Abstract
Heterochromatin protein 1 (HP1) was discovered as a protein essential for maintaining the silent transcriptional status of genes located within or close to centromeric regions of Drosophila chromosomes. Mammals express three variants of HP1 and of these, HP1α is a direct homolog of Drosophila HP1. The prevailing view states that HP1 is a structural component of heterochromatin and is essential for compact DNA packaging. HP1 contains a chromodomain that binds to di- and tri-methylated lysine 9 of histone H3. Additionally, it contains a chromoshadow domain that allows HP1 to dimerize and interact with other proteins. HP1 is thought to form “bridges” between neighboring rows of nucleosomes in heterochromatin. In mammalian cells, a significant portion of HP1α is located in the centromeric regions of chromosomes. In this study, we show that the majority of HP1α is removed from centromeres upon heat shock. This occurs without a loss of H3K9 trimethylation and does not correlate with a decompaction of centromeres. Furthermore, HP1α is not degraded and remains bound to chromatin. Therefore, it is likely that HP1α is simply redistributed to euchromatic regions. We propose that this redistribution is essential for reversal of the transcriptional status of euchromatic and heterochromatic compartments.
Key words: heterochromatin, heat shock, HP1α, centromeres, DNA packaging
Introduction
Heterochromatin protein 1 (HP1) was first discovered in Drosophila as a dominant suppressor of position-effect variegation (PEV) and was later found to participate in the formation of compact heterochromatin.1,2 Later, a family of highly conserved HP1 proteins was discovered in eukaryotes. This family includes HP1α, HP1β, HP1γ and several other proteins (reviewed in refs. 3 and 4). Although initial studies demonstrated the role of HP1 in the formation of heterochromatin, especially in centromeric regions,5,6 it is becoming increasingly evident that HP1 has multiple functions and is also present in actively transcribed euchromatic regions.7 HP1 also plays a role in centromeric sister chromatid cohesion,8,9 telomere maintenance10 and DNA repair.11–13 It is possible that the three major variants of HP1 (α, β, γ) display a certain degree of functional specialization, with HP1α as the most important variant for formation of centromeric heterochromatin.14–18 Mammalian HP1α is a direct homolog of Drosophila HP1. All members of the HP1 family possess two characteristic domains: the amino-terminal chromodomain and the carboxy-terminal chromoshadow domain. The HP1 chromodomain specifically recognizes methylated lysine 9 in the N-terminal tail of histone H3,19,20 while the chromoshadow domain enables HP1 to both dimerize and to interact with other proteins.21–23 Being bound to chromatin via interaction of chromodomain with H3K9me, HP1 can recruit many proteins that participate in heterochromatin formation including the H3K9 histone methyltransferase24 and DNA methyltransferases.25 The current model of heterochromatin formation suggests a cycle of recruitment of the H3-K9 methyltransferase by HP1 and methylation of H3-K9 in neighboring nucleosomes with a subsequent recruitment of additional HP1 molecules.24 Immunostaining of HP1α has shown that it localizes to large masses of condensed chromatin, which appear to be the centromeric regions of interphase chromosomes. Indeed, the HP1α-specific antibodies predominately stain centromeres in metaphase spreads.5,14 The supposition that HP1 mediates suppression of transcription via packaging of nucleosomal arrays into compact, higher-order structures is based on the original observation that the protein is essential for manifestation of PEV. However, it is difficult to prove that compact packaging of chromatin is the major cause of transcriptional repression. It is believed that HP1 plays a structural role in establishing condensed heterochromatin, but the exact mechanisms of this process are far from understood. Previous work proposed that an HP1-dimer could bridge neighboring nucleosomes, thus stabilizing a higher-order chromatin structure.26,27 However, the role of HP1 as an essential structural component of compact heterochromatin remains experimentally unproven.26,27 It is possible that the ability of HP1 to recruit different co-repressors is of primary importance for gene silencing, while the compact packaging of DNA into heterochromatin does not, by itself, constitute a serious obstacle for transcription. Indeed, it has been shown that heterochromatic compartments contain a number of transcribed genes.28 The rapid exchange rate of HP1 does not support the hypothesis that HP1 directly contributes to the maintenance of stable, higher-order chromatin structure.29 The presence of HP1 might be necessary for heterochromatin formation but not for maintenance once it has been established. Here, we show that under heat shock conditions, HP1α is released from the centromeric regions of human chromosomes. Interestingly, this displacement of HP1α does not correlate with a loss of histone H3 lysine 9 trimethylation in centromeres or decompaction of centromeric heterochromatin. Furthermore, the displaced HP1α is not degraded and is redistributed to different genomic loci. Collectively, these data suggest that the role of HP1α in gene silencing is not directly connected to the formation of dense, higher-order chromatin structures, but rather it is based on other molecular mechanisms, such as imposing a restriction on core nucleosome dynamics.30
Results
HP1α is released from centromeric regions of interphase chromosomes under heat shock conditions.
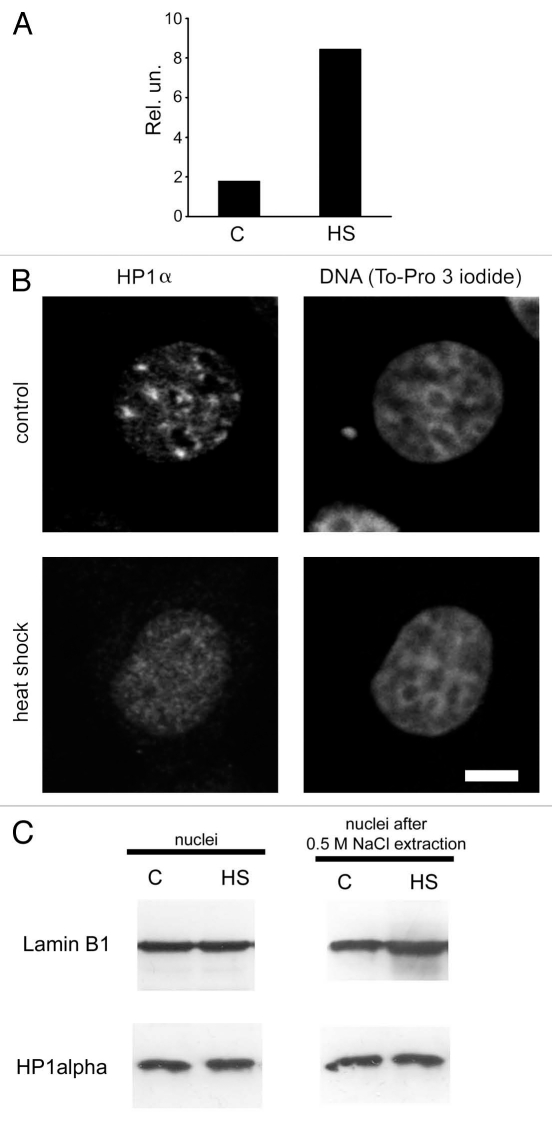
A cultured breast cancer cell line, MCF-7, was chosen as our experimental model. First, we had to determine if the conditions used represented a functional heat shock. To do this, we measured the mRNA levels of heat shock protein 70 (hsp70) in control cells and in cells stressed by being placed at 45°C for 30 min. Quantitative reverse transcriptase—PCR analysis showed that expression of hsp70 increased in response to the treatment used (Fig. 1A). Next, we immunostained human MCF-7 cells with an antibody against HP1α. This experiment indicated that a significant portion of the HP1α present in control cell nuclei was localized to large masses of condensed chromatin (compare HP1α staining of nuclei with To-Pro staining; Fig. 1B, line “control”). Based on previous observations,5,14,31 we assumed that the areas preferentially stained by the HP1α-specific antibody were clustered regions of centromeric heterochromatin. To verify this, we stained nuclei with antibodies against CENP-A, the centromeric variant of histone H3.32 The regions of preferential accumulation of the HP1α were confirmed as centromeres by co-localization of the CENP-A antibody to the same regions (Fig. 2A, line “control”). We next immunostained with an antibody against HP1α MCF-7 cells subjected to heat shock (30 min at 45°C). Surprisingly, HP1α did not form any foci in these cells, but was instead distributed almost evenly throughout the entire nuclear volume (Figs. 1B and 2A). However, the densely packed heterochromatic regions were still clearly visible upon either To-Pro or DAPI staining of the heat-shocked cells (Figs. 1B and 2A, line “heat shock”). Furthermore, such regions contained histone variant CENP-A (Fig. 2A, line “heat shock”). Thus, it seems reasonable to assume that the compact structure of the centromeres was preserved in heat shocked cells, despite the fact that HP1α was delocalized from the centromeres. It should be mentioned that the number and the size of CENP-A foci remained unchanged under heat shock conditions (see Sup. Table S1). The decrease in HP1α levels in the centromeric regions was not unique to MCF-7 cells. Similar results were obtained when human lymphoid Jurkat cells were examined (see Sup. Fig. S1). There are several possible fates of the HP1α that is delocalized from centromeres. One possibility is that it is relocated to other genomic loci that are temporarily inactivated upon heat shock. An equally likely possibility is that HP1α is simply degraded in response to heat shock. To determine which of these actually occurs, we compared the overall amount of HP1α present in normal and heat shocked cells by western blot analysis. Additionally, we compared the amount of chromatin-bound HP1α that remained in the nuclei of normal and stressed cells after medium salt extraction (0.5 M NaCl). We used nuclear lamin B1 as an internal control for these experiments. The results suggest that HP1α is not degraded in heat-shocked cells (Fig. 1C, left part). Furthermore, the proportion of HP1α bound to chromatin remained unchanged following heat shock (Fig. 1C, right part). These data suggest that heat shock caused a redistribution of HP1α from centromeres to other genomic loci. We next examined whether the centromeric localization of HP1α can be re-established if heat shocked cells are cultivated under normal conditions. In this experiment, heat shocked cells were incubated at 37°C for 1, 3, 6, 12 and 20 h after heat shock. Cells from each time point were immunostained with an antibody against HP1α. The results show that centromeric localization of HP1 is restored after 20 h of normal culture conditions following heat shock (see Suppl. Fig. S2 and S3).
Figure 1.
Redistribution of HP1α within the nucleus under heat shock conditions. (A) qRT-PCR analysis of hsp70 gene expression in control (untreated, C) and heat shock-treated (HS) MCF-7 cells. RNA extracted from treated and non-treated cells was reverse transcribed and the cDNA obtained was analyzed using a SYBR Green-based quantitative PCR approach. Amplification levels of hsp70 cDNA were normalized to the amplification level of GAPDH cDNA. Results of one representative experiment are shown. (B) Nuclear distribution of HP1α in control (untreated) and heat shock-treated cells. MCF-7 cells were immunostained with a mouse monoclonal antibody against HP1α and visualised by Alexa Fluor 488-conjugated anti-mouse IgG. DNA was stained with To-Pro 3 iodide fluorescent dye. Images were collected using a Leica laser scanning confocal microscope. Only one representative section is shown in each case. Bar scale: 5 µm. (C) Western blot analysis of HP1α in the nuclei (non-extracted and extracted with 0.5 M NaCl) of control (untreated, C) and treated with heat shock (HS) MCF-7 cells. Lamin B1 was used as a loading control.
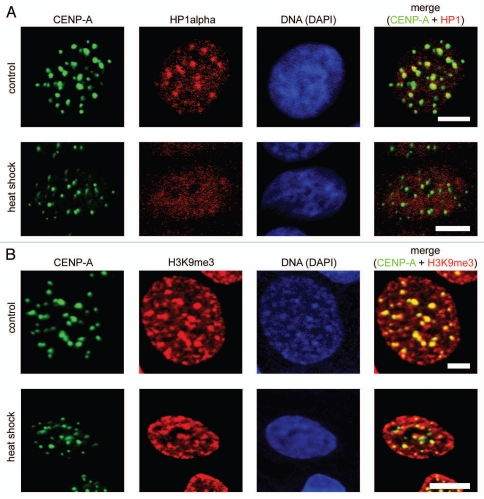
Figure 2.
Redistribution of HP1α under heat shock conditions did not lead to decompaction of centromeric DNA. (A) Human MCF-7 cells, both untreated and heat shock-treated, were double immunostained with a human antibody against CENP-A (column 1) and a mouse monoclonal antibody against HP1α (column 2). Human and mouse primary antibodies were visualized by incubation with secondary antibodies conjugated to FITC and Alexa Fluor 555, respectively. Co-localization of CENP-A foci with HP1α is shown as yellow in the merged images (column 4). (B) Human MCF-7 cells, both untreated and heat shock-treated, were double immunostained with a human antibody against CENP-A (column 1) and a rabbit polyclonal antibody against histone H3 tri-methylated at lysine 9 (H3K9me3, column 2). Human and rabbit primary antibodies were visualized by incubation with secondary antibodies conjugated to FITC and Alexa Fluor 594, respectively. Co-localization of CENP-A foci with H3K9me3 is shown as yellow in the merged images (column 4). In (A and B) DNA was stained with DAPI fluorescent dye (column 3). Images were collected using a Zeiss LSM 510 META NLO multiphoton microscope. Only one representative section is shown in each case. Bar scale: 5 µm.
Displacement of HP1 from centromeric heterochromatin does not correlate with demethylation of H3 lysine 9. HP1 contains a chromodomain that recognizes di- and tri-methylated lysine 9 in histone H3 (H3K9me2 and H3K9me3).19 The interaction between HP1 and H3K9me3 is thought to be essential for the propagation and stability of heterochromatin.33 Recently, it was shown that eukaryotic cells possess enzymes capable of removing methyl groups from H3 lysine 9.34,35 Thus, we were interested in determining if H3K9 trimethylation was temporarily removed from centromeric areas in response to heat shock. Control and heat shocked MCF-7 cells were fixed and stained with an antibody against H3K9me3. It has been reported previously that these antibodies preferentially stain compact masses of pericentromeric heterochromatin.36 Indeed, in both control and heat shocked cells we observed preferential staining of compact spots of centromeric heterochromatin, as concluded from preferential staining of the same areas with an antibody against CENP-A (Fig. 2B). In control cells, antibodies against HP1α also localized to this areas (Fig. 2A). Importantly, in heat shocked cells, the staining of pericentromeric heterochromatin with antibodies against tri-methylated lysine 9 of histone H3 remained unchanged despite the fact that Hp1a was not detected in these areas (Fig. 2A and B). The overall amount of the histone H3 containing tri-methylated lysine 9 also remained about the same in control and heat-shocked MCF-7 cells, as shown by western blot analysis (see Suppl. Fig. S4). Therefore, it is clear that H3K9 trimethylation is not, by itself, sufficient to maintain HP1α bound to the chromatin.
Removal of HP1α from pericentromeric chromatin does not affect the compaction of DNA in this region.
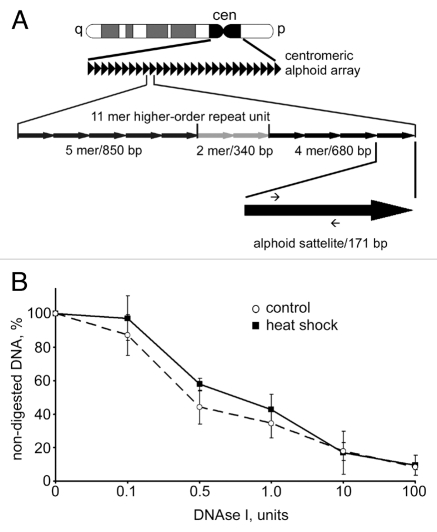
The results shown in Figures 1 and 2 suggest that removal of HP1α from centromeric regions does not affect compaction of DNA. Indeed the spots of densely packed (intensely stained by either To-Pro or DAPI) DNA co-localize with the spots stained by an antibody against CENP-A and appear to be of the same size in both control and heat shocked MCF-7 cells. To verify this, we compared the DNase I sensitivity of centromeric repeats in normal and heat shocked MCF-7 cells using a quantitative PCR-based approach. The method is based on the supposition that upon digestion of nuclei with increasing amounts of DNase I the speed of degradation of a short test amplicon will be inversely correlated with the level of DNA packaging. Our previous study demonstrated that this approach permitted us to discriminate between actively transcribed and repressed genes.37 Moreover, we tested this approach on human MCF-7 cells using as genomic models inactive in these cells erythroid-specific (β-globin) and active house-keeping (GAPDH) genes and found again that we can detect the expected difference in sensitivity of these genes to DNase I (see Sup. Fig. S5). To estimate DNase I sensitivity of centromeric repeats we designed the test amplicon within the D1Z7 repeat. Human centromeres consist of megabases of alphoid satellite DNA, a repeat family that contains ∼171 bp monomers38 (see Fig. 3A). The D1Z7 alphoid repeat used in this study is present within centromeric regions of several human chromosomes, including chromosomes 1, 5 and 19.39,40 We measured the DNase I sensitivity of this amplicon in both control and heat shocked MCF-7 cells. The data show that the DNase I sensitivity of the test amplicon does not change as a result of heat shock (Fig. 3B), indicating that the removal of HP1α from centromeric heterochromatin does not cause decompaction of these regions.
Figure 3.
DNAse I sensitivity of centromeric DNA did not change upon heat shock. (A) A scheme representing the principal organization of centromeric alphoid arrays (for details see text). Positions of primers used for qPCR are depicted as small arrows on the black arrow that represents an individual 171 bp alphoid satellite. (B) Dynamics of digestion of alphoid satellite DNA by DNase I in untreated MCF-7 cells (dashed line) and heat shocked MCF cells (black line). Aliquots (50 ng) of genomic DNA extracted from nuclei of MCF-7 cells digested with increasing amounts of DNase I were subjected to SYBR Green-based quantitative PCR analysis. The Ct values obtained were converted to DNA concentration using a standard curve (data not shown). DNase I sensitivity was expressed as a percentage of preserved template for amplification of an alphoid satellite test fragment (y-axis) and is plotted for varying DNase I concentrations (0–100 U; x-axis). The data shown are an average of four independent experiments. Error bars represent the standard deviation for each concentration.
Discussion
Under heat shock conditions, cells undergo dramatic changes, such as drastic modification of gene expression, an alteration of the epigenetic status of large genomic areas, among others. Therefore, heat shocked cells are a good model for studying the structural and functional dynamics of chromatin. It has been well established that although the transcription of many genes is repressed upon heat shock, certain genes (e.g., heat shock proteins) are transcriptionally induced. Moreover, it was recently shown that heat shock might lead to the transcriptional activation of some constitutive heterochromatic regions, such as satellite III, which is located in the pericentromeric regions of certain human chromosomes.41,42 One of the trans-acting factors known to guide massive transcriptional repression in mammalian cells is heterochromatin protein 1 (HP1).43 Thus, we decided to examine the nuclear dynamics of HP1α, a direct homolog of Drosophila HP1,44 in human cells following heat shock. It was demonstrated that under these conditions the major part if not all of HP1α was removed from centromeres and redistributed over euchromatic compartment. Most important, the compact packaging of centromeric heterochromatin seemed preserved after HP1α removal. The results of this study, along with previously published results,45 strongly suggest that HP1α is not necessary to maintain a compact state of pericentromeric heterochromatin. It should be noted that Drosophila HP1 (a homolog of mammalian HP1α) was identified as an essential component of heterochromatin, based on the studies of suppression of position effect variegation.1,2 Direct evidence for the necessity of HP1 for compact DNA packaging has never been presented.26 However, studies have shown that the recruitment of HP1 to a genomic region was sufficient to initiate assembly of a heterochromatin domain.46 Thus, we propose that HP1 is necessary to form a nucleation center that initiates heterochromatin formation and that HP1 acts as a transcriptional repressor in assembled heterochromatin, rather than as a structural component.47 Indeed, it is becoming increasingly evident that the long-standing model postulating that heterochromatin represses transcription simply through highly compact DNA packaging is an oversimplification. First of all, some genes located in pericentromeric heterochromatin are transcribed.48 Second, it has been shown that HP1 directly blocks promoters by preventing assembly of pre-initiation complexes.49,50 In addition, HP1 can repress transcription by recruiting different co-repressors, including histone deacetylases and DNA methylases.47,51 If HP1α is necessary for repression of centromeric genes, then removal of this protein from centromeres as a result of heat shock should activate these genes. Indeed, it was reported that the transcription of centromeric repeats was stimulated under heat shock conditions.41,42 Furthermore, derepression of heterochromatic genes under heat shock conditions seems to be directly controlled by heat shock factor 1;52 taking into account our present data, it is logical to assume that this derepression is caused by removal of HP1α from the pericentromeric regions. Thus, the important question becomes—what happens to the HP1α that is removed from pericentromeric regions under heat shock conditions? We have demonstrated that it is not simply destroyed. An intriguing possibility is that HP1α is moved from pericentromeric heterochromatin to euchromatic regions to suppress transcription of specific euchromatic genes. In other words, upon heat shock, re-localization of HP1α may be a mechanism responsible for the reversal of the transcriptional status of the euchromatic and the heterochromatic compartments.
We have demonstrated that under heat shock conditions the HP1α is removed from centromeres without demethylation of histone H3 at position K9. This finding is in agreement with the results of several previously published studies53,54 demonstrated that H3K9 di- and trimethylation is not sufficient to keep HP1 bound to nucleosomes. It has been reported that the chromoshadow domain of HP1 interacts with histone H3 at a region located inside the nucleosomal barrel, at the entry/exit point of the nucleosome.30 In vitro, HP1α interacts with the histone fold of the histones H3 and H1.27 Interaction of HP1α with both tri-methylated lysine 9 of histone H3 and the globular part of histone H3 is regulated by phosphorylation. A previous study showed that HP1α was removed from centromeric heterochromatin upon phosphorylation of serine 10 of histone H3 (H3S10)55,56 and by phosphorylation of tyrosine 41 of the same histone (H3Y41) by Januse kinase 2 (JAK2).57 The fact that JAK2 is expressed in MCF-7 cells,58 as well as indirect evidence for its activation in response to heat shock,59 allows us to speculate that the release of HP1α in response to heat shock may be JAK2-dependent.
Materials and Methods
Antibodies.
A rabbit polyclonal antibody against lamin B1 and a mouse monoclonal antibody against HP1α were purchased from Abcam. A rabbit polyclonal antibody against histone H3 tri-methylated at lysine 9 (H3K9me3) was purchased from Active Motif. The human autoimmune antibody against CENP-A and a FITC-conjugated anti-human IgG were generous gifts from Dr. O. Zatsepina (Institute of Bioorganic Chemistry RAS, Moscow, Russia). The secondary antibodies conjugated to either Alexa Fluor 488, Alexa Fluor 555 or FITC were purchased from Molecular Probes/Invitrogen; the horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG were purchased from Amersham/GE Healthcare.
Cell culture and treatments.
The human breast cancer cells (line MCF-7) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum. The human lymphoid cells (the Jurkat line) were grown in RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum. Heat shock of exponentially growing cells was performed by placing cells at 45°C for 30 min. For “recovery” experiments, the cells treated by heat shock were maintained at 37°C for 1, 3, 6, 12 or 20 h.
Immunofluorescence.
For immunostaining, MCF-7 cells were grown on microscope slides; an aliquot of the suspension of Jurkat cells was centrifuged onto silane-coated microscope slides (Sigma) at 800 rpm for 5 min in a cytocentrifuge Cytospin 4 (Thermo Electron Corp.). All samples were fixed and permeabilized in CSK buffer [10 mM PIPES (pH 7.0), 100 mM NaCl, 1.5 mM MgCl2, 300 mM sucrose and 1.2 mM phenylmethylsulphonilfluoride (PMSF)] supplemented with 1% paraformaldehyde (PFA) and 2.5% Triton X-100 for 20 min at room temperature before staining. The fixed cells were washed three times (5 min each) in phosphate-buffered saline [PBS, 7 mM Na2HPO4, 1.5 mM KH2PO4, (pH 7.4), 137 NaCl, 2.7 mM KCl]. After washing, the cells were pre-incubated with 1% BSA (bovine serum albumin) in PBS for 1 h and then incubated with antibodies in PBS supplemented with 0.2% BSA for 45 min at room temperature in a humid chamber. In control experiments, the cells were incubated either without antibodies or with rabbit (or mouse) gamma globulins (Jackson ImmunoResearch) in a buffer containing 0.2% BSA. After incubation, the cells were washed three times (5 min each) with PBS supplemented with 0.2% BSA and 0.05% Tween 20. Then, primary antibodies bound to antigens were visualised using Alexa Fluor 488-, Alexa Fluor 555- or FITC-conjugated secondary antibodies. DNA was stained with a fluorescent dye, 4,6-diamino-2-phenylindole (DAPI) or To-Pro 3 iodide, for 10 min at room temperature. The results of immunostaining were analyzed using a Leica DRMB fluorescence microscope equipped with a charge-coupled device camera, a Leica TCS SP2 laser scanning confocal microscope or a Zeiss LSM 510 META NLO multiphoton microscope.
CENP-A foci analysis was performed using ImageJ software (rsb.info.nih.gov/ij). The Student's t test was used to evaluate the differences in number of foci per cell and foci size in control and heat-shocked cells.
Preparation of nuclear extracts.
Exponentially growing MCF-7 cells were lysed by incubation in cell lysis buffer (10 mM NaCl, 20 mM HEPES (pH 7.6), 1.5 mM MgCl2, 1 mM ZnSO4, 20% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) supplemented with Protease Inhibitor Cocktail (Roche) for 2 min on ice. After centrifugation, the pelleted nuclei were collected and shaken for 60 min on ice in nuclear extraction buffer (0.5 M NaCl, 20 mM HEPES (pH 7.6), 1.5 mM MgCl2, 1 mM ZnSO4, 20% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) supplemented with Protease Inhibitor Cocktail (Roche). After centrifugation (7,000x g for 10 min), nuclear extracts were stored at −70°C. For NaCl extraction, MCF-7 cells were collected by centrifugation and washed twice with PBS. The cells were then incubated for 30 min on ice in permeabilization buffer (10 mM PIPES, (pH 7.8), 0.5% Triton X-100, 100 mM NaCl, 0.3 M sucrose, 0.2 mM PMSF and 3 mM MgCl2). After incubation, the cells were washed three times with TM buffer (50 mM Tris-HCl (pH 7.5), 3 mM MgCl2). An equal volume of TM buffer supplemented with 1 M NaCl was added (final concentration of NaCl was 0.5 M). After a 20 min incubation on ice, the pellets were washed twice with the above buffers and resuspended in TM buffer. Protein concentration was measured on a Qubit Fluorometer (Invitrogen) or a NanoDrop 8000 (Thermo Scientific Corp.).
Histone extraction.
Exponentially growing MCF-7 cells (5 × 106) were washed with PBS and lyzed by incubation in lysis buffer (1x PBS, 0.5% Triton X-100, 0.2 mM EDTA) supplemented with Protease (Roche) and Phosphatase (Sigma) Inhibitor Cocktails for 10 min at 4°C. After centrifugation (6,500 × g for 10 min at 4°C) the pellets were gently resuspended in ice-cold 0.2 N HCl and incubated for 12 h on ice. After centrifugation (6,500x g for 10 min at 4°C) the supernatants were collected and stored at −70°C. Protein concentration was measured using Bradford reagent (Sigma) according to the manufacturer's instructions.
Immunoblotting.
Aliquots (20 µg) of each sample were separated by 12–15% SDS-PAGE and blotted onto polyvinylidene difluoride membranes (Hybond-P, Amersham Biosciences). The membranes were blocked overnight in 5% dry milk in PBS containing 0.1% Tween 20 (PBS-T) and incubated for 1 h with a primary antibody diluted in PBS containing 0.02% Tween 20 and 5% dry milk. After three washes with PBS-T, the membranes were incubated for 1 h with secondary antibodies (horse-radish peroxidase-conjugated anti-rabbit IgG) in PBS containing 0.02% Tween 20 and 5% dry milk. The immunoblots were visualized using an Amersham ECL kit. For data presentation, the films were scanned and processed with Adobe Photoshop CS software.
For histones separation aliquots (10 µg) of each sample were redissolved in electrophoresis sample buffer (0.1 N HCl, 0.5 β-mercaptoethanol, 0.02% Pyranin Y, 9 M urea) and electrophoresed (180 V) on 20% polyacrilamide gels containing 0.9 N acetic acid and 2.5 M urea. Proteins were transferred to polyvinylidene difluoride membranes (Hybond-P, Amersham Biosciences) in transfer buffer containing 0.1% acetic acid and 10% methanol. Immunoblotting was carried out as described for nuclear extracts.
Extraction of RNA and quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis.
RNA was extracted from cells using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. All RNA samples were further treated with DNase I (Fermentas) to remove residual DNA. RNA (1 µg) was reverse transcribed in a total volume of 20 µL for 1 h at 42°C using 0.4 µg random hexamer primers and 200 U reverse transcriptase (Fermentas) in the presence of 20 U of ribonuclease inhibitor (Fermentas). The cDNA obtained was analysed by SYBR Green I-based quantitative polymerase chain reaction (qPCR) using the CFX-96 PCR detection system (Bio-Rad). A PCR mixture in a 20 µL volume contained 50 mM Tris-HCl (pH 8.6), 50 mM KCl, 1.5 mM MgCl2, 0.1% Tween-20, 0.5 µM of each primer, 0.2 mM of each dNTP, 0.5 mM SYBR Green I (Syntol), 0.75 U of Hot Start Taq Polymerase (Sibenzyme) and 25 ng of cDNA template. Each PCR was performed in quadruplicate and the corresponding results were averaged. The sequences of the primers are presented in Supplementary Table S2.
DNase I sensitivity assay.
Exponentially growing MCF-7 cells were washed twice in DMEM, resuspended in 5 ml of ice-cold lysis buffer (10 mM PIPES (pH 7.8), 100 mM NaCl, 0.3 M sucrose, 3 mM MgCl2, 0.5% TritonX-100, 0.5 mM CuSO4 and 0.2 mM PMSF) and incubated on ice for 15 min. The nuclei were pelleted, washed with the DNase I digestion buffer (40 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM CaCl2, 0.2 mM PMSF), resuspended in the same buffer, pre-incubated at 37°C for 2 min and digested with DNase I (10 U/µL, Roche) at 0–100 U/100 µL at 37°C for 30 min. The reaction was stopped by the addition of EDTA to 10 mM. Genomic DNA was isolated by phenol/chloroform extraction. The DNA samples were quantified using the NanoDrop 8000 (Thermo Scientific Corp.). SYBR Green-based quantitative PCRs were performed on 50 ng aliquots of digested genomic DNA using the CFX-96 PCR detection system (Bio-Rad). The PCRs were performed in 20 µL reaction volumes, which included 50 mM Tris-HCl (pH 8.6), 50 mM KCl, 1.5 mM MgCl2, 0.1% Tween-20, 0.5 µM of each primer, 0.2 mM of each dNTP, 0.5 mM SYBR Green I (Syntol), 0.75 U of Hot Start Taq Polymerase (Sibenzyme) and 50 ng of DNA. Each PCR was performed in quadruplicate and the corresponding results were averaged. The sequences of the primers are presented in Supplementary Table S2.
Acknowledgements
The work of A.K.V. and S.V.R. was supported by the Ministry of Science and Education of the Russian Federation (grant 02.740.11.0289), the Presidium of the Russian Academy of Sciences (MCB grant) and by the Russian Foundation for Basic Research (RFBR grant 08-04-00048-a). The work of O.L.K. was supported by a grant from the President of the Russian Federation for Young Scientists (MK-311.2010.4).
Supplementary Material
References
- 1.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 4.Kwon SH, Workman JL. The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol Cells. 2008;26:217–227. [PubMed] [Google Scholar]
- 5.Wreggett KA, Hill F, James PS, Hutchings A, Butcher GW, Singh PB. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Ma A, Chow CM, Horsley D, Brown NR, Cowell IG, et al. Conservation of heterochromatin protein 1 function. Mol Cell Biol. 2000;20:6970–6983. doi: 10.1128/mcb.20.18.6970-6983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit E, Greil F, van Steensel B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 9.Inoue A, Hyle J, Lechner MS, Lahti JM. Perturbation of HP1 localization and chromatin binding ability causes defects in sister-chromatid cohesion. Mutat Res. 2008;657:48–55. doi: 10.1016/j.mrgentox.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, et al. HP1 controls telomere capping, telomere elongation and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Ayoub N, Jeyasekharan AD, Venkitaraman AR. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle. 2009;8:2945–2950. [PubMed] [Google Scholar]
- 12.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 13.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, Hwang KK, Worman HJ, Courvalin JC, Eissenberg JC. Expression and functional analysis of three isoforms of human heterochromatin-associated protein HP1 in Drosophila. Chromosoma. 2001;109:536–544. doi: 10.1007/s004120000113. [DOI] [PubMed] [Google Scholar]
- 16.Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, et al. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 17.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norwood LE, Grade SK, Cryderman DE, Hines KA, Furiasse N, Toro R, et al. Conserved properties of HP1(Hsalpha) Gene. 2004;336:37–46. doi: 10.1016/j.gene.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 20.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 21.Smothers JF, Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol Cell Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 23.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh PB, Georgatos SD. HP1: facts, open questions and speculation. J Struct Biol. 2002;140:10–16. doi: 10.1016/s1047-8477(02)00536-1. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 28.Dimitri P, Caizzi R, Giordano E, Carmela Accardo M, Lattanzi G, Biamonti G. Constitutive heterochromatin: a surprising variety of expressed sequences. Chromosoma. 2009;118:419–435. doi: 10.1007/s00412-009-0211-y. [DOI] [PubMed] [Google Scholar]
- 29.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 30.Lavigne M, Eskeland R, Azebi S, Saint-Andre V, Jang SM, Batsche E, et al. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5:1000769. doi: 10.1371/journal.pgen.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minc E, Allory Y, Courvalin JC, Buendia B. Immunolocalization of HP1 proteins in metaphasic mammalian chromosomes. Methods Cell Sci. 2001;23:171–174. doi: 10.1007/978-94-010-0330-8_18. [DOI] [PubMed] [Google Scholar]
- 32.Smith MM. Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- 33.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, et al. Loss of the Suv39 h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 34.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 35.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klochkov DB, Gavrilov AA, Vassetzky YS, Razin SV. Early replication timing of the chicken alpha-globin gene domain correlates with its open chromatin state in cells of different lineages. Genomics. 2009;93:481–486. doi: 10.1016/j.ygeno.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Finelli P, Antonacci R, Marzella R, Lonoce A, Archidiacono N, Rocchi M. Structural organization of multiple alphoid subsets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18 and 19. Genomics. 1996;38:325–330. doi: 10.1006/geno.1996.0635. [DOI] [PubMed] [Google Scholar]
- 40.Pironon N, Puechberty J, Roizes G. Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19 and 21. BMC Genomics. 2010;11:195. doi: 10.1186/1471-2164-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, et al. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008;36:423–434. doi: 10.1093/nar/gkm1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169–174. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Saunders WS, Chue C, Goebl M, Craig C, Clark RF, Powers JA, et al. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J Cell Sci. 1993;104:573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 45.Mateos-Langerak J, Brink MC, Luijsterburg MS, van der Kraan I, van Driel R, Verschure PJ. Pericentromeric heterochromatin domains are maintained without accumulation of HP1. Mol Biol Cell. 2007;18:1464–1471. doi: 10.1091/mbc.E06-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, et al. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 49.Vassallo MF, Tanese N. Isoform-specific interaction of HP1 with human TAFII130. Proc Natl Acad Sci USA. 2002;99:5919–5924. doi: 10.1073/pnas.092025499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smallwood A, Black JC, Tanese N, Pradhan S, Carey M. HP1-mediated silencing targets Pol II coactivator complexes. Nat Struct Mol Biol. 2008;15:318–320. doi: 10.1038/nsmb.1385. [DOI] [PubMed] [Google Scholar]
- 51.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eymery A, Souchier C, Vourc'h C, Jolly C. Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells. Exp Cell Res. 2010;316:1845–1855. doi: 10.1016/j.yexcr.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Eskeland R, Eberharter A, Imhof A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol Cell Biol. 2007;27:453–465. doi: 10.1128/MCB.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meehan RR, Kao CF, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164–3174. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mateescu B, England P, Halgand F, Yaniv M, Muchardt C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 2004;5:490–496. doi: 10.1038/sj.embor.7400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 57.Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Behera R, Kumar V, Lohite K, Karnik S, Kundu GC. Activation of JAK2/STAT3 signaling by osteopontin promotes tumor growth in human breast cancer cells. Carcinogenesis. 2010;31:192–200. doi: 10.1093/carcin/bgp289. [DOI] [PubMed] [Google Scholar]
- 59.Cheng MB, Zhang Y, Zhong X, Sutter B, Cao CY, Chen XS, et al. Stat1 mediates an auto-regulation of hsp90beta gene in heat shock response. Cell Signal. 2010;22:1206–1213. doi: 10.1016/j.cellsig.2010.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.