Abstract
The field of computational protein design has experienced important recent success. However, the de novo computational design of high-affinity protein–ligand interfaces is still largely an open challenge. Using the Rosetta program, we attempted the in silico design of a high-affinity protein interface to a small peptide ligand. We chose the thermophilic endo-1,4-β-xylanase from Nonomuraea flexuosa as the protein scaffold on which to perform our designs. Over the course of the study, 12 proteins derived from this scaffold were produced and assayed for binding to the target ligand. Unfortunately, none of the designed proteins displayed evidence of high-affinity binding. Structural characterization of four designed proteins revealed that although the predicted structure of the protein model was highly accurate, this structural accuracy did not translate into accurate prediction of binding affinity. Crystallographic analyses indicate that the lack of binding affinity is possibly due to unaccounted for protein dynamics in the ‘thumb’ region of our design scaffold intrinsic to the family 11 β-xylanase fold. Further computational analysis revealed two specific, single amino acid substitutions responsible for an observed change in backbone conformation, and decreased dynamic stability of the catalytic cleft. These findings offer new insight into the dynamic and structural determinants of the β-xylanase proteins.
Keywords: β-xylanase fold, computational protein design, protein–ligand interface, protein dynamics, Rosetta
Introduction
The ability to rationally design proteins through computational methods has long been a goal of biotechnology and pharmaceutical researchers. The development of widely applicable, repeatable and accurate rational protein design methods is expected to enable the development of protein-based therapeutics for human medical applications and improved enzymatic processes essential in industry and manufacturing. The market for clinical protein therapeutics, some $94 billion in 2010, is expected to grow to half of total prescription drug sales by 2014 (Strohl and Knight, 2009), and industrial use of engineered proteins will soon reach over $5 billion per year (Arora et al., 2009).
Computational protein design has experienced important success in recent years, with significant achievements in the design of novel enzymes (Jiang et al., 2008; Röthlisberger et al., 2008; Gerlt and Babbitt, 2009), biocatalysts (Kaplan and DeGrado, 2004; Damborsky and Brezovsky, 2009), antivirals (Flower et al., 2003; Shi et al., 2007), protein–protein interfaces (Das and Baker, 2008; Karanicolas and Kuhlman, 2009; Sammond et al., 2010), diagnostics (Sodee et al., 2000; Taillefer et al., 2000) and novel protein folds (Kuhlman et al., 2003). However, a particular aspect of computational protein design that has proved more difficult is the design of protein–ligand interfaces, particularly the design of proteins capable of tightly binding small molecules and peptides (Hayden, 2009; Schreier et al., 2009a).
The goal of the current study was to develop and experimentally validate computational tools and protocols for designing high-affinity protein–ligand interfaces using the Rosetta protein design program (http://www.Rosettacommons.org/). The protein design functionality of the Rosetta program has demonstrated prior success at designing enzymes (Jiang et al., 2008; Röthlisberger et al., 2008; Siegel et al., 2010), altering the specificity of protein–protein interactions (Humphris and Kortemme, 2007a; Mandell and Kortemme, 2009; Sammond et al., 2010), creating novel protein folds never before seen in nature (Kuhlman et al., 2003) and predicting protein–peptide specificity (Sood and Baker, 2006). Here we set out to expand the application of Rosetta to the design of a de novo, high-affinity interface to a small peptide ligand.
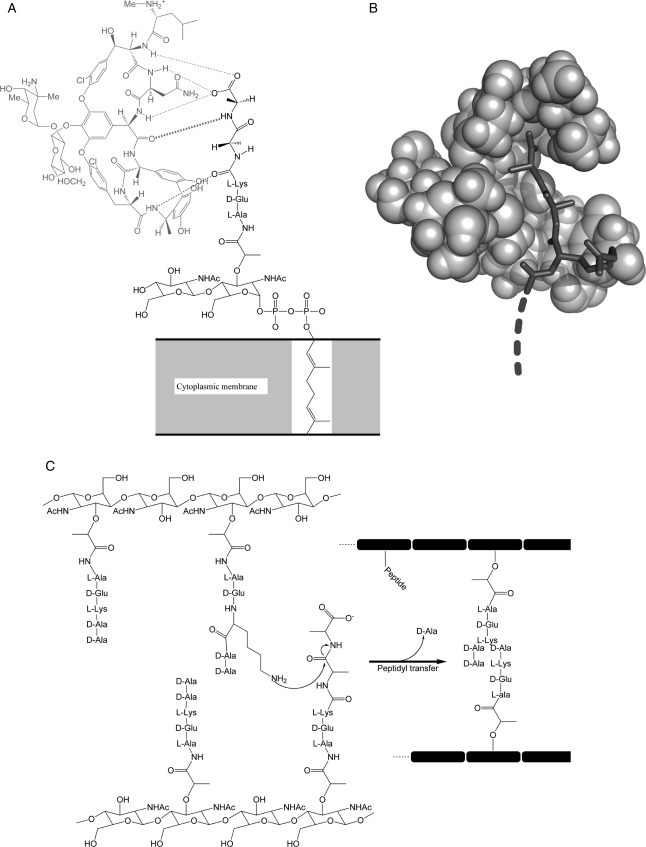
The target ligand system we chose for our proof-of-concept study was the d-alanine-d-alanine (d-ala-d-ala) C-terminal dipeptide of the peptidoglycan precursor from Staphylococcus aureus. These d-ala-d-ala peptides are critical to S.aureus cell wall biosynthesis and are the primary target for the glycopeptide vancomycin, an antibiotic of last resort for treating multiple-resistant Gram-positive infections (Boneca and Chiosis, 2003). Vancomycin acts by binding and sequestering the d-ala terminus of the peptidoglycan precursor (Fig. 1A) preventing its incorporation into the bacterial cell wall (Fig. 1C). This compromises the integrity of the bacterial cell wall, rendering it vulnerable to lysis due to normal osmotic pressure changes (Loll and Axelsen, 2000). Some bacteria acquire resistance to vancomycin by replacing this C-terminal dipeptide with a d-alanine-d-lactate (d-ala-d-lac) moiety (Cui et al., 2006).
Fig. 1.
The d-ala-d-ala peptidoglycan and vancomycin's mode of action. (A) Vancomycin (light gray) forms five critical hydrogen bonds to terminal d-ala-d-ala residues of the S.aureus peptidoglycan precursor anchored in the cytoplasmic membrane. (B) Space-filling model showing how vancomycin binds and sequesters the terminal d-ala peptides, thus preventing the peptidyl transfer cross-linking (C) of glycopeptide chains essential for cell wall biosynthesis.
We attempted to use Rosetta to perform the de novo design of the family 11 endo-1,4-β-xylanase from Nonomuraea flexuosa [protein data bank (PDB) ID 1m4w] to replicate the binding and sequestration mode of action of the vancomycin antibiotic. This protein was chosen due to its available 2.1Å resolution 3D coordinates, thermostability, expression and production characteristics, molecular mass, and the geometry and size of its enzymatic cleft (Hakulinen et al., 2003). We were encouraged that previously successful Rosetta enzyme design work had been performed using this protein, proving its feasibility as a scaffold for computational design (Jiang et al., 2008; Röthlisberger et al., 2008).
In the course of Rosetta computations, the scaffold protein's enzymatic cleft is mutated in silico to form an interface capable of binding to the target d-ala-d-ala or d-ala-d-lac dipeptides (Fig. 1A and B). Following computations, the designed protein sequences were produced in the laboratory and assayed for binding to the target dipeptides using multiple, complementary methods.
Unfortunately, none of the designed proteins demonstrated high-affinity (Kd < 100 μM) binding to their target ligands. Subsequent structure determination of four of the Rosetta designed proteins revealed conformational changes in the protein backbone and altered protein dynamics as significant contributing factors to the lack of observed ligand binding affinity. Our results make the experimental xylanase mutant structures determined in the course of the study available to the scientific community. The results presented here can also be utilized as a benchmark case for the further development of computational design algorithms.
Materials and methods
Selection of thermostable scaffold protein
To identify protein scaffolds suitable for Rosetta design, a search of the PDB was conducted for proteins with high-resolution crystallographic structures (<2.5 Å), no structurally important metal atoms, a molecular weight <50 kDa and a binding surface or pocket of the appropriate geometry to accommodate a dipeptide ligand. Preference was given to thermostable proteins under the assumption that their robustness would allow more extensive design mutations without destabilizing the overall protein fold.
The PDB file of the selected scaffold was prepared for Rosetta design by the removal of all redundant protein chains and non-proteinacious molecules, including crystallographic water and reagent molecules. All ligand atoms were removed, and any ‘anisou’ or alternate atom positions or side-chain rotamers were discarded, retaining only the 3D coordinates and identities of protein main-chain and side-chain atoms.
Ligand model and generation of ligand ensemble
The d-ala-d-ala dipeptide ligand moiety consists of 25 atoms—12 heavy atoms and 12 hydrogen atoms of the d-ala-d-ala terminus of the target glycopeptide, plus the carbonyl carbon of the preceding lysine residue comprising the peptido linkage. A d-ala-d-lac ligand representing a resistant form of the S.aureus glycopeptide was generated by substituting the C-terminal amide nitrogen of the d-ala-d-ala ensemble with oxygen (Supplementary data, Fig. S1). To account for potential conformational flexibility of the dipeptide, an ensemble of conformers was created using the Molecular Operating Environment (MOE) software. The ensemble was populated by systematically rotating the backbone phi/psi angles of the target peptide in 10° increments, then removing all conformers not possessing ‘allowed’ beta-sheet Ramachandran angles for d-amino acids. Each conformer was then output as an individual .pdb file. Design calculations were performed with a representative conformer ensemble of 225 d-ala-d-ala and 225 d-ala-d-lac dipeptide structures.
Rosetta computations
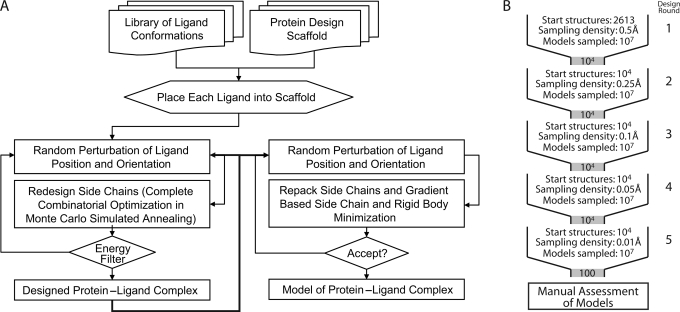
De novo computational design and ligand docking of the chosen scaffold with the target ligand ensemble was performed using the RosettaLigand module of Rosetta version 2.3 (Meiler and Baker, 2006). RosettaLigand utilizes a Monte Carlo/Metropolis simulated annealing search algorithm to dock the ligand molecule with three translational and two rotational degrees of freedom. Simultaneously, RosettaLigand designs the protein scaffold by varying the identities of the amino acids comprising the binding interface (Fig. 2A). The knowledge-based energy function combines van der Waals (VDW) attractive and repulsive interactions, hydrogen bonding energy, a desolvation penalty and pair-wise electrostatics (Kuhlman and Baker, 2000), as well as side-chain rotamer probabilities derived from the PDB (Dunbrack and Cohen, 1997).
Fig. 2.
Diagram of computational protocols and strategies. (A) Flowchart of Rosetta computational process showing the multi-step, iterative nature of the Rosetta design and scoring procedures. Only models that achieve specified minimum energies are accepted and output. (B) Schematic of design protocol. At each cycle, starting structures are used to create a large number of designs, which then undergo filtering before being carried to the next cycle. In each of the five cycles, the sampling density is increased by reducing the design perturbation parameters. After the final round of design, the output models are manually assessed to determine the best overall candidate designs.
All peptide conformations were placed manually into the ligand binding site. In an iterative protocol, RosettaLigand simultaneously optimizes ligand position and protein sequence. During computations, ligand position and orientation are randomly perturbed before all interface residues are re-designed to optimize protein–ligand interactions. This ‘dock-design’ protocol is repeated five times. Following each round of dock-design, 10000 of the 100000 models generated were selected based on predicted ligand binding energy normalized by the number of mutations from wild-type, degree of ligand burial, ligand hydrogen-bond donor/acceptor saturation and egress of the N-terminal extension of the glycopeptide ligand. These best scoring 10000 models were then used as starting points in the following round of dock-design computations (Fig. 2B). At each successive round, perturbation of the initial ligand position and orientation was narrowed, leading to increased conformational search density from round to round. While the first round allowed for complete ligand reorientation and movement of up to 5 Å, the final round limited movement to 5° and 0.5 Å. The protocol uses a softened repulsive VDW scoring potential to smooth the energy landscape. After five dock-design iterations, predicted ligand binding energies plateaued and the amino acid sequences of designed proteins converged. In a final step, 10000 models were energy minimized using hard-repulsive VDW scoring potentials to discriminate the best protein sequences based on predicted ligand binding energy. This process allowed for minimal ligand movement and optimization of side-chain conformations.
Selection of designed mutant proteins for expression
The resulting protein designs were clustered according to binding pose and sequence, and the top scoring models of each sequence group were ranked according to predicted binding energy. Interestingly, the best scoring models shared the same principal binding mode and a subset of mutations. Models were then analyzed at atomic detail on a residue-by-residue basis, examining for hydrogen bonding geometries, hydrophobic packing, burial of polar groups and binding pocket access/occlusion. Additional filtering of the models for each of the 1m4w scaffold designs was performed to accommodate egress of the N-terminal extension of the glycopeptide target. The best nine models for each target ligand were chosen for experimental evaluation of predicted ligand binding (Table I). Later, three additional point mutants of the design 1m4w_6 were created (see below).
Table I.
Characteristics of the 1m4w protein designs
| AA position | WT | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | v48 | w20 | w20v48 | SS-type | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | W | W | W | W | W | W | R | R | R | R | R | W | W | Strand | Finger |
| 46 | N | R | R | R | R | R | L | L | L | F | L | L | L | ||
| 48 | V | F | F | F | V | K | L | W | F | I | V | L | V | ||
| 72 | N | Y | Y | Y | Y | Y | N | N | N | N | N | N | N | Loop | |
| 74 | Y | W | F | I | L | L | R | R | R | R | R | R | R | Strand | |
| 87 | E | S | S | S | S | S | S | S | S | S | S | S | S | Palm | |
| 89 | Y | Y | Y | Y | Y | Y | H | H | H | H | H | H | H | ||
| 120 | W | T | T | T | T | T | W | W | W | W | W | W | W | Thumb | |
| 121 | R | H | H | H | H | H | R | R | R | R | R | R | R | ||
| 124 | A | V | V | V | V | V | A | A | A | A | A | A | A | ||
| 133 | F | H | H | H | H | H | Y | Y | Y | Y | Y | Y | Y | ||
| 135 | Q | S | S | S | S | S | Q | Q | Q | Q | Q | Q | Q | ||
| 176 | E | I | I | I | I | I | E | E | E | E | E | E | E | Finger | |
| # Mutations | 0 | 11 | 11 | 11 | 10 | 11 | 7 | 7 | 7 | 7 | 6 | 6 | 5 | ||
| Ligand | — | lac | lac | lac | lac | lac | ala | ala | ala | ala | ala | ala | ala | ||
| Ebind (r.e.u) | — | 19.9 | 19.9 | 19.9 | 20.2 | 19.8 | 17.6 | 17.4 | 17.2 | 17.1 | 12.9 | 13.3 | 15.4 | ||
| Affinity* (kcal/mol) | — | −7.4 | −7.4 | −7.4 | −7.6 | −7.4 | −6.2 | −6.0 | −5.9 | −5.9 | −3.5 | −3.8 | −4.9 | ||
| PDB ID | 1M4W | 3MF6 | 3MF9 | 3MFC | 3MFA |
Amino acid identities at given sequence positions for wild-type 1m4w plus 12 designed mutants. Designation of each 1m4w_‘X’ protein at top. Gray type denotes mutated amino acids. Secondary structure and protein region of mutations shown at far right. Number of mutations from wild-type, ligand target (d-ala-d-ala or d-ala-d-lac), computed RosettaLigand energy of binding in Rosetta energy units (r.e.u), RosettaLigand predicted affinity (in kcal/mol from the method of Meiler and Baker, 2006) and PDB IDs for the deposited structures are at the bottom.
Maximally efficient gene synthesis strategy
A hierarchical strategy for gene construction of the nine mutant proteins was devised to minimize mutational primers and reaction steps (Supplementary data, Fig. S2A). Genes were assembled using recursive polymerase chain reaction (PCR) (Stemmer et al., 1995) from E.coli codon-optimized oligonucleotides designed using the Gene2Oligo web server (http://berry.engin.umich.edu/gene2oligo/) (Rouillard et al., 2004). Once assembled, the genes were cloned into a T7-driven pET29b expression vector. Point mutations were introduced using Quickchange™ (Stratagene). All constructs were confirmed by DNA sequencing.
Expression and purification of designed proteins
Proteins were expressed in BL21(DE3) pLysS cells (Stratagene). Cells were grown in LB media supplemented with kanamycin at 37°C until an optical density (600 nm) of 0.4–0.6 was reached. The cells were then transferred to 16°C. After 30 min, the samples were induced with isopropyl-β-d-thiogalactopyranoside to a final concentration of 150 µM and grown for ∼14 h. Cells were then harvested by centrifugation.
Cells were lysed by French-press in 25 mM HEPES, 100 mM NaCl, 5 mM imidazole, 5% glycerol and pH 7.6–7.8 buffer containing protease inhibitor cocktail (Roche). A single-step purification protocol using TALON™ cobalt-affinity resin (Clontech) was sufficient to obtain >95% purity as assessed by SDS-PAGE. Following purification, proteins were immediately dialyzed into a buffer containing 25 mM HEPES, 100 mM NaCl and 5% glycerol at pH 7.6–7.8.
Molecular weights were confirmed by MALDI-MS on a PerSeptive Biosystems Voyager-DE STR instrument. Protein aggregation state and solution properties were assessed by dynamic light scattering using a DynaPro ProteinSolutions molecular sizing instrument (Wyatt Technology Corporation). Proper protein folding was confirmed by circular dichroism (CD) using a Jasco J-810 spectropolarimeter and 1D NMR on a Bruker Avance 600 MHz spectrometer.
15N-labeled proteins for NMR were obtained by expression in M9 minimal media with 15NH4Cl as the sole nitrogen source. For X-ray diffraction and NMR structural characterization, proteins were purified by cobalt affinity chromatography as described above, followed by size-exclusion chromatography using a HiLoad 16/60 Superdex 75 gel filtration column (GE Healthcare). This additional purification step gave >99% purity as assessed by SDS-PAGE.
Peptides for protein–ligand binding studies
Peptides were purchased from Genscript. N-terminally acylated l-lys-d-ala-d-ala tripeptide or l-lys-d-ala-d-lac was used in isothermal titration calorimetry (ITC) and NMR titrations. Three dansylated peptides were used for fluorescence studies: (dansyl)-l-lys-d-ala-d-ala peptide with the dansyl label covalently linked to the N-terminal nitrogen; l-lys-(dansyl)-d-ala-d-ala peptide with dansyl label attached to the lysine ɛ-amino group, and (dansyl)-AEEAE-l-lys-d-ala-d-ala with a pentapeptide linker that separates the target peptide from the dansyl group.
All assays were carried out in 100 mM NaCl, 25 mM HEPES and 5% glycerol aqueous buffer at pH 7.7 unless otherwise noted. Protein concentrations were measured at 280 nm using a Shimadzu UV-mini 1240 spectrophotometer and calculated extinction coefficients (ExPASy ProtParam server http://www.expasy.ch/tools/protparam.html).
Fluorescence anisotropy
Fluorescence anisotropy (FA) titrations were carried out at 25°C using a T-format PTI Quantamaster 2000-7SE spectrofluorometer equipped with excitation and emission polarizers. The fluorescence emission intensities parallel and perpendicular to the vertically polarized excitation light were analyzed to determine the steady-state anisotropy values for each point in the titration. During the titrations, the concentration of dansyl-labeled peptide ligand was held constant while increasing concentrations of protein were added. Dansylated samples were excited at 340 nm and the fluorescence emission signal was monitored at 520 nm with both excitation and emission slit widths set to 1 mm.
NMR chemical-shift perturbation assay
NMR experiments were performed using a Bruker Avance 600 MHz spectrometer equipped with a cryoprobe. 1H–15N heteronuclear single quantum coherence (HSQC) spectra were acquired with 15N-labeled proteins at 200–600 µM in 25 mM HEPES, pH 7.6–7.8, 100 mM NaCl and 2.5% glycerol H2O/10% D2O. A series of 15N–1H HSQC spectra were acquired of protein titrated with 0, 1, 5 and 10 molar equivalents of peptide at 298 K. Data were processed using Topspin 2.0b (Bruker) and analyzed with Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Isothermal titration calorimetry
ITC experiments were performed at 30°C using a MicroCal VP-ITC instrument. Unlabeled peptide was titrated into the cell containing 0.6–1.1 mM protein in 100 mM NaCl, 25 mM HEPES, 5% glycerol and pH 7.6–7.8 buffer. Ligand concentrations were 15–20 times the molar concentration of the protein.
Crystallization of proteins derived from model 1m4w_6
Crystallization screens of designed 1m4w_6 as well as three derivative point mutants (Table I) were built from Hampton research HR2-130 Crystal Screen HT reagents using a Thermo Fisher Scientific MaxCell™ crystallization workstation incorporating a MicroLab Starlet™ liquid-handling robot (Hamilton Corporation, Reno, NV, USA) and a Mosquito™ nanoliter drop-setting robot (TTP LabTech, Oxford, UK). All screening was performed using 96-well MRC plates (Hampton Research) and experiments were visualized and recorded using a Thermo Fisher Scientific Rhombix™ Tablestore automated imaging system. Protein was concentrated to 10 mg/ml in 100 mM NaCl, 25 mM HEPES, 5% glycerol and pH 7.8 buffer. Initial hits from the robotic screen were optimized in 24-well sitting-drop plates using individual Hampton Research Optimize reagents.
Diffraction data collection and processing
Complete data sets were acquired in-house using a Bruker Microstar rotating-anode X-ray generator and a Bruker Proteum PT135 CCD area detector. Crystals were maintained at 100 K using a Bruker Kryo-Flex cryostat. Data collection sweeps were optimized using Cosmo (Bruker AXS, 2008) software and data integrated and scaled using SADABS (Bruker AXS, 2008) and XPREP (Bruker AXS, 2008) in the PROTEUM2 package (Bruker AXS, 2008). The cryoprotectant used was the crystallization buffer supplemented with 30% ethylene glycol.
Additional X-ray diffraction data were collected at Southeast Regional Collaborative Access Team, beamline 22-ID, Advanced Photon Source, Argonne National Laboratory using a MAR165 CCD area detector. A total of 360 frames with a 0.5° oscillation angle were collected at 100 K using a wavelength of 1.00Å and a crystal-to-detector distance of 150 mm.
Data processing and structure refinement
Diffraction data were phased by molecular replacement with the program MOLREP (Vagin and Teplyakov, 1997), using the 1m4w coordinates obtained from the PDB or Rosetta designed models. Molecular replacement phases were then used to initiate automated model building with the program, Arp/wArp (Langer et al., 2008). Model refinement was performed using REFMAC5 (Murshudov et al.,1997) with iterated manual fitting using COOT (Emsley and Cowtan, 2004). All data analysis and refinement were performed using the CCP4 package (Collaborative Computational Project, Number 4, 1994) and ccp4i gui (Potterton et al., 2003).
Results
Scaffold selection
We began by attempting to identify a suitable protein scaffold for our de novo protein–peptide interface design effort. The 1m4w is a thermophilic endo-1,4-β-xylanase (EC 3.2.1.8) from N.flexuosa with a crystal structure determined at 2.10 Å resolution (Hakulinen et al., 2003). Its β-jelly-roll topology of two twisted beta-sheets forms a large cleft where enzymatic activity occurs, typical to family 11 xylanases. The protein does not naturally interact with peptide ligands, instead binding large polysaccharides on its outer surface, while residues inside the cleft catalyze the glycosidic cleavage of xylanose subunits. The overall molecular weight of ∼22 kDa, the size and geometry of its enzymatic cleft were well suited to a de novo re-design strategy. Additionally, the thermostable nature of 1m4w was expected to allow a more extensive re-design of residues in the binding cleft without significant destabilization of the protein backbone.
RosettaLigand computations
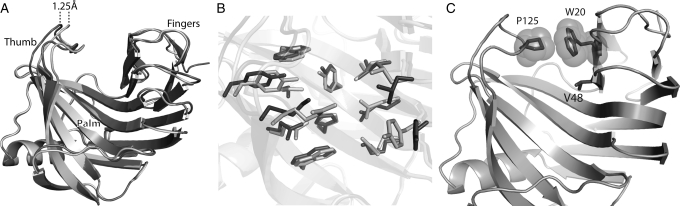
The RosettaLigand module of the Rosetta suite of programs was used to accommodate the non-standard d-ala and d-lac ligands during design of the protein–peptide interface. The goal of RosettaLigand dock-design computation is to identify the smallest set of mutations to the native scaffold protein sequence, which also provides the highest-affinity binding to the target dipeptide ligands. The best scoring nine sequences possessing binding energies of at least −1.5 Rosetta energy units (r.e.u.) per amino acid mutation from wild-type were selected for laboratory expression and assay (Table I; Supplementary data, Fig. S2A). Each of the nine proteins is 197 amino acids in length and displays a unique combination of between 7 and 11 mutations. All of the mutations are located in the catalytic cleft on the inside of the concave jelly-roll protein fold, in one of three regions that directly interact with the ligand. These regions are referred to as the ‘thumb’, ‘palm’ or ‘finger’ (see Fig. 3A) (Hakulinen et al., 2003). The nine selected protein designs were labeled sequentially as 1m4w_1 through 1m4w_9 (Table I).
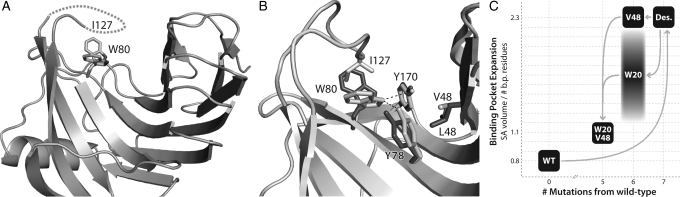
Fig. 3.
Backbone opening of the binding pocket and prediction of interface rotamer conformations between 1m4w_6-predicted model (light gray) and X-ray structure (dark gray). (A) Cartoon representation of the model and X-ray structure showing the 1.25 Å shift in the backbone configuration of the ‘thumb’ region. (B) Detailed comparison of the residues comprising the ligand interface. Most of the residue side chains are superimposable, while several are out of position due to the altered backbone conformation. Only two side-chain rotamers assume substantially different conformations from prediction. (C) Residues identified as directly responsible for binding pocket opening. W20–P125 (shown with VDW spheres) form a hydrophobic interaction between ‘thumb’ and ‘fingers’ at the top of the binding pocket, while V48 lies lower in the ‘palm’ of the protein.
During the design process, many of the residues in the catalytic site of the 1m4w enzyme were altered in favor of the new peptide binding function, thus eliminating the proteins' native catalytic functionality. The wide and deep catalytic cleft of the protein was transformed by the design process into a tightly fitting binding pocket, closely contacting the target d-ala-d-ala or d-ala-d-lac dipeptide ligands on all sides except the N-termini, thus allowing for egress of the un-modeled remainder of the glycopeptide (Fig. 4A; Supplementary data, Fig. S1). Predicted binding energies for the initial nine RosettaLigand protein designs ranged from −17 to −20r.e.u. (Table I). Previous studies by Meiler and Baker (2006) found that Rosetta energy units correspond to experimentally determined binding energies with a correlation of 0.63. Using the Meiler and Baker method, the Rosetta energies for the initial nine chosen designs correspond to a predicted free energy of binding of −5.82 to −7.50 ± 1.9 kcal/mol and a Kd of 54 ± 34 to 3 ± 2 µM. Additionally, good hydrophobic packing of both ligand methyl groups and strong binding of the carboxyl terminus were common features in each of the nine protein designs.
Fig. 4.
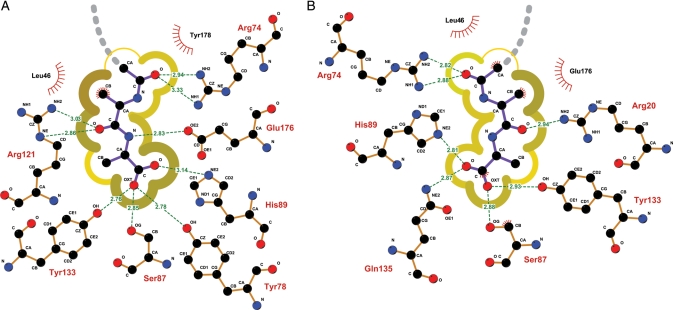
Detailed schematic of ligand interface. (A) RosettaLigand-predicted interface of 1m4w_6 showing individual residues and H-bonds involved in binding, and the degree of solvent accessibility to the ligand. Darker yellow, thicker lines indicate low exposed surface area; lighter, thinner lines indicate more solvent exposure. Gray dashed line denotes the path of the unmodeled portion of glycopeptide ligand. (B) Detail of the X-ray determined 1m4w_6 apo interface with ligand re-docked. Note the decrease in number of H-bonds and increase in degree of solvent exposure. Solvent accessibility was computed with NACCESS (Hubbard and Thornton, 1993) using a probe radius of 1.4 Å and visualized with LigPlot (Wallace et al., 1995).
Expression characteristics and solution properties of designed proteins
Expression of the RosettaLigand designed proteins proceeded as outlined in the Materials and methods section. All of the 1m4w designed proteins expressed well, yielding between 7 and 12 mg/l induction. All 1m4w proteins were found to express >50% soluble, with most being >75% soluble. Dynamic light scattering and size-exclusion chromatography of each of the expressed proteins indicated that the 1m4w designs existed in solution as homogeneous, monomeric species.
Far-UV CD spectra of the 1m4w designed proteins indicated secondary structure composition similar or identical to wild-type (Supplementary data, Fig. S3A). One-dimensional NMR results confirmed that all of the 1m4w proteins were well folded and stable. Additionally, the 1m4w designed proteins exhibited a high degree of stability and resistance to proteolysis. Samples left at room temperature for several weeks following purification showed no signs of degradation.
Assay of predicted binding affinity of designed proteins
Following computational design and expression of the chosen interface designs, binding assays were performed to validate the predicted affinities. Unfortunately, none of the designed proteins tested in this study yielded evidence of specific, high-affinity binding to their target peptide. We thus conclude that the RosettaLigand interface designs were not successful.
Using FA, several of the 1m4w designs indicated low- to moderate-affinity binding, with Kd values between 367 and 449 µM (Supplementary data, Fig. S3B). Non-specific, background binding affinities for the 1m4w designs during FA measurements were observed to be at or above 850 µM. These negative results for high-affinity binding were later confirmed by ITC and NMR spectroscopy.
Structure determination of 1m4w_6
To determine a cause for the lack of observed binding among the designed proteins, a high-resolution X-ray diffraction structure of 1m4w6 was determined. The optimal crystallization buffer contained 0.1 M NaCl, 1.125M ammonium sulfate, 0.1M Bis-Tris pH 5.5, 3% Jeffamine M600 pH 7.0 and at 20°C produced diffracting, single, rod-shaped crystals of up to 150 µm × 450 µm (Fig. S4). The final conditions differed significantly from that used for the wild-type 1m4w structure (Hakulinen et al., 2003). Data sets were collected for 1m4w_6 crystals in the apo form to a resolution of 1.28Å. Refinement statistics for the structure of the 1m4w_6 designed mutant are listed in Table II.
Table II.
Crystallographic statistics for the four deposited 1m4w-derived structures
| 1m4w_6 | 1m4w_6w20 | 1m4w_6v48 | 1m4w_6w20v48 | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength, Å | 1.00 | 1.5418 | 1.5418 | 1.5418 |
| Resolution (outer shell), Å | 55.30–1.28 (1.34–1.28) | 38.48–1.69 (1.79–1.69) | 49.01–1.70 (1.79–1.70) | 55.32–1.63 (1.73–1.63) |
| Rmerge,a % | 7.6 (53.3) | 8.6 (40.2) | 8.9 (29.6) | 4.6 (21.1) |
| Mean I/sigma(I) | 54.89 (3.52) | 23.22 (3.63) | 28.48 (3.34) | 26.44 (3.01) |
| Completeness, % | 99.8 (96.4) | 99.7 (97.9) | 100.0 (100.0) | 88.5 (48.1) |
| Redundancy | 9.70 (5.5) | 18.78 (6.77) | 21.80 (12.06) | 7.53 (1.22) |
| Unique observations | 62177 (4534) | 28769 (4289) | 28204 (3957) | 28568 (2549) |
| Refinement | ||||
| Rcryst/Rfree, %b | 18.07/19.37 | 17.62/21.62 | 16.40/20.38 | 18.42/22.63 |
| No. of protein atoms | 1169 | 1077 | 1155 | 1157 |
| No. of solvent waters | 386 | 438 | 404 | 366 |
| Bond length RMSD, Å | 0.030 | 0.026 | 0.028 | 0.013 |
| Bond angle RMSD, ° | 2.235 | 1.952 | 1.954 | 1.274 |
| Avg. protein B, Å2 | 12.476 | 17.679 | 15.363 | 19.194 |
| Ramachandran plot, %c | ||||
| Most favored | 88.3 | 89.5 | 89.0 | 86.3 |
| Allowed | 10.5 | 9.9 | 9.7 | 12.4 |
| Generously allowed | 1.2 | 0.6 | 1.3 | 1.2 |
| Disallowed | 0.0 | 0.0 | 0.0 | 0.0 |
Outer resolution bin statistics are given in parentheses.
aRmerge = Shkl(Si|Ihkl,i − <Ihkl>))/Shkl,i<Ihkli>, where Ihkl,i, is the intensity of an individual measurement of the reflection with Miller indices h, k and l, and <Ihkl> is the mean intensity of that reflection.
bRcryst = S||Fobs, hkl| − |Fcalc, hkl||/|Fobs, hkl|, where |Fobs, hkl| and |Fcalc, hkl| are the observed and calculated structure factor amplitudes. Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
cCalculated with the program PROCHECK.
Structural analysis of 1m4w_6
Using the newly obtained high-resolution 3D structure of the designed 1m4w_6 protein, a comparative structural analysis was performed. The most identifiable difference between the 1m4w_6 experimental structure (PDB ID 3mf6) and RosettaLigand-predicted 1m4w_6 model is an expansion of the binding pocket. This expansion occurs through a 1.25Å outward movement of the protein ‘thumb’ region when compared with the original 1m4w structure (Fig. 3A). Moreover, the solvent accessible (SA) surface area of the pocket increases 2.5 times, while normalized SA volume expands by a factor of 2.3 (Fig. 5C). Although flexibility of residue side chains within the pocket partially compensate for this ‘opening’ relative to prediction, a significant enlargement of the binding pocket is observed. The all-atom root mean square deviation (RMSD) for the whole protein is 0.61 Å, but rises to 0.96 Å within the binding pocket (Fig. 3B). Notably, interface residues that contribute most to RMSD are also those possessing the highest crystallographic temperature factors (B-factors). The expansion of the binding pocket disrupts interactions observed in the computational model. When the ligand is re-docked into the crystallographic structure, only 8 of 11 predicted hydrogen bond interactions are able to assume correct bonding geometry, while the ratio of ligand surface area in VDW contact with protein decreased from 0.79 to 0.63 (Fig. 4B). Thus, we hypothesized that the lack of observed ligand binding affinity was due to the expansion of the binding pocket and resulting disruption of predicted binding contacts.
Fig. 5.
Structural determinants of β-xylanase ‘thumb’ destabilization. (A) Loss of resolvable electron density in the ‘thumb’ region is caused by an alternate confirmation of W80 in the protein ‘palm’. (B) A ‘domino’ effect of altered side-chain packing results from the substitution of wild-type (light gray) V to designed (dark gray) L at position 48. This added steric bulk pushes Y78 out of H-bonding position, which then allows W80 to adopt an alternative conformation that clashes with I127 and disrupts the hydrophobic packing of the two, thus destabilizing the ‘thumb’ loop. (C) Chart showing the relative degree of binding pocket expansion for each sequence substitution. Wild-type (WT) and W20V48 proteins display a closed conformation, while the designed (Des.) and V48 substitutions result in an ‘open’ conformation. The W20 mutant, due to ‘thumb’ destabilization, dynamically inhabits a range of conformations between ‘open’ and ‘closed’.
Rosetta analysis of 1m4w_6
To investigate the hypothesis that binding pocket enlargement was responsible for the lack of detected binding, a detailed analysis of residue-level energy contributions to binding affinity was performed comparing the 1m4w_6 experimental and predicted structures. In comparing the two structures, RosettaLigand calculations showed a modest but clear loss of binding affinity as pocket backbone opening increased, as indicated by several of the contributing energy terms (Supplementary data, Table SI). For example, the total number of residues involved in the hydrogen bonding network between ligand and protein decreased from 8 to 6, while the number of total hydrogen bonds dropped from 11 to 8. Correspondingly, the total hydrogen bond energy worsened from −8.1 to −5.3r.e.u. while VDW packing was significantly reduced from −14.5 to −10.3r.e.u. Similarly, solvation and electrostatic interaction energies worsened as pocket expansion increased. A weighted composite Rosetta binding energy score for the protein–ligand system decreased from −17.2 to −12.9r.e.u. From this analysis, we concluded that RosettaLigand can discriminate between the binding energies of a wild-type backbone configuration and that of an enlarged binding pocket, and that this energy differential could potentially explain the lack of experimentally observed ligand binding.
Additional analysis of pair-wise Rosetta energies revealed a potentially significant contributor to the backbone opening of the ‘thumb’ region: a Trp20 to Arg mutation that disrupts an interaction with Pro125 in wild-type 1m4w. This hydrophobic interaction in the wild-type protein appears to stabilize the ‘thumb’ loop in a ‘closed’ configuration and help keep the binding pocket laterally compact (Fig. 3C). Additionally, the mutation of Val48 to the sterically bulkier Leu in 1m4w_6 further acts as a wedge to prop open the binding pocket in the ‘palm’ region at a position of mechanical advantage (Fig. 3C), causing added strain within the interface. Evolutionary evidence for the crucial function of these residues can be seen from a sequence alignment of 1m4w with its nearest 250 homologues. In all 250, the Trp20, Pro125 and Val48 are either strictly or highly conserved.
Structure-guided re-design of 1m4w_6
Using the information gleaned from RosettaLigand computational analysis, a structure-guided re-design of the 1m4w_6 protein was performed to test the hypotheses that (i) the observed lack of binding affinity was due primarily to the unintended expansion of the binding pocket and resulting disruption of the predicted binding interactions, and (ii) either or both of two identified mutations (Arg20 and Leu48) from wild-type were largely responsible for the opening of the ‘thumb’ region and expansion of the binding pocket.
Three separate mutants were made starting from the 1m4w_6 sequence, by reverting Arg20, Leu48 and a double reversion of both residues to the wild-type amino acid identities (Table I). These newly designed proteins were used to identify the individual and cumulative contributions by each mutation to the backbone conformational change seen in the 1m4w_6 design. Reverting these mutations, it was hoped, would restore the binding pocket to the predicted (wild-type) geometry, thus conferring the originally predicted ligand binding affinity.
Following site-directed mutagenesis and expression of the revertant mutants (see Materials and methods) ligand binding assays for each of the three 1m4w_6-derived proteins were performed using FA and ITC. None of the re-designed 1m4w_6-derived mutants displayed observable binding affinities above those obtained from the original 1m4w_6 design. Using FA, the 1m4w_6w20, 1m4w_v48 and the 1m4w_6w20v48 displayed Kd values of 672, 536 and 392 µM, respectively (Supplementary data, Fig. S3C).
To understand the lack of binding affinity among the three 1m4w_6-derived revertant mutants, structure determination by X-ray crystallography was again performed. Using close-grid screens around the 1m4w_6 crystallization conditions, high-quality, diffracting crystals were obtained for the 1m4w_6v48, 1m4w_6w20 and 1m4w_6w20v48 constructs (PDB IDs 3mf9, 3mfc and 3mfa, respectively). Multiple single crystals formed in several buffers centered around wells containing 0.1M NaCl, 1.25M ammonium sulfate, 0.1M Bis-Tris pH 5.5, 3.5% Jeffamine M600 pH 7.0 at 20°C. Complete data sets down to 1.6–1.7Å were obtained for the three protein constructs (Table II). The data sets for all three proteins were phased by molecular replacement using MOLREP and models built using the Apr/warp software suite (see Materials and methods). Attempts to obtain ligand bound co-crystals were unsuccessful. All protein structures obtained were in the apo configuration.
Structural analysis of 1m4w_6 re-designed proteins
High-resolution structures of the re-designed 1m4w_6 derived revertant mutants revealed the relative contributions of the respective mutations to backbone conformation and binding pocket opening. In agreement with RosettaLigand prediction and part (ii) of our hypothesis, the double revertant mutant 1m4w_6w20v48 possessed a native-like ‘closed’ conformation, while the backbone of the 1m4w_6v48 mutant displayed an ‘open’ configuration largely unchanged from 1m4w_6 (Fig. 5C). The backbone RMSD of 1m4w_6w20v48 was 0.38Å from wild-type, while 1m4w_6v48 was similar to the 1m4w_6 crystallographic structure. Unexpectedly, the ‘thumb’ region of the 1m4w_6w20 mutant was not resolvable due to lack of electron density, indicating a high degree of mobility (Fig. 5A).
Discussion
The intent of this study was to explore computational methods for designing de novo high-affinity protein–peptide interfaces. The protein designs described above did not achieve our goal of high-affinity binding to their target peptides. Nonetheless, four high-resolution structures of endo-1,4-β-xylanase-derived proteins yielded important insights into the structural dynamics of family 11 xylanase proteins.
Experimental design
Our hypothesis at the outset of this study was that RosettaLigand was capable of de novo design of a high-affinity protein–peptide interface to a non-standard dipeptide ligand. Experimental testing of our original nine protein–peptide interface designs yielded negative results for high-affinity ligand binding, thus failing to prove this hypothesis. Subsequent structure determination and detailed analysis of one of the designs, 1m4w_6, led to our second hypothesis that backbone opening and expansion of the designed ligand binding pocket, caused by specific mutations, resulted in the disruption of predicted binding contacts and consequent lack of ligand affinity. It was hoped that by reverting these specific residues to wild-type, the ligand binding pocket would ‘re-close’, thus allowing the predicted ligand binding interactions to form and bind the target dipeptide with high affinity.
Testing the second hypothesis by expression and assay of three re-designed proteins yielded similar negative results for ligand binding. Structure determination and analysis of the three proteins yielded further important insights. Our hypothesis was incorrect in predicting that ‘re-closing’ of the binding pocket would result in high-affinity ligand binding. While an expanded, ‘open’ geometry of the binding pocket may contribute to a lack of high-affinity binding, a closed geometry, as seen in the structure of the double revertant mutant 1m4w_6w20v48, is not sufficient to confer high-affinity ligand binding.
However, part of the second hypothesis was shown to be true. The two specific residues identified by a detailed Rosetta energy analysis comparing the predicted and experimentally determined structures of 1m4w_6 were indeed responsible for the binding pocket expansion, and reverting these residues to wild-type restored the predicted geometry of the binding pocket. We speculate that changes in the configurational dynamics of the protein as seen in crystallographic B-factors may be partly responsible for the lack of high-affinity ligand binding. However, confirmation of this hypothesis remains outside the scope of our experimental data. An equally likely contributor to failure may be shortcomings in the Rosetta energy function, in particular its solvation energy function or treatment of water molecules.
RosettaLigand can accurately predict both the fine and large-scale structure of designed proteins and protein–ligand interfaces
Figure 3B compares the position of each side-chain atom for residues that comprise the binding pocket between predicted and experimentally attained 1m4w_6 structures. We see that even with the ‘opening’ of the binding pocket due to expansion of the ‘thumb’ region backbone, the majority of side chains assume their predicted conformations. Furthermore, even with this ‘thumb’ region backbone shift, the RMSD of all the side-chain atoms in the unliganded 1m4w_6 binding pocket is 0.96Å. This level of accuracy improves still further when the ‘thumb’ region backbone re-adopts the native ‘closed’ conformation, as in the structure of 1m4w_6w20v48, where the residues comprising the unliganded binding pocket attain an RMSD of 0.63Å.
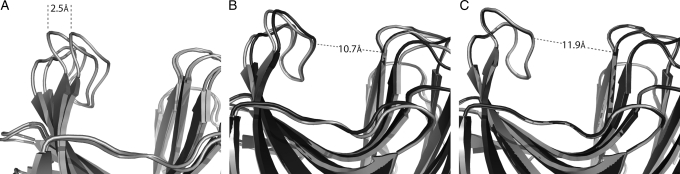
As described in the Results section, we tested RosettaLigand's ability to predict the backbone changes observed in the mutant proteins. The original protocol intentionally prevented the protein backbone from adapting in response to mutations introduced during design. The decision to use a fixed-backbone protocol initially was made to increase speed of the calculations and was based on the erroneous assumption that a thermophilic protein scaffold such as 1m4w would be unlikely to experience significant conformational change from the mutation of a small number of residues in the enzymatic cleft. When subsequently using protocols able to accommodate backbone flexibility, RosettaLigand is quantitatively able to predict the shift in backbone configuration when the destabilizing Trp20 and Val48 mutations are alternately included or removed. If the respective mutations for the ‘open’ 1m4w_6 and ‘re-closed’ 1m4w_6w20v48 are substituted onto the other's backbone coordinates, flexible-backbone relaxation protocols in RosettaLigand can accurately recover the backbone conformation observed in the experimental structures and account for binding pocket expansion (Fig. 6). When the Trp20 and Val48 mutations are introduced onto a native ‘closed’ backbone configuration, the pocket expands to that seen in the 1m4w_6 structure (Fig. 6C). When the mutations are removed, the backbone ‘re-closes’ to the native 1m4w configuration (Fig. 6B). Had we adopted a flexible-backbone protocol during our initial design calculations, it is likely that opening of the 1m4w_6 design would have been predicted accurately.
Fig. 6.
RosettaLigand flexible-backbone protocols can recapitulate backbone conformational shift. (A) 2.5 Å magnitude shift in backbone conformation between the ‘closed’ and ‘opened’ confirmations of the 1m4w wild-type and designed protein, respectively. (B) When the W20 and V48 sequence positions are substituted onto an ‘open’ backbone conformation (light gray), RosettaLigand, using flexible-backbone protocols, recovers the ‘closed’ configuration (dark gray). (C) Likewise, substituting R20 and L48 onto a ‘closed’ backbone will result in a ‘re-open’ conformation.
We thus conclude that RosettaLigand is able to predict the structure of the 1m4w designs to near atomic resolution of both the binding interface and the protein as a whole, and that the modeling of backbone conformational changes is important when designing protein–peptide interfaces.
Accurate structure prediction of the designed proteins did not translate into binding affinity
Although RosettaLigand can accurately predict large-scale changes in backbone configuration observed in the designed protein structures, the computational protocols employed in this study are significantly limited at addressing complex protein dynamics and potential entropic factors of ligand binding. Rosetta scoring and binding energy calculations are performed using a single, static, atomic representation of protein and ligand. Although recent advances in flexible backbone and relaxation functionality within Rosetta have expanded its ability to address structural fluctuation during design (Davis and Baker, 2009), the ability to fully predict the effects of dynamics at a protein–ligand interface remains limited.
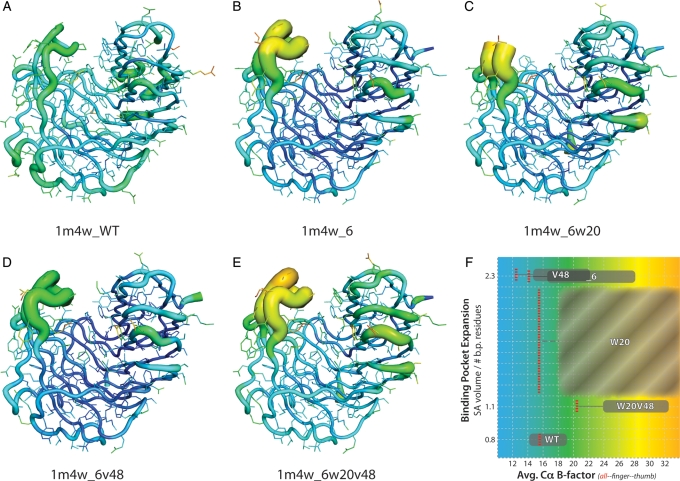
Analysis of the crystallographic data from all four of the determined 1m4w mutants when compared with wild-type 1m4w indicates both a significant increase in the mobility of the loop forming segments of the proteins' ‘thumb’ region and an overall increase in the B-factors of the protein backbone comprising the ligand binding pocket. It is interesting to note that even after the reversion mutations of the 1m4w_6w20v48 protein allowed the ‘re-closing’ of the ligand binding pocket to wild-type dimensions, the global B-factors of the protein, and more significantly those of the ‘thumb’ and ‘finger’ regions which comprise the two sides of the binding cleft, remain elevated an average of >60% (Fig. 7). These elevated B-factors suggest a fundamental alteration in the dynamics of the protein as a whole (Rueda et al., 2007) that could significantly impact the energetics of ligand binding.
Fig. 7.
Visualization of crystallographic B-factors for wild-type and four 1m4w mutant proteins. (A–E) Backbone and residue side chains colored and sized by B-factor values for wild-type 1m4w and X-ray determined structures. Red/thick = higher B-factor, blue/thin = lower B-factor. (F) The average B-factor values (x-axis) as a function of binding pocket volume (y-axis) for each protein (WT = 1m4w; _6 = 1m4w_6; V48 = 1m4w_6v48; W20 = 1m4w_6w20; W20V48 = 1m4w6w20v48). Note that while the average B-factor value for the entire protein (all) decreases for some of the designs, the ‘thumb’ and ‘finger’ B-factors are increased for all designed structures. This suggests a fundamental shift in the overall dynamics of the protein. Also note that the binding pocket volume for 1m4w_6w20 (C) is shown as a value range in (F) due to lack of electron density in the ‘thumb’ region. The binding pocket volume of 1m4w_6v48 and 1m4w_6 are equal. B-factor values for the whole protein (red, dashed line), ‘finger’ region (left extent of gray box) and ‘thumb’ region (right extent of gray box).
Increased dynamic mobility of the ‘thumb’ region specifically can be observed in all four designed structures when compared with wild-type (Fig. 7). These B-factors are 1.5- to 2.0-fold higher than in the wild-type 1m4w. In the case of the 1m4w_6w20 mutant, the lack of electron density in the ‘thumb’ loop is indicative of increased mobility. This ‘thumb’ region contributes ∼40% of the ligand interface surface area and 5 of 11 of predicted hydrogen bonds to the ligand. Thus, this observed change in dynamics in the 1m4w ‘thumb’ region is hypothesized to be a contributing factor to the lack of observed ligand binding.
Beyond the implications of altered proteins dynamics, standard RosettaLigand design protocols rely on a bulk, non-explicit solvation term (Lazaridis and Karplus, 1999) to represent water molecules in and around the binding interface. Entropic factors of binding-pocket desolvation are not well addressed by an implicit solvation term (Gilson and Zhou, 2007). Examination of the four X-ray structures reveal 9–11 ordered water molecules within the binding pocket. Due to the increased importance of predicting individual atomic interactions in the design of high-affinity interfaces, the explicit modeling of water molecules is desirable for successful design of protein–ligand interfaces (Amadasi et al., 2006; Thilagavathi and Mancera, 2010). Although recent extensions to Rosetta now allow explicit interfacial waters to be modeled, this functionality did not exist at the time this study commenced.
Ligand and scaffold selection are important determinants of design success
A dipeptide ligand composed of small, non-polar amino acids is a difficult target for a proof-of-concept experiment and was intended to push the boundaries of RosettaLigand technology. This, however, may have been overly ambitious. A larger, more apolar ligand possessing greater VDW surface area and opportunity for charge–charge interactions would be preferred in future work. Also, it remains an open question as to whether the selection of a ‘d' peptide target ligand, while theoretically equivalent to ‘l' amino acids from a chemical and computational standpoint, may have negatively contributed to the difficulty in achieving high-affinity binding (Sela and Zisman, 1997; Yamada and Kera, 1998).
More important to the potential success of protein–ligand interface design are the dynamics and conformational stability of a design scaffold protein. As found here, even highly stable, thermophilic proteins with melting temperatures well above 100°C (Sunna et al., 2000) potentially possess dynamic modes that can negatively impact high-affinity interface design due to increased entropic penalties for ligand binding. The dynamics of the endo-1,4-β-xylanase fold, as noted in recent work by Vieira et al., indicate that the 1m4w ‘thumb’ is inherently mobile in solution at elevated in situ temperatures (Vieira et al., 2009). Evidence for intensified ‘thumb’ and binding site dynamics can be seen in the crystallographic B-factors of each of the four designed protein structures. The relatively small number of mutations (in the case of 1m4w_6w20, only six) necessary to cause significant destabilizing dynamics was unanticipated for a thermophilic protein. This dynamic propensity is an undesirable trait in a protein scaffold when attempting to design a well-defined, stable, high-affinity interface. Deliberate care is advisable when choosing a de novo design scaffold, and particular attention should be given to protein dynamic modes. In this respect, scaffolds that have been extensively classified by NMR, small-angle X-ray scattering molecular dynamic simulations or other methods that yield information on protein dynamics are preferred.
The high-resolution structures of RosettaLigand interface designs reveal critical structural and dynamic determinants of β-xylanase proteins
The most notable feature of the 1m4w_6 designed protein when compared with the wild-type 1m4w protein scaffold is the radial expansion of the binding pocket defined by the ‘thumb’, ‘palm’ and ‘finger’ regions (Fig. 3A). A similar degree of expansion is also observed in the 1m4w_6v48 derivative of 1m4w_6, where Leu at position 48 has been reverted to wild-type Val. These two designs share a common mutation of Trp to Arg at position 20, which disrupts a critical hydrophobic contact between ‘finger’ (W20) and ‘thumb’ (P125), resulting in expansion of the binding pocket (Fig. 3C).
Necessary but not sufficient for closure of the binding pocket of 1m4w_6 and its derivatives is the restoration of the hydrophobic contact between residues Trp20 and Pro125. This interaction is crucial to maintaining a closed geometry under crystallization conditions. At higher temperatures near 100°C where this enzyme has evolved to function (Hakulinen et al., 2003), this interaction may be important in regulating the dynamics and enzyme kinetics of the 1m4w protein. That this Trp–Pro interaction is highly conserved across multiple species indicates that it is likely a key structural, dynamic and kinetic determinant common to family 11 xylanases.
While the hydrophobic Trp20–Pro125 interaction is necessary, it is not sufficient to allow stable closing of the binding pocket. The destabilization and consequent lack of electron density observed in the crystal structure of 1m4w_6w20 results from a clash of an alternative configuration of Trp80 in the ‘palm’ with Ile127 in the loop that forms the ‘thumb’ (Fig. 5A). This clash is in turn due to the altered packing of Tyr78, which is directly caused by the added steric bulk of the Ile48 mutation in the ‘fingers’. It is this ‘domino’ effect leading from I48 > Y78 > W80 > I127 (‘fingers’ to ‘palm’ to ‘thumb’) that breaks the contact between Trp20 and Pro125, thereby resulting in added mobility of the ‘thumb’ loop (Fig. 5B). Thus, although reversion of position 20 to the wild-type Trp is necessary for binding pocket closing, it is not in and of itself sufficient. The designed Leu at position 48 must also be reverted to wild-type Val to result in a ‘closed’ pocket configuration (Fig. 5C).
It is intriguing that the effects of a single, conservative substitution at a spatially distal amino acid position can have such a pronounced effect on the stability of a thermophilic protein at relatively low temperature, i.e. that the additional bulk of a single methylene group is transmitted from one side of the protein to the other, through three (bulky) amino acid side chains, to destabilize a large tertiary structural element at well below physiologic temperature. This suggests that the amino acid sequence of the 1m4w protein, even in the protein core (palm region), is finely tuned to accommodate this dynamic mobility. This further suggests that the increased dynamic mobility of the ‘thumb’ region due to mutations introduced during design mimics the effect of increased temperature. These mutations might therefore be thought of as having enabled high-temperature, native-like dynamics at low temperatures.
The continuing challenge of de novo protein–peptide interface design
While the lack of success experienced in the course of this particular study may or may not be attributable to factors such as scaffold selection, unanticipated protein dynamics or the lack of explicitly modeled interfacial waters, it is important to note that progress in the field of de novo ligand interface design as a whole has lagged significantly behind other areas of de novo protein design. Not long ago, it was considered by some to be a solved problem, but retractions in several key papers (Check Hayden, 2008) have led to the conclusion that the design of high-affinity protein–ligand interfaces is one of the fundamental areas of basic protein function that remains an open problem (Schreier et al., 2009b).
Rosetta has proven adept at such challenging tasks as design of novel protein folds (Kuhlman et al., 2003), altered recognition and cleavage specificity of a DNA endonuclease (Ashworth et al., 2010), and even the design of enzymes with catalytic modes not found in nature (Jiang et al., 2008; Röthlisberger et al., 2008; Siegel et al., 2010). Protein–protein interfaces have been re-designed for altered and multiple specificity (Joachimiak et al., 2006; Humphris and Kortemme, 2007b), while Rosetta and other techniques have successfully re-designed protein–peptide interfaces for altered specificity and increased affinity (Sood and Baker, 2006; Cortajarena et al., 2008; Jackrel et al., 2009).
What is it that makes de novo design of protein–ligand interfaces so difficult, and why would de novo interface design be significantly more challenging than the re-design of a protein–peptide interface, or the design of a novel enzyme? While a completely satisfactory answer to these questions has yet to be established, one contributing factor could be protein dynamics. The requirement to design and manipulate dynamics may set a higher bar for the de novo design of ligand binding. Unfortunately, protein dynamics is also one of the most difficult and least tractable problems for current protein design programs.
De novo protein design by definition entails establishing entirely new functionality in a protein. It requires an ability to recreate and manipulate all properties of a protein necessary for a given function. Conversely, re-design, where basic protein functionality is retained but relies on conserved intrinsic properties of the protein important to its function. Such conserved intrinsic properties could include protein dynamic modes conducive to ligand binding. Similarly, re-design of protein–protein specificity may benefit from conserved functionality and dynamics, as well as having the added advantage of a larger interface surface area and number of potential interactions to offset small errors in the design algorithms. Such small errors may have a larger impact in ligand interface design where each of a small number of interactions must be optimal for tight interaction.
Yet surely the creation of novel catalytic function in the de novo design of enzymes (Jiang et al., 2008; Röthlisberger et al., 2008; Siegel et al., 2010) requires no less precision and accuracy than the design of ligand binding. What has allowed these efforts to succeed where interface design has yet to? A partial answer may lay in the nature of enzyme function. In this case, the precise geometry of the catalytic mechanism is critical, and facilitating this geometry can be thought of as binding the chemical transition state. However, the timescale on which transition state re-binding occurs is extremely short, on the order of 10−12 s, when compared with high-affinity ligand binding interactions which must be maintained for seconds or longer (Zhang et al., 2008). Furthermore, recent studies suggest that the chemical reaction in enzyme catalysis is insensitive to global protein dynamics, which instead affect only enzyme kinetics (Pisliakov et al., 2009; Kamerlin and Warshel, 2010). In this light, it is notable that all of the successful enzyme designs cited above were performed using a naturally occurring enzyme as a design scaffold (some even used 1m4w) and that all of these designed enzymes possess relatively poor kinetic properties, even after undergoing multiple rounds of directed evolution to improve efficiency (Jiang et al., 2008; Röthlisberger et al., 2008; Siegel et al., 2010). The implication of these observations match the findings of this study, which found that Rosetta was capable of designing interfaces with a high degree of structural/geometric accuracy—as would be needed to stabilize a catalytic transition state intermediate—but lacked the ability to account for or design protein dynamic modes necessary for binding or efficient kinetics. While these speculations are far from conclusive with the small amount of evidence presented here, it is an intriguing line of thought that may warrant further attention in future studies.
Conclusion
Our attempts at using the RosettaLigand program to design in silico a high-affinity protein–peptide interface to a bacterial dipeptide target were unsuccessful. Twelve proteins using 1m4w as a design scaffold were assayed for binding to their intended target. No high-affinity binding was detected for any of these 12 designs.
We have proposed several potential contributors to this apparent lack of success, including overambitious target peptide selection and the lack of explicitly modeled interfacial water molecules. However, possibly the most significant negative contributor to the study outcome may be the unappreciated nature and extent of dynamics inherent to the design scaffold protein.
We have shown that RosettaLigand is able to predict the structure of designed interfaces to near-atomic resolution, and of large-scale protein conformational changes due to mutations introduced during the design process. However, accurate structure prediction did not translate into high-affinity ligand binding. We therefore conclude that the computational design of proteins that tightly bind small molecules remains possibly a greater challenge than the design of enzymes. While computational enzyme design requires accurate structural prediction of catalytic residues, tight substrate binding is not needed for success.
In addition to the lessons and caveats learned above concerning protein design applications, we have also gained new information regarding structural and functional determinants of family 11 endo-1,4-β-xylanase proteins. Specifically, the four high-resolution X-ray structures complement prior reports of the dynamics of the ‘thumb’ region of in family 11 xylanases, as well as reveal new insights into individual amino acids involved in the structural and functional dynamics of the β-xylanase protein fold. These xylanase structures may also serve as benchmark systems for future computational design protocols that model protein–peptide or protein–small molecule interfaces.
Supplementary data
Funding
This work was supported by Defense Advanced Research Projects Agency, Protein Design Project. A.M. was supported by National Institutes of Health 5 T90 DA022873. K.K. was supported by National Institutes of Health 1F31DA024528. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Supplementary Material
References
- Amadasi A., Spyrakis F., Cozzini P., Abraham D.J., Kellogg G.E., Mozzarelli A. J. Mol. Biol. 2006;358:289–309. doi: 10.1016/j.jmb.2006.01.053. doi:10.1016/j.jmb.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Arora N., Banerjee A.K., Mutyala S., Murty U.S. Bioinformation. 2009;3:446–453. doi: 10.6026/97320630003446. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/19759868 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth J., Taylor G.K., Havranek J.J., Quadri S.A., Stoddard B.L., Baker D. Nucl. Acids Res. 2010;38:5601–5608. doi: 10.1093/nar/gkq283. doi:10.1093/nar/gkq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca I.G., Chiosis G. Exp. Opin. the. Targets. 2003;7:311–328. doi: 10.1517/14728222.7.3.311. doi:10.1517/14728222.7.3.311. [DOI] [PubMed] [Google Scholar]
- Check Hayden E. Nature. 2008;453:275–278. doi:10.1038/453275a. [Google Scholar]
- Cortajarena A.L., Yi F., Regan L. ACS Chem. Biol. 2008;3:161–166. doi: 10.1021/cb700260z. doi:10.1021/cb700260z. [DOI] [PubMed] [Google Scholar]
- Cui L., Iwamoto A., Lian J.-Q., et al. Antimicrob. Agents Chemother. 2006;50:428–38. doi: 10.1128/AAC.50.2.428-438.2006. doi:10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damborsky J., Brezovsky J. Curr. Opin. Chem. Biol. 2009;13:26–34. doi: 10.1016/j.cbpa.2009.02.021. doi:10.1016/j.cbpa.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Das R., Baker D. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. doi:10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Baker D. J. Mol. Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. doi:10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Dunbrack R.L., Cohen F.E. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. doi:10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. doi:10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flower D.R., McSparron H., Blythe M.J., et al. Novartis Found. Symp. 2003;254:102–120. ; discussion 120–125, 216–222, 250–252. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14712934 . [PubMed] [Google Scholar]
- Gerlt J.A., Babbitt P.C. Curr. Opin. Chem. Biol. 2009;13:10–18. doi: 10.1016/j.cbpa.2009.01.014. doi:10.1016/j.cbpa.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M.K., Zhou H.-xiang. Annu. Rev. Biophys. Biomol. Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. doi:10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- Hakulinen N., Turunen O., Janis J., Leisola M., Rouvinen J. Eur. J. Biochem. 2003;270:1399–1412. doi: 10.1046/j.1432-1033.2003.03496.x. doi:10.1046/j.1432-1033.2003.03496.x. [DOI] [PubMed] [Google Scholar]
- Hayden E.C. Nature. 2009;461:859. doi: 10.1038/461859a. doi:10.1038/461859a. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J., Thornton J.M. ‘NACCESS’, Computer Program. 1993 Department of Biochemistry and Molecular Biology, University College London doi:10.1371/journal.pcbi.0030164. [Google Scholar]
- Humphris E.L., Kortemme T. PLoS Comput. Biol. 2007a;3:e164. doi: 10.1371/journal.pcbi.0030164. doi:10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphris E.L., Kortemme T. PLoS Comput. Biol. 2007b;3:e164. doi: 10.1371/journal.pcbi.0030164. doi:10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackrel M.E., Valverde R., Regan L. Protein Sci. 2009;18:762–774. doi: 10.1002/pro.75. doi:10.1002/pro.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Althoff E.A., Clemente F.R., et al. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. doi:10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachimiak L.A., Kortemme T., Stoddard B.L., Baker D. J. Mol. Biol. 2006;361:195–208. doi: 10.1016/j.jmb.2006.05.022. doi:10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Kamerlin S.C.L., Warshel A. Proteins. 2010;78:1339–1375. doi: 10.1002/prot.22654. doi:10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J., DeGrado W.F. Proc. Natl Acad. Sci. USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. doi:10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanicolas J., Kuhlman B. Curr. Opin. Struct. Biol. 2009;19:458–463. doi: 10.1016/j.sbi.2009.07.005. doi:10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B., Baker D. Proc. Natl Acad. Sci. USA. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10984534 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B., Dantas G., Ireton G.C., Varani G., Stoddard B.L., Baker D. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. doi:10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Nat. Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. doi:10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T., Karplus M. Proteins Struct. Funct. Genet. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. doi:10.1002/(SICI)1097-0134(19990501)35:2<133::AID-PROT1><133::AID-PROT1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Loll P.J., Axelsen P.H. Annu. Rev. Biophys. Biomol. Struct. 2000;29:265–289. doi: 10.1146/annurev.biophys.29.1.265. [DOI] [PubMed] [Google Scholar]
- Mandell D.J., Kortemme T. Nat. Chem. Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. doi:10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- Meiler J., Baker D. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. doi:10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. doi:10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pisliakov A.V., Cao J., Kamerlin S.C.L., Warshel A. Proc. Natl Acad. Sci. USA. 2009;106:17359–17364. doi: 10.1073/pnas.0909150106. doi:10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E., Briggs P., Turkenburg M., Dodson E. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12832755 . [DOI] [PubMed] [Google Scholar]
- Röthlisberger D., Khersonsky O., Wollacott A.M., et al. Nature. 2008;453:190–195. doi: 10.1038/nature06879. doi:10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Rouillard J.-marie, Lee W., Truan G., Gao X., Zhou X., Gulari E. Nucleic Acids Res. 2004;32:W176–W180. doi: 10.1093/nar/gkh401. doi:10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda M., Ferrer-Costa C., Meyer T., et al. Proc. Natl Acad. Sci. USA. 2007;104:796–801. doi: 10.1073/pnas.0605534104. doi:10.1073/pnas.0605534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammond D.W., Eletr Z.M., Purbeck C., Kuhlman B. Proteins. 2010;78:1055–1065. doi: 10.1002/prot.22631. doi:10.1002/prot.22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier B., Stumpp C., Wiesner S., Höcker B. Proc. Natl Acad. Sci. USA. 2009a;106:18491–18496. doi: 10.1073/pnas.0907950106. doi:10.1073/pnas.0907950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier B., Stumpp C., Wiesner S., Höcker B. Proc. Natl Acad. Sci. USA. 2009b;106:18491–18496. doi: 10.1073/pnas.0907950106. doi:10.1073/pnas.0907950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela M., Zisman E. FASEB J. 1997;11:449–456. doi: 10.1096/fasebj.11.6.9194525. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9194525 . [DOI] [PubMed] [Google Scholar]
- Shi L., Sings H.L., Bryan J.T., et al. Clin. Pharmacol. Ther. 2007;81:259–264. doi: 10.1038/sj.clpt.6100055. doi:10.1038/sj.clpt.6100055. [DOI] [PubMed] [Google Scholar]
- Siegel J.B., Zanghellini A., Lovick H.M., et al. Science. 2010;329:309–313. doi: 10.1126/science.1190239. doi:10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodee D.B., Malguria N., Faulhaber P., Resnick M.I., Albert J., Bakale G. Urology. 2000;56:988–993. doi: 10.1016/s0090-4295(00)00824-4. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11113745 . [DOI] [PubMed] [Google Scholar]
- Sood V.D., Baker D. J. Mol. Biol. 2006;357:917–927. doi: 10.1016/j.jmb.2006.01.045. doi:10.1016/j.jmb.2006.01.045. [DOI] [PubMed] [Google Scholar]
- Stemmer W.P., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7590320 . [DOI] [PubMed] [Google Scholar]
- Strohl W.R., Knight D.M. Curr. Opin. Biotechnol. 2009;20:668–672. doi: 10.1016/j.copbio.2009.10.012. doi:10.1016/j.copbio.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Sunna A., Gibbs M.D., Chin C.W., Nelson P.J., Bergquist P.L. Appl. Environ. Microbiol. 2000;66:664–670. doi: 10.1128/aem.66.2.664-670.2000. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10653733 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillefer R., Edell S., Innes G., Lister-James J. J. Nucl. Med. 2000;41:1214–1223. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10914912 . [PubMed] [Google Scholar]
- Thilagavathi R., Mancera R.L. J. Chem. Inf. Model. 2010;50:415–421. doi: 10.1021/ci900345h. doi:10.1021/ci900345h. [DOI] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. J. Appl. Crystallogr. 1997;30:1022–1025. doi:10.1107/S0021889897006766. [Google Scholar]
- Vieira D.S., Degrève L., Ward R.J. Biochim. Biophys. Acta. 2009;1790:1301–1306. doi: 10.1016/j.bbagen.2009.04.017. doi:10.1016/j.bbagen.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7630882 . [DOI] [PubMed] [Google Scholar]
- Yamada R., Kera Y. EXS. 1998;85:145–155. doi: 10.1007/978-3-0348-8837-0_9. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9949873 . [DOI] [PubMed] [Google Scholar]
- Zhang X., DeChancie J., Gunaydin H., et al. J. Org. Chem. 2008;73:889–899. doi: 10.1021/jo701974n. doi:10.1021/jo701974n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.