Abstract
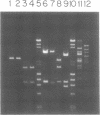
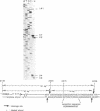
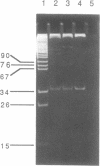
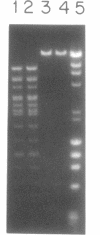
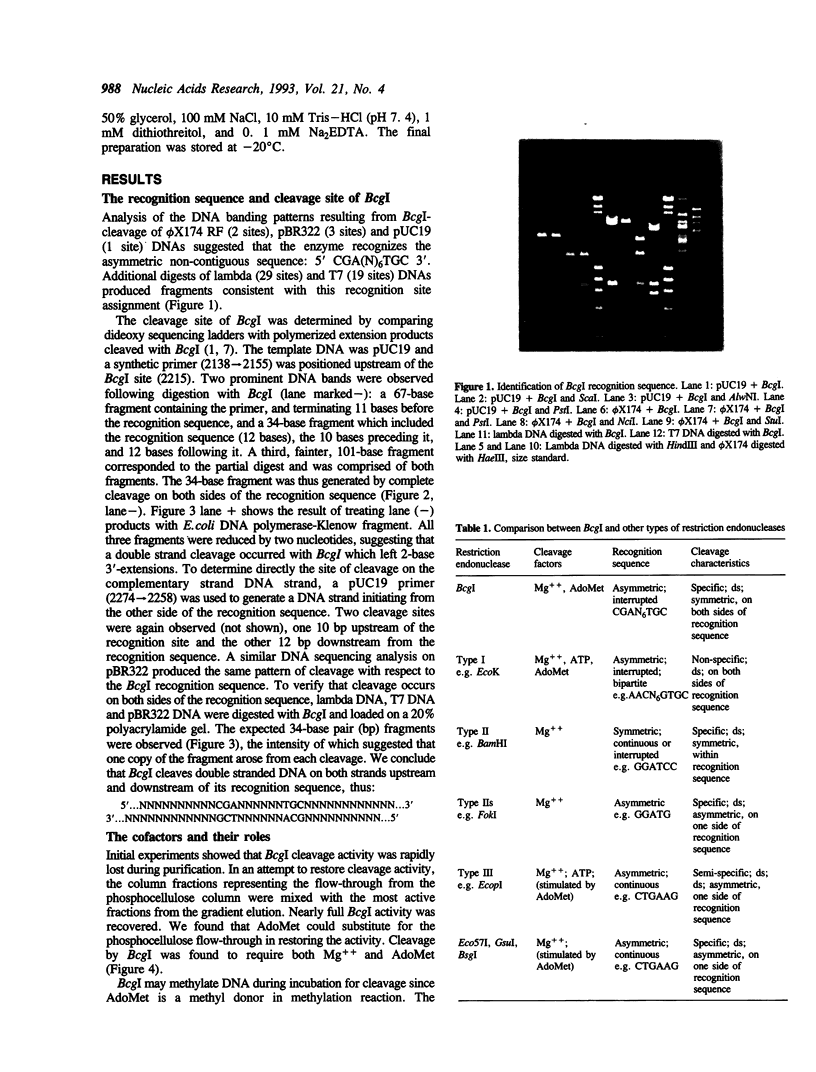
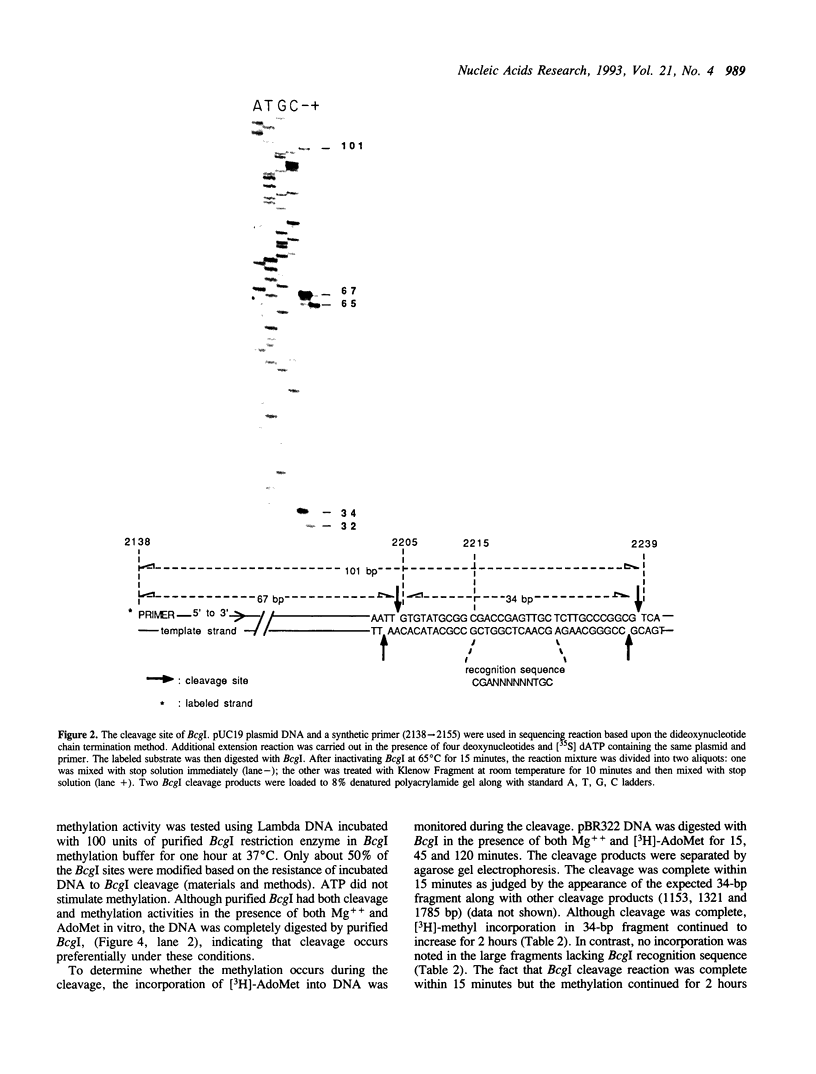
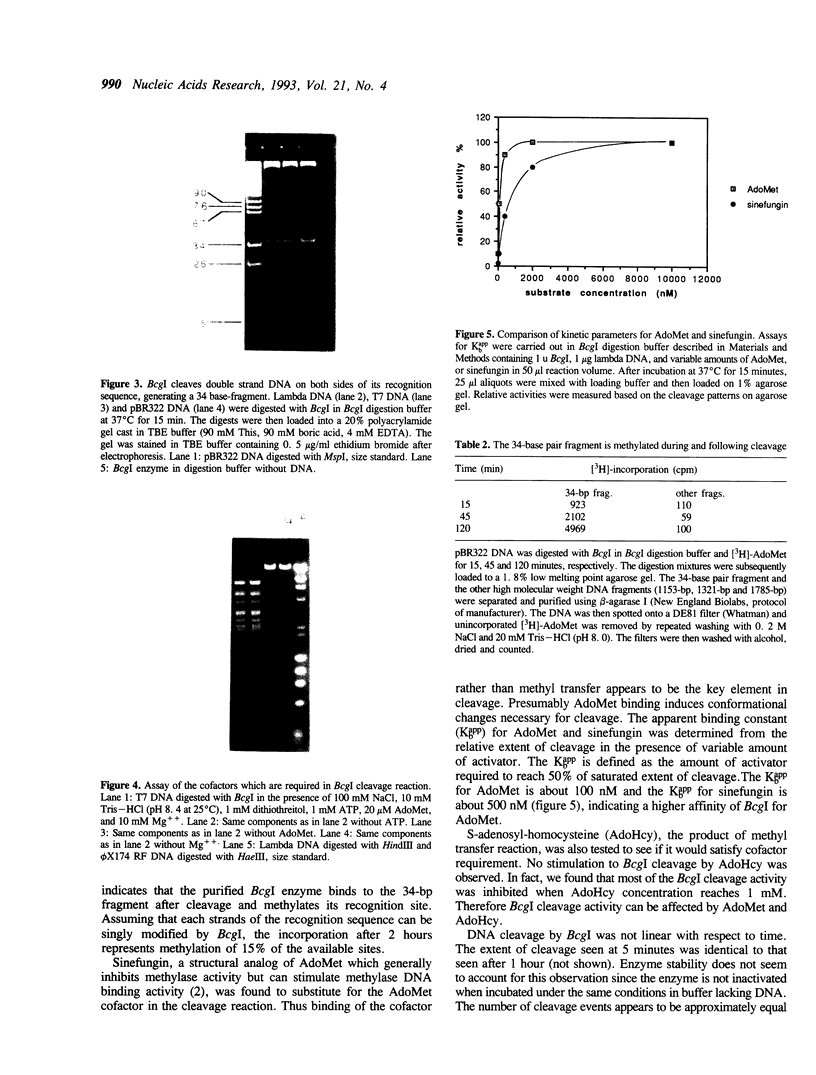
We have purified and characterized a new restriction endonuclease, BcgI, which has properties unlike those of the three recognized classes of restriction enzymes. BcgI was isolated from Bacillus coagulans, and it recognizes the sequence CGAN6TGC. BcgI cleaves double stranded DNA on both strands upstream and downstream of the recognition sequence, so that the recognition sequence is released as a 34-base pair fragment with 2-base 3'-extensions. Mg++ and S-adenosylmethionine are required for cleavage. Sinefungin, a structural analogue of AdoMet which generally inhibits methylase activity, can replace AdoMet in the cleavage reaction. The apparent binding constant (Kappd) for AdoMet is about 100 nM, while the KappD for sinefungin is about 500 nM.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown N. L., Hutchison C. A., 3rd, Smith M. The specific non-symmetrical sequence recognized by restriction endonuclease MboII. J Mol Biol. 1980 Jun 15;140(1):143–148. doi: 10.1016/0022-2836(80)90360-5. [DOI] [PubMed] [Google Scholar]
- Friedman S. Binding of the EcoRII methylase to azacytosine-containing DNA. Nucleic Acids Res. 1986 Jun 11;14(11):4543–4556. doi: 10.1093/nar/14.11.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orekhov A. V., Rebentish B. A., Debabov V. G. Novaia sait-spetsificheskaia éndonukleaza iz Streptomyces--SGRII. Dokl Akad Nauk SSSR. 1982;263(1):217–220. [PubMed] [Google Scholar]
- Petrusyte M., Bitinaite J., Menkevicius S., Klimasauskas S., Butkus V., Janulaitis A. Restriction endonucleases of a new type. Gene. 1988 Dec 25;74(1):89–91. doi: 10.1016/0378-1119(88)90259-4. [DOI] [PubMed] [Google Scholar]
- Piatrushite M. P., Bitinaite Iu B., Kershulite D. R., Menkiavichius S. Iu, Butkus V. V. Restriktsionnye éndonukleazy novogo tipa. Dokl Akad Nauk SSSR. 1987;295(5):1250–1253. [PubMed] [Google Scholar]
- Roberts R. J., Macelis D. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1992 May 11;20 (Suppl):2167–2180. doi: 10.1093/nar/20.suppl.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Roizes G., Charlieu J. P., Marcais B., Bellis M. Use of the DNA fragments generated by a restriction enzyme (Bcg I) for the construction of overlapping clone libraries. DNA Seq. 1991;2(1):65–67. doi: 10.3109/10425179109008442. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]