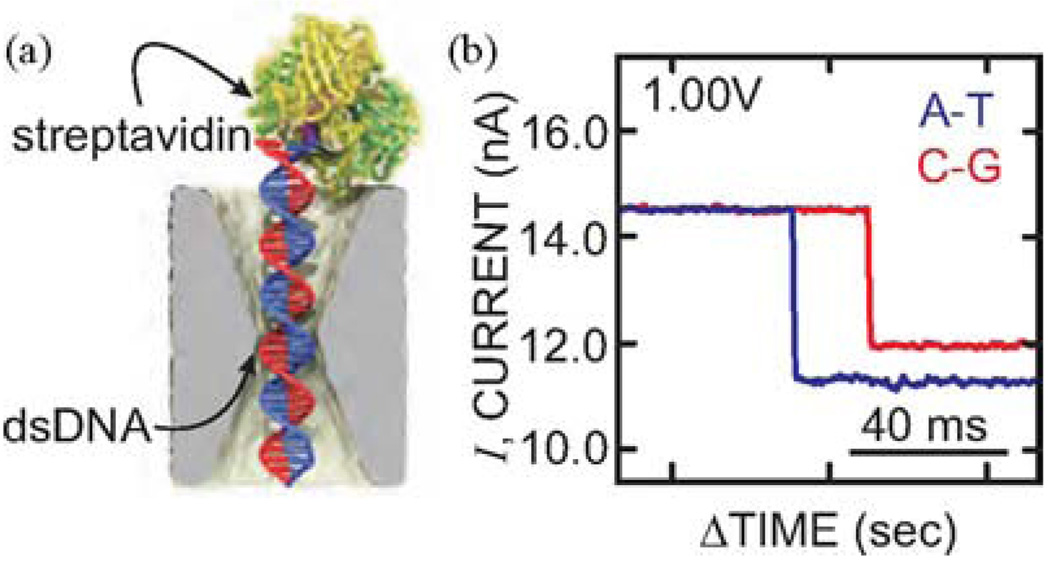
We attribute these separate peaks to resolved C-G/G-C and A-T/T-A bps, respectively. This assertion is corroborated by even longer duration measurements of blockade currents associated with streptavidin bound, 100 bp long, C-G and A-T biotinylated duplexes trapped by the electric field in a pore in a configuration represented schematically in Fig. 6(a). Streptavidin has an extraordinary affinity for biotin, which we leveraged to measure the blockade current for a trapped biotin–DNA in a 2.5 × 2.3 ± 0.2 nm pore immersed in 1 M KCl. At low voltage, the duration of a blockade is interminable—the blockade ends only if the voltage is manually reversed and the DNA is impelled out of the pore, streptavidin and all. However, the dwell time is an exponentially decreasing function of the applied voltage. In our analysis of the blockades due to the two variants, C-G and A-T, we focused on dwell times at the peak of the distribution or longer and on traces with essentially the same open pore current measured after intervening flushes and cleans. Fig. 6(b) shows a typical current blockade observed at 1.0 V for dwell times > 10 s. Clearly, it is easy to discriminate what is initially at low voltage C-G base pairs stretched in the pore from the smaller blockade current associated with A-T—the difference is 533 ± 98 pA at 1 V. It seems that bases can easily be discriminated under these conditions over a range of voltage. Apparently, the increase in molarity (10×) and larger voltage (10×) exaggerate the effect of stretching on the blockade current.
Fig. 6.
Streptavidin bound biotin DNA trapped by the electric field in a nanopore. (a) Model of biotinylated dsDNA bound to streptavidin in a 2.6 nm × 2.1 nm cross section pore in a 23 nm thick membrane at very low electric field. (b) Current blockade measured at 1.0 V transmembrane bias in 1M KCl in a 2.6 × 2.1 nm pore associated with what is initially C-G and A-T duplexes biotinylated to streptavidin. The difference in blockade current can be used to discriminate bases.
Using the stretching transition, we have recently shown that it is possible to trap a single dsDNA molecule in a nanopore <3nm in diameter by first applying a voltage larger than the threshold, forcing the molecule into the pore, and then dropping the voltage below the threshold value. If the electric field is then rapidly switched to a value below threshold during the translocation, the DNA translocation proceeds very slowly, presumably due to the increased energy required for the transition to S-form. In the inset to Fig. 5(c), an MD representation of a dsDNA molecule trapped in a pore in this way is shown. Using this method, it is possible to trap a single λ-DNA molecule for tens of seconds in the pore; compared to the milliseconds previously demonstrated—this represents a velocity change of five orders of magnitude. This simple result opens the door to more comprehensive measurements of DNA sequences. Moreover, MD simulations of these experiments reveal that, when trapped, dsDNA is stretched in the pore constriction in a way which preserves the natural inclination of the base pairs, the angle they make relative to the helical axis [see Fig. 7(a)] [75]. Fig. 7(b) shows the cumulative number of base-pairs passing through the pore as a function of time. With voltages above the threshold, the DNA translocates easily, but below the threshold, it oscillates in place, trapped by a harmonic potential with a spring constant of ∼7 nN/nm.
Fig. 7.
Trapping dsDNA in a synthetic nanopore. (a) Snapshot of dsDNA trapped in a 2.0-nm diameter pore in a 0.1-M KCl solution. Simulations like these were used to determine the sensitivity of the ionic current blockade to the type and orientation of the confined base pair. The DNA preserved its canonical B-form structure outside the pore constriction, whereas in the constriction it is stretched. (b) MD simulations showing the number of base pairs permeating the pore at four different biases. (Inset) Histogram of the displacement of the base pair nearest the center of the membrane at 0 V. The solid line shows the expected distribution of a harmonic trap with a 7.2 nN/nm spring constant.
There are other potential methods to control the translocation of DNA within the pore. For example, Polonsky et al. suggested using electrodes embedded in the pore to control the translocation, i.e., generating a voltage based potential well inside the pore to control translocation [78]. Another potential method is by pulling on a bead attached to a piece of DNA. Using an optically trapped bead to pull back on the DNA, Trepagnier et al. have slowed the translocation of DNA by a factor of ∼200 (150 bp/ms) and even been able to “floss” the DNA back and forth through the pore, allowing for repeated measurements [79]. Others have used a similar setup using magnetic tweezers— which can apply larger forces but with less precision [80].
VII. Nanopore—Challenges
One of the most severe limitations affecting the implementation of high-throughput single molecule sequencing using a nanopore is associated with bandwidth and noise in the measurement of the blockade current. While it is possible to differentiate one long strand of homonucleotide DNA from another type by measuring the level of the blocking current, the rapid rate of translocation through the pore, coupled with the narrow bandwidth and noise (2–10 pA-rms) in the measurement, frustrates discrimination with single nucleotide resolution in a polynucleotide. However, a cursory examination of the data from Fig. 5(c) and (d) reveals the challenge for sequencing DNA with a nanopore.
Two properties of the trapped dsDNA make sequencing possible. First, the ions passing through the pore are forced to come in contact with nucleotides in the constriction. Second, because trapping the DNA allows current data to be acquired over long time intervals while the same nucleotides are held near the constriction, the sequence-dependent current values can be averaged for greater accuracy. So, the molecular configuration determines how the ions passing through the pore come into contact with the nucleotides, while the translocation kinetics affect the time interval in which the same nucleotides are held in the constriction as the data is acquired.
The bandwidth and noise performance of the measurement system represent severe limitations for high-throughput sequencing with a solid-state nanopore. As a first attempt to reconcile the SNR required for single molecule detection with the bandwidth, Smeets et al. [81] has analyzed the electrical characteristics of a nanopore, representing it by an equivalent lumped element circuit consisting of a resistance associated with the pore [shown in Fig. 8(a)] Rp in parallel with the total membrane capacitance (including parasitics), Cm, and a series resistance Rel associated predominately with the electrolyte. According to this model, a change in the pore resistance affects the (Fourier transform of the) membrane voltage vm according to
| (1a) |
| (1b) |
| (1c) |
so that the voltage across the membrane has a pole at fp = 1/2πRe1Cm and that the transient response characterized by υm (t) = υin e −t/CmRel, which has a characteristic response time of τ = CmRel ∼ 1 µs for a 12–15 nm thick membrane, with Rel ∼ 10–100 kΩ for 10–100 mM KCl, which corresponds to a bandwidth of Δf < 40 kHz. Considering the typical velocity of long DNA fragments through a nanopore (1 bp/10 ns), a response time < 10 ns is required to capture a current transient associated with a single base pair.
Fig. 8.
(a) Simplified lumped element model from Smeets et al. [81]. (b) Magnitude of the pore current as a function of frequency measured through two different membranes: one with a 3-nm pore in a nitride layer 12-nm thick; and another with a 5-nm pore in a 30-nm thick nitride layer with a polyimide coating along with corresponding fits to the data (solid lines) (c) Noise power spectra of two nanopores measured in (b). From bottom to top, a 300-MW resistor, a 5-nm pore in polyimide covered Si3N4 membrane and a 3-nm pore in 12-nm Si3N4 membrane. The low frequency 1/f noise (red line), the high frequency dielectric noise (magenta line) along with the amplifier noise (orange) are analyzed for a 5-nm pore in polyimide-coated membrane. The fit to the total noise is shown in green. (d) RMS current noise as a function of bandwidth for the same two pores. The capacitive noise predominates at high frequency. Adapted from Ref. [82].
The large membrane capacitance, found prevalently in reports on both α-HL and solid-state nanopores, also has a deleterious effect on the SNR. We have measured the magnitude of the current, the power spectrum density, and the rms noise for both a 12-nm silicon nitride membrane and a 30-nm composite nitride/polyimide membrane with the same area [see Fig. 8(b)–(d)] [82]. Fig. 8(b) shows the current frequency responses in 1-M KCl electrolyte associated with two different nitride thicknesses 12 and 200 nm on a silicon substrate. In Fig. 8(b), the 12 nm membrane has a 3.3 × 4.8 ± 0.2 nm cross-section pore in it, while the 30-nm membrane with polyimide on top has a 1.7 × 2.8 ± 0.2 nm pore. Generally, we find that the frequency response of the current through the membrane consists of two components: one associated with the conductance through the pore that predominates at low frequency and is manifested by zero-slope versus frequency; and another due to the displacement current associated with the membrane capacitance and associated parasitics. While both depend linearly on the applied voltage, the displacement current increases with frequency, which is why the current grows so large at high frequency.
The noise power spectra measured in 1-M KCl are shown in Fig. 8(c) along with the spectrum of a 300-MΩ resistor, a value comparable to the resistance of the 2.2-nm pore. We analyzed the noise into three components: thermal, 1/f, and dielectric noise. We expect that the thermal noise spectral density associated with the pore resistance, St = 4kBT/R, to be negligible over the band <100 kHz since the noise from the resistor is below all of the nanopore spectra. At low frequencies, we observed that the noise power density is inversely proportional to the frequency, which is indicative of excess or 1/f, noise. Its noise power spectrum is modeled by S1/f = I2A/fβ = I2(α/Nc)/fβ, where I is the current through the device, α is the Hooge parameter (an empirically determined proportionality constant that depends on the type and concentration of charge carriers, Nc is the total number of current carriers, f is the frequency, and β is an exponent that is typically unity. Fig. 8(d) illustrates that 1/f noise becomes negligible at frequencies >1 kHz, and the spectrum exhibits linear frequency dependence up to about 50 kHz. According to the figure, the noise in the range 100 Hz–50 kHz is the dominate contribution to the rms-current noise—it is exponentially larger than the 1/f component. The linear frequency dependence coupled with the lack of a dependence of the noise in this part of the spectrum on the electrolyte concentration indicates dielectric noise with a spectrum of the form: SD = 4kBTDCD(2π f), where kB is Boltzmann’s constant, T is the absolute temperature, and D and CD are the loss tangent and the effective capacitance of the dielectric material.
Preliminary MD simulations, extrapolated from our experiments, indicate that to discriminate between C-G and G-C base pairs on ds-DNA, we demand at minimum that ΔI ∼3 pA for a 2.0-nm pore (in 100-mM KCl). (This signal is not optimized, e.g., it seems likely that the signal will be larger with increased electrolyte concentration and a slit geometry.) Therefore, for signal-to-noise >2, we need peak-to-peak noise <1.5 pA or an rms value of Irms ∼1.5 pA/8 = 0.2 pA. If dielectric noise associated with the capacitance predominates, then , where D is the dielectric loss constant (D ∼0.2 for our membranes). For a bandwidth Δf ∼ 1 kHz, we estimate that DCm ∼ 1 pF is required to discriminate between base pairs, which is a factor ten times smaller than the typical membrane capacitance. On the other hand, for a $1000 genome, corresponds to an estimated throughput of 330 000 bp/s, which translates to a capacitance of 10 fF for the same noise current specification. A 10 fF parallel plate capacitance in a 10-nm thick silicon nitride would have an area of about 1 µm × 1 µm, which is accessible with current semiconductor microfabrication technology.
VIII. Conclusion
Sequencing a single molecule of DNA using a nanopore is a revolutionary concept because it combines the potential for long read lengths (>5 kbp) with high speed (1 bp/10 ns), while obviating the need for costly procedures like PCR amplification due to the exquisite single molecule sensitivity. The prospects for implementing this concept seem bright. The cost savings from the removal of required reagents, coupled with the speed of nanopore sequencing places the $1000 genome within grasp. But challenges remain, i.e., high fidelity reads demand stringent control over both the molecular configuration in the pore and the translocation kinetics. The molecular configuration determines how the ions passing through the pore come into contact with the nucleotides, while the translocation kinetics affect the time interval in which the same nucleotides are held in the constriction as the data is acquired. Proteins like α-HL and its mutants offer exquisitely precise self-assembled nanopores and have demonstrated the facility for discriminating individual nucleotides but it is currently difficult to design protein structure ab initio, which frustrates tailoring a pore for sequencing genomic DNA. Nanopores in solid-state membranes have been proposed as an alternative, because of the flexibility in fabrication and ease of integration into a sequencing platform. Preliminary results have shown that with careful control of the dimensions of the pore and the shape of the electric field, control of DNA translocation through the pore is possible. Furthermore, discrimination between different basepairs of DNA may be feasible. Thus, a nanopore promises inexpensive, reliable, high-throughput sequencing, which could thrust genomic science into personal medicine.
Acknowledgment
The authors would like to gratefully acknowledge contributions from their collaborators K. Schulten, S. Sligar, J. P. Leburton, Q. Zhao, and B. Luan.
This work was supported by the National Institutes of Health under Grant R01 HG003713A and Grant PHS 5 P41-RR05969 and by the National Science Foundation under Grants TH 2008-01040 ANTC and PHY0822613. The supercomputer time was provided through TeraGrid via a Large Resources Allocation Grant MCA055S028. The review of this paper was arranged by Associate Editor C. Zhou.
Biographies

Winston Timp (M’06) received the B.S. degrees in electrical engineering in 2001, the B.Sc. degree in biochemistry, chemistry, and physics from the University of Illinois at Urbana-Champaign, Urbana, in 2002, and the M.S. and Ph.D. degrees in electrical engineering from the Massachusetts Institute of Technology, Cambridge, in 2005 and 2007, respectively, working under P. Matsudaira at the Whitehead Institute for Biomedical Research.
He is currently a Postdoctoral Fellow at the Center for Epigenetics, Department of Medicine, Johns Hopkins University, Baltimore, MD. His research interests include biophysics, cell signaling, and epigenetics.

Utkur M. Mirsaidov received the Ph.D. degree in physics from the University of Texas at Austin, in 2005 working under Prof. John Markert.
He did his Postdoctoral work with Gregory Timp at the Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign. His research interests include single molecule biophysics, bio-imaging and biosensors.

Deqiang Wang received the B.S. degree in the Department of Electrical Engineering from University of Jilin, Jilin, P.R. China, in 2001 and the Ph.D. degree from the Institute of Microelectronics, Chinese Academy of Sciences, in 2006, under the supervision of Prof. Tianchun Ye and Prof. Changqing Xie.
Currently, he is a Postdoctoral Research Associate in the Beckman Institute for Advanced Science and Technology, University of Illinois, Urbana-Champaign. His main research interest is single-molecule nanopore detection. His research activities also include e-beam and X-ray lithography, micro and nano fabrication, X-ray optical elements.
Jeff Comer, photograph and biography not available at the time of publication.
Aleksei Aksimentiev, photograph and biography not available at the time of publication.

Gregory Timp (M’94–F’07) received the Ph.D. degree in electrical engineering from Massachusetts Institute of Technology, Cambridge, in 1984 under the supervision of M. Dresselhaus.
From 1984 to 1986, he was a Postdoctoral Fellow in low temperature transport and nanostructure physics at IBM with A. Fowler. In 1986, he joined Bell Laboratories, where he was engaged in the research on nanostructure physics. As part of one collaboration, he investigated low temperature transport in electron waveguides and high mobility nanostructures, which was so short that the transport is ballistic. In another effort, he explored the use of optical traps and laser focusing of single atoms for lithography applications. From 2000 to 2009, he was a Member in the Department of Electrical and Computer Engineering and the Beckman Institute for Advanced Science and Technology at the University of Illinois, Urbana. He is currently the Keough-Hesburgh Professor of electrical engineering and biological sciences at the University of Notre Dame, Notre Dame, IN, where he has been involved in the research on the boundary between biology and nanoelectronics. His research interests include using nanopore sensors to detect the electronic structure of biomolecules and using optical tweezers to manipulate nanoparticles and living cells into large arrays, and super high frequency >30 GHz circuit design using radio frequency MOSFETs.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Winston Timp, Email: timp0@alum.mit.edu, Center for Epigenetics, Department of Medicine, Johns Hopkins University, Baltimore, MD21205 USA.
Utkur M. Mirsaidov, Email: mirsaidov@gmail.com, National University of Singapore, Singapore 117543.
Deqiang Wang, Email: dewang@illinois.edu, University of Notre Dame, South Bend, IN 46556 USA.
Jeff Comer, Email: jeffcomer@gmail.com, Beckman Institute, Urbana, IL 61801 USA.
Aleksei Aksimentiev, Email: aksiment@illinois.edu, Department of Physics, University of Illinois at Urbana-Champaign, Urbana, IL 61801 USA.
Gregory Timp, Email: gtimp@nd.edu, University of Notre Dame, South Bend, IN 46556 USA.
References
- 1.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;vol. 409(no. 6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Bejerano G, Haussler D, Blanchette M. Into the heart of darkness: Large-scale clustering of human non-coding DNA. Bioinformatics. 2004;vol. 20(no. 1):i40–i48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- 3.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature. 2007;vol. 447(no. 7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;vol. 312(no. 5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JKV, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PVK, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;vol. 318(no. 5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;vol. 304(no. 5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;vol. 316(no. 5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 8.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum. Mol. Genet. 2006;vol. 15:R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- 9.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;vol. 92(no. 5):255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Sanger F, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;vol. 74(no. 12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.3130 and 3130xl Genetic Analyzers. Foster City, CA: Applied Biosystems, Inc.; 2005. [Google Scholar]
- 12.Kumagai H, Utsunomiya S, Nakamura S, Yamamoto R, Harada A, Kaji T, Hazama M, Ohashi T, Inami A, Ikegami T. Large-scale microfabricated channel plates for high-throughput, fully automated DNA sequencing. Electrophoresis. 2008;vol. 29(no. 23):4723–4732. doi: 10.1002/elps.200800301. [DOI] [PubMed] [Google Scholar]
- 13.Marziali A, Akeson M. New DNA sequencing methods. Annu. Rev. Biomed. Eng. 2001;vol. 3:195–223. doi: 10.1146/annurev.bioeng.3.1.195. [DOI] [PubMed] [Google Scholar]
- 14.Shendure J, Mitra R, Varma C, Church G. Advanced sequencing technologies: Methods and goals. Nat. Rev. Genet. 2004;vol. 5(no. 5):335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 15.Blazej R, Kumaresan P, Mathies R. Microfabricated bioprocessor for integrated nanoliter-scale sanger dna sequencing. Proc. Nat. Acad. Sci. 2006;vol. 103(no. 19):7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paegel BM, Blazej RG, Mathies RA. Microfluidic devices for dna sequencing: Sample preparation and electrophoretic analysis. Curr. Opin. Biotechnol. 2003;vol. 14(no. 1):42–50. doi: 10.1016/s0958-1669(02)00004-6. [DOI] [PubMed] [Google Scholar]
- 17.Drmanac S, Kita D, Labat I, Hauser B, Schmidt C, Burczak JD, Drmanac R. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat. Biotechnol. 1998;vol. 16(no. 1):54–58. doi: 10.1038/nbt0198-54. [DOI] [PubMed] [Google Scholar]
- 18.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd D, Johnson D, Luo S, McCurdy S, Foy M, Ewan M. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;vol. 18(no. 6):630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 19.Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum. Mutat. 2002;vol. 19(no. 5):479–485. doi: 10.1002/humu.10078. [DOI] [PubMed] [Google Scholar]
- 20.Mitra RD, Shendure J, Olejnik J, Edyta Krzymanska O, Church GM. Fluorescent in situ sequencing on polymerase colonies. Anal. Biochem. 2003;vol. 320(no. 1):55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 21.Branton D, Deamer D, Marziali A, Bayley H, Benner S, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;vol. 26(no. 10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasianowicz J, Brandin E, Branton D, Deamer D. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Nat. Acad. Sci. 1996;vol. 93(no. 24):13 770–13 773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 2009;vol. 27(no. 9):847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;vol. 24(no. 3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Gupta PK. Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;vol. 26(no. 11):602–611. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, DiMeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. Single-molecule DNA sequencing of a viral genome. Science. 2008;vol. 320(no. 5872):106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 27.Coulter W. Means for counting particles suspended in a fluid. U.S. Patent. 1953;2(656):508. [Google Scholar]
- 28.Lederer H, May RP, Kjems JK, Baer G, Heumann H. Solution structure of a short DNA fragment studied by neutron-scattering. Eur. J. Biochem. 1986;vol. 161(no. 1):191–196. doi: 10.1111/j.1432-1033.1986.tb10141.x. [DOI] [PubMed] [Google Scholar]
- 29.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;vol. 5(no. 10):1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinland B, Pluen A, Sturm J, Weill G. Persistence length of single-stranded dna. Macromolecules. 1997;vol. 30(no. 19):5763–5765. [Google Scholar]
- 31.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 1999;vol. 77(no. 6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA. 2000;vol. 97(no. 3):1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heng JB, Ho C, Kim T, Timp R, Aksimentiev A, Grinkova YV, Sligar S, Schulten K, Timp G. Sizing DNA using a nanometer-diameter pore. Biophys. J. 2004;vol. 87(no. 4):2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JL, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003;vol. 2(no. 9):611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 35.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E. 2005;vol. 71(no. 5):051903-1–051903-10. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 36.Gyarfas B, Olasagasti F, Benner S, Garalde D, Lieberman KR, Akeson M. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 angstrom resolution. ACS Nano. 2009;vol. 3(no. 6):1457–1466. doi: 10.1021/nn900303g. [DOI] [PubMed] [Google Scholar]
- 37.Wilson NA, Abu-Shumays R, Gyarfas B, Wang H, Lieberman KR, Akeson M, Dunbar WB. Electronic control of DNA polymerase binding and unbinding to single dna molecules. ACS Nano. 2009;vol. 3(no. 4):995–1003. doi: 10.1021/nn9000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurt N, Wang HY, Akeson M, Lieberman KR. Specific nucleotide binding and rebinding to individual DNA polymerase complexes captured on a nanopore. J. Amer. Chem. Soc. 2009;vol. 131(no. 10):3772–3778. doi: 10.1021/ja809663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benner S, Chen RJA, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat. Nanotechnol. 2007;vol. 2(no. 11):718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen P, Gu JJ, Brandin E, Kim YR, Wang Q, Branton D. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 2004;vol. 4(no. 11):2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg HC. Random Walks in Biology. Princeton, NJ: Princeton Univ. Press; 1993. [Google Scholar]
- 42.Nair PR, Alam MA. Performance limits of nanobiosensors. Appl. Phys. Lett. 2006;vol. 88(no. 23):233120-1–233120-3. [Google Scholar]
- 43.Viovy JL. Electrophoresis of dna and other polyelectrolytes: Physical mechanisms. Rev. Mod. Phys. 2000;vol. 72(no. 3):813–872. [Google Scholar]
- 44.Luan B, Aksimentiev A. Electro-osmotic screening of the DNA charge in a nanopore. Phys. Rev. E. 2008;vol. 78(no. 2) doi: 10.1103/PhysRevE.78.021912. article 021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freund JB. Electro-osmosis in a nanometer-scale channel studied by atomistic simulation. J. Chem. Phys. 2002;vol. 116(no. 5):2194–2200. [Google Scholar]
- 46.Fologea D, Uplinger J, Thomas B, McNabb DS, Li JL. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;vol. 5(no. 9):1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wanunu M, Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007;vol. 7(no. 6):1580–1585. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- 48.Clarke J, Wu H, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore dna sequencing. Nat. Nanotechnol. 2009;vol. 4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 49.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single dna molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc. Natl. Acad. Sci. U.S.A. 2008;vol. 105(no. 50):720–725. doi: 10.1073/pnas.0808296105. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golovchenko J. private communication. 2009 Mar. [Google Scholar]
- 51.Chang H, Kosari F, Andreadakis G, Alam M, Vasmatzis G, Bashir R. DNA-mediated fluctuations in ionic current through silicon oxide nanopore channels. Nano Lett. 2004;vol. 4(no. 8):1551–1556. [Google Scholar]
- 52.Comer J, Dimitrov V, Zhao Q, Timp G, Aksimentiev A. Microscopic mechanics of hairpin DNA translocation through synthetic nanopores. Biophys. J. 2009;vol. 96(no. 2):593–608. doi: 10.1016/j.bpj.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aksimentiev A, Heng JB, Timp G, Schulten K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys. J. 2004;vol. 87(no. 3):2086–2097. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan R, Karnik R, Yue M, Li DY, Majumdar A, Yang PD. DNA translocation in inorganic nanotubes. Nano Lett. 2005;vol. 5(no. 9):1633–1637. doi: 10.1021/nl0509677. [DOI] [PubMed] [Google Scholar]
- 55.Smeets RMM, Keyser UF, Krapf D, Wu MY, Dekker NH, Dekker C. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2006;vol. 6(no. 1):89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 56.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri M. Recognizing a single base in an individual DNA strand: A step toward DNA sequencing in nanopores. Angew. Chem. Int. Ed. 2005;vol. 44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. From the cover: Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl. Acad. Sci. U.S.A. 2009;vol. 106(no. 19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo KF, Ala Nissila T, Ying SC, Bhattacharya A. Sequence dependence of DNA translocation through a nanopore. Phys. Rev. Lett. 2008;vol. 100(no. 5):058101.1–058101.4. doi: 10.1103/PhysRevLett.100.058101. [DOI] [PubMed] [Google Scholar]
- 59.Meller A, Branton D. Single molecule measurements of DNA transport through a nanopore. Electrophoresis. 2002;vol. 23(no. 16):2583–2591. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 60.Furini S, Domene C, Rossi M, Tartagni M, Cavalcanti S. Model-based prediction of the α-hemolysin structure in the hexameric state. Biophys. J. 2008;vol. 95(no. 5):2265–2274. doi: 10.1529/biophysj.107.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;vol. 274(no. 5294):1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 62.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: Direct identification of ribonucleoside and deoxyribonucleoside 5’-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J. Amer. Chem. Soc. 2006;vol. 128(no. 5):1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 63.Likharev KK. Single-electron devices and their applications. Proc. IEEE. 1999 Apr.vol. 87(no. 4):606–632. [Google Scholar]
- 64.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001;vol. 412(no. 6843):166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 65.Siwy Z, Fulinski A. Fabrication of a synthetic nanopore ion pump. Phys. Rev. Lett. 2002;vol. 89(no. 19):198103–198106. doi: 10.1103/PhysRevLett.89.198103. [DOI] [PubMed] [Google Scholar]
- 66.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2003;vol. 2(no. 8):537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 67.Ohshiro T, Umezawa Y. Complementary base-pair-facilitated electron tunneling for electrically pinpointing complementary nucleobases. Proc. Natl. Acad. Sci. U.S.A. 2006;vol. 103(no. 1):10–14. doi: 10.1073/pnas.0506130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sigalov G, Comer J, Timp G, Aksimentiev A. Detection of DNA sequences using an alternating electric field in a nanopore capacitor. Nano Lett. 2008;vol. 8(no. 1):56–63. doi: 10.1021/nl071890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagerqvist J, Zwolak M, Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;vol. 6(no. 4):779–782. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zwolak M, Di Ventra M. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 2005;vol. 5(no. 3):421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 71.Chang SA, He J, Lin LS, Zhang PM, Liang F, Young M, Huang S, Lindsay S. Tunnel conductance of watson-crick nucleoside base pairs from telegraph noise. Nanotechnology. 2009;vol. 20(no. 18):185102-1–185102-7. doi: 10.1088/0957-4484/20/18/185102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He J, Lin LS, Liu H, Zhang PM, Lee M, Sankey OF, Lindsay SM. A hydrogen-bonded electron-tunneling circuit reads the base composition of unmodified DNA. Nanotechnology. 2009;vol. 20(no. 7):075102-1–075102-8. doi: 10.1088/0957-4484/20/7/075102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meunier V, Krstic PS. Enhancement of the transverse conductance in DNA nucleotides. J. Chem. Phys. 2008;vol. 128(no. 4):041103-1–041103-4. doi: 10.1063/1.2835350. [DOI] [PubMed] [Google Scholar]
- 74.Zhao Q, Comer J, Dimitrov V, Yemenicioglu S, Aksimentiev A, Timp G. Stretching and unzipping nucleic acid hairpins using a synthetic nanopore. Nucleic Acids Res. 2008;vol. 36(no. 5):1532–1541. doi: 10.1093/nar/gkm1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: An extensible molecule. Science. 1996;vol. 271(no. 5250):792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 76.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. The electromechanics of DNA in a synthetic nanopore. Biophys. J. 2006;vol. 90(no. 3):1098–1106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mirsaidov U, Timp W, Zou X, Dimitrov V, Schulten K, Feinberg AP, Timp G. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophys. J. 2009;vol. 96(no. 4):L32–L34. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Polonsky S, Rossnagel S, Stolovitzky G. Nanopore in metal-dielectric sandwich for DNA position control. Appl. Phys. Lett. 2007;vol. 91(no. 15):153103-1–153103-3. [Google Scholar]
- 79.Trepagnier E, Radenovic A, Sivak D, Geissler P, Liphardt J. Controlling DNA capture and propagation through artificial nanopores. Nano Lett. 2007;vol. 7(no. 9):2824–2830. doi: 10.1021/nl0714334. [DOI] [PubMed] [Google Scholar]
- 80.Peng H, Ling X. Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology. 2009;vol. 20(no. 18):185101-1–185109-8. doi: 10.1088/0957-4484/20/18/185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smeets R, Keyser U, Dekker N, Dekker C. Noise in solid-state nanopores. Proc. Natl. Acad. Sci. U.S.A. 2008;vol. 105(no. 2):417–421. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dimitrov V, Mirsaidov U, Wang D, Sorsch T, Mansfield W, Miner J, Klemens F, Cirelli R, Yemenicioglu S, Timp G. Nanopores in solid-state membranes engineered for single molecule detection. Nanotechnology. 2010;vol. 21(no. 6) doi: 10.1088/0957-4484/21/6/065502. article 065502. [DOI] [PubMed] [Google Scholar]