Abstract
For the improved production of vaccines and therapeutic proteins, a detailed understanding of the metabolic dynamics during batch or fed-batch production is requested. To study the new human cell line AGE1.HN, a flexible metabolic flux analysis method was developed that is considering dynamic changes in growth and metabolism during cultivation. This method comprises analysis of formation of cellular components as well as conversion of major substrates and products, spline fitting of dynamic data and flux estimation using metabolite balancing. During batch cultivation of AGE1.HN three distinct phases were observed, an initial one with consumption of pyruvate and high glycolytic activity, a second characterized by a highly efficient metabolism with very little energy spilling waste production and a third with glutamine limitation and decreasing viability. Main events triggering changes in cellular metabolism were depletion of pyruvate and glutamine. Potential targets for the improvement identified from the analysis are (i) reduction of overflow metabolism in the beginning of cultivation, e.g. accomplished by reduction of pyruvate content in the medium and (ii) prolongation of phase 2 with its highly efficient energy metabolism applying e.g. specific feeding strategies. The method presented allows fast and reliable metabolic flux analysis during the development of producer cells and production processes from microtiter plate to large scale reactors with moderate analytical and computational effort. It seems well suited to guide media optimization and genetic engineering of producing cell lines.
Electronic supplementary material
The online version of this article (doi:10.1007/s00449-010-0502-y) contains supplementary material, which is available to authorized users.
Keywords: Growth phases, Cell culture, Kinetics, Production, Mammalian cell, Metabolic engineering
Introduction
Cell culture products represent currently the most important and most promising source for new biopharmaceuticals. The majority of recombinant biopharmaceuticals are produced by mammalian cell culture and this fraction is steadily increasing [1–3]. The projected sales volume for therapeutic proteins by 2010 is USD 70 billion [4, 5]. Commonly employed production cell lines are, e.g., Chinese Hamster Ovary (CHO) cells or murine lymphoid cells. These production systems must be capable of generating high quantities of proteins to enable the administration of high doses required in clinical application [6–8]. A number of approaches were applied to optimize product yields [9], e.g., cell cycle arrest and control of apoptosis [10–13] or addition of nutrient supplements [14–16]. However, there is still room for improvement. New production cell lines are entering the market due to demands for higher product quality (e.g., glycosylation), efficiency, productivity and robustness. Cell lines created from primary cells represent promising new production systems. These cell lines overcome typical limitations of other cells as for example serum requirements, attachment dependence and eliminate also the repeated safety testing required for each new batch of primary tissue [17]. Concerning manufacturing of therapeutic proteins, engineered human cell lines may have in particular another unique advantage over standard producer cell lines, as for example CHO cells, because of their genuine human post-translational modification machinery. The cell line AGE1.HN represents such a new human cell line which was immortalized and engineered for apoptosis resistance and can be used for production of recombinant proteins but also for viruses and viral vectors. However, further targeted optimization would be promoted by a profound knowledge of growth and metabolism of the cells. In order to find optimized cultivation conditions and targets for metabolic engineering for improved production metabolic flux analysis seems most suited since it represents the functional end point of genome, proteome and metabolome interactions [18, 19]. This methodology has revealed valuable insights into the metabolism of several mammalian cell systems [20–24]. A requirement for reliable stationary metabolic flux analysis is metabolic steady state that is usually not reached due to complex growth requirements. It seems generally possible to optimize the medium such that metabolic steady state is reached during a longer period [25]. However, if the metabolism of cells in a conventional medium should be investigated, dynamic changes of the cellular metabolism commonly observed in cell culture [26] have to be monitored and considered. One way to deal with this challenge is to focus just on the exponential growth phase of the cells where in some cases metabolic steady state can be observed [22]. Another possibility is to divide the growth of the cells in different metabolic phases and calculate the flux distribution for each phase [26]. The third possibility is to compute instantaneous flux distributions during the whole cultivation [27]. Most detailed information can be obtained using 13C-substrate labeling techniques. This can be accomplished either assuming intracellular steady state of metabolites [28] or analyzing intracellular metabolite and isotopomer dynamics. The latter requires much more sophisticated analytical methods and is much more laborious both for analysis as well as modeling and computation [29, 30].
In this study we quantitatively analyze dynamics of growth and metabolism of the new human cell line AGE1.HN using a dynamic flux analysis method. The presented method of metabolic flux analysis is well suited to monitor and investigate metabolic shifts caused by environmental changes that occur during batch cultivation of mammalian cells. It does not require any kinetic model and is therefore widely applicable. Estimated flux dynamics were compared to results obtained from stationary metabolic flux analysis in different metabolic phases. We analyzed parental AGE1.HN cells in this study to gain detailed quantitative knowledge and understanding of growth behavior, cellular metabolism as well as its dynamics. The results are therefore expected to be useful for rational development of optimal growth media, for further optimization and engineering of the cell line to obtain higher cell densities in a process but also for all derived producer cells and for the design of other producer cell lines.
Materials and methods
Cell line
The AGE1.HN cell line was developed from primary cells from a human brain tissue sample. The primary cells were immortalized with an expression plasmid containing the adenoviral E1 A and B genes of human adenovirus type 5 driven by the human pGK and the endogenous E1B promoter, respectively. While the E1A induces progression of the cell cycle, the E1B proteins prevent apoptosis caused by E1A through interaction with P53 and bax. The cell line was further modified to express the structural and regulatory protein pIX from human adenovirus type 5. This modification changes cell metabolism, enhances productivity for secreted proteins and modulates susceptibility to a variety of viruses. The AGE1.HN cells express marker genes representative for cells of neuronal lineage and lack expression of glial markers. The cell line was adapted to robust growth in suspension in both serum/protein-free and chemically defined culture media and has been proven to grow efficiently in shaker, spinner, roller and stirred tank bioreactors. The entire development work for the generation of the AGE1.HN line was carried out in a dedicated cell culture suite and the complete development history was recorded. The documentation for all media and supplements used in the AGE1.HN cell line development is in compliance with the stipulations of EMEA 410/01. The AGE1.HN cell line is proprietary to ProBioGen AG (Berlin, Germany) covered by patent application 06807777.5-2406.
Cell culture and experimental procedure
The cells were cultured in baffled shake flasks (Corning, NY, USA) at 37 °C on a shaker (2 inches orbit, ES-X, Kühner, Basel, Switzerland) enclosed in a 5% CO2 supplied, humidified (80%) incubator (Mytron, Heilbad Heiligenstadt, Germany). The pre-culture was carried out in a 250 ml baffled shake flask in 42-Max-UB-medium which is serum-free and was especially developed for the AGE1.HN cell line. The medium composition is presented in Table S1. The cells were centrifuged (800 l min−1, 7 min, 22 °C, Megafuge, Heraeus Instruments, Hanau, Germany) and the supernatant discarded. The pellet was resuspended in 42-Max-UB-medium without glucose and glutamine. Four 125-ml baffled shake flasks were inoculated yielding an initial cell density of about 4 × 105 cells ml−1. Glucose and glutamine were finally added. The final culture volume was 54 ml. Samples (1.6 ml) were taken every day. 500 μl were directly used for cell counting. The rest was centrifuged (1,500 l min−1, 5 min, 22 °C, Megafuge, Heraeus Instruments, Hanau, Germany), the supernatant transferred into fresh tubes and frozen (−20 °C). The analysis of several cultivation parameters was carried out using an automated cell culture analyzer (Cedex AS20, Innovatis, Bielefeld, Germany) which determines cell density, viability (Trypan Blue exclusion method), size, morphology and aggregation rate. Dissolved oxygen was always above 80% in the applied setup which was determined in an additional cultivation using the OXY-4 mini system (PreSens-Precision Sensing GmbH, Regensburg, Germany) described earlier [31].
Quantification of metabolites
Glucose, lactate and pyruvate in the supernatant were analyzed using high pressure liquid chromatography (HPLC) as described previously [22]. Quantification of proteinogenic amino acids was performed by another HPLC method [32]. The measured metabolite concentrations as well as cell density were corrected for evaporation using Eq. 1. The determined evaporation rate k E in the applied setup was 0.0356 h−1.
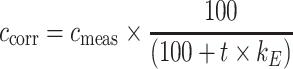 |
1 |
c corr represents the corrected concentration and c meas the measured concentration.
Glutamine hydrolysis was observed to be a first order process having a rate constant k G of 0.00198 h−1 in the applied medium (pH 7.4, 37 °C). The actual glutamine uptake rate r Gln was calculated employing polynomials of degree 5 in Matlab 7.5 (The Mathworks, Natick, MA, USA) using
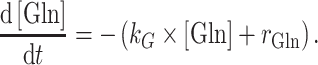 |
2 |
Metabolic network
A metabolic network model (Fig. 1) was set up using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database for Homo sapiens as well as other published metabolic network information [33]. The enzymes that were included into the model are given in Table S4 of the supplementary material section. The stoichiometric matrix of the model had the dimensions 41 × 70 and a rank of 41. It is presented in the supplementary material (Table S5).
Fig. 1.
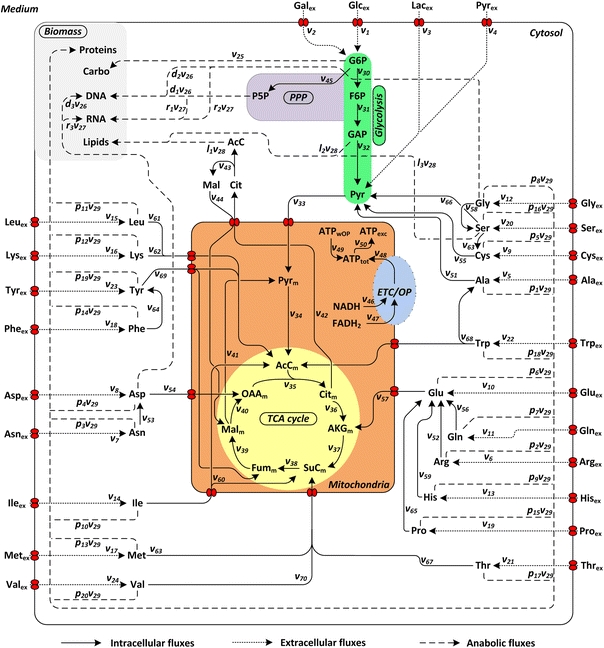
Metabolic network used for metabolic flux analysis. PPP pentose phosphate pathway, TCA tricarboxylic acid; ETC electron transport chain, OP oxidative phosphorylation, Carbo carbohydrates, Glc glucose, Gal galactose, Lac lactate, Pyr pyruvate, G6P glucose 6-phosphate, P5P pentose 5-phosphate, F6P fructose 6-phosphate, GAP glyceraldehyde 3-phosphate, AcC acetyl coenzyme A, Cit citrate, AKG α-ketoglutarate, SuC succinyl coenzyme A, Fum fumarate, Mal malate, OAA oxaloacetate, ATP adenosine triphosphate, ATP OP ATP from oxidative phosphorylation, ATP wOP ATP without oxidative phosphorylation, ATP tot total ATP, ATP exc ATP excess, NADH nicotinamide adenine dinucleotide, FADH 2 flavin adenine dinucleotide, standard abbreviations for amino acids. Indices: m mitochondrial, ex extracellular
Central energy metabolism
The main pathways of the energy metabolism, glycolysis, oxidative decarboxylation, TCA cycle, electron transport chain and oxidative phosphorylation are represented. Due to the possible action of nicotinamide nucleotide transhydrogenases and the fact that for some reactions it is not known whether NADH or NADPH take part, NADH and NADPH were lumped together. The excess of NAD(P)H and FADH2 from all reactions considered in the model was calculated. For oxidative phosphorylation, it was assumed that per NAD(P)H and FADH2 2.5 ATP and 1.5 ATP are produced, respectively. The excess of ATP (ATPtot) was calculated considering the demand or production of all reactions. However, the energy needed for transport of metabolites and maintenance was not included in the ATP-balance.
Pentose phosphate pathway
Pentose phosphate pathway/glycolysis split cannot be resolved by metabolite balancing alone and was assumed to be just responsible for nucleic acid synthesis.
Amino acid metabolism
The metabolism of proteinogenic amino acids is represented by selected degradation pathways. In cases where several pathways are possible, the most probable and suitable pathway was chosen (Table S4 of supplementary material; Fig. 1).
Other reactions
Since it is not possible to distinguish between the reactions catalyzed by the enzymes phosphoenolpyruvate carboxykinase, malic enzyme (malate dehydrogenase, oxaloacetate decarboxylating) and pyruvate carboxylase with the method applied in this study, just one reaction from malate to pyruvate was included which represents the sum of the fluxes through these enzymes.
Synthesis of biomass
The biomass fluxes are represented as five fluxes to the major macromolecules of the cell. Cytosolic acetyl coenzyme A, which is needed for the synthesis of fatty acids and lipids, is assumed to be solely derived from citrate that is transported through the citrate/malate shuttle and converted through the citrate lyase to acetyl coenzyme A and malate. Further precursors that were considered for the synthesis of lipids are glyceraldehyde 3-phosphate and serine. Carbohydrates were taken as polyglucose and are therefore derived from glucose 6-phosphate. The precursors considered for nucleic acid synthesis are ribose 5-phosphate, glycine and aspartate. For proteins, a certain amount of each proteinogenic amino acid is needed per cell which was calculated using the amino acid composition of total cellular protein determined in this study (described below).
Transport of metabolites
As mentioned above the energy demand for transport was not included in the ATP balance. From calculated fluxes, a rough estimate could however be derived using literature knowledge, e.g., about amino acid transport [34]. The mitochondrial transporters included in the model are the pyruvate carrier, citrate/malate shuttle, glutamate/aspartate shuttle and the ADP/ATP transporter [35, 36]. For further metabolites entering the mitochondria, no specific transport mechanisms were assumed.
Anabolic demand of AGE1.HN
Macromolecules
The applied macromolecular composition of the cells was: proteins, 70.6%; lipids, 9.2%; carbohydrates, 7.1%; DNA, 1.4%; RNA, 5.8%; rest/ash, 5.9%. The lipid composition was determined once using 4.9 × 108 cells, harvested at a cell density of 3.4 × 106 cells ml−1 (viability 97%) using a published protocol [22]. The other parts of the composition were taken from Bonarius et al. [21].
Amino acid composition
The amino acid composition of total cellular protein was determined using samples of 1–2 × 107 cells that were washed twice with PBS to remove amino acids from the medium. For hydrolysis 6 M HCl (280 μl/107 cells) was used and incubated for 24 h at 110 °C. The solution was neutralized using 6 M NaOH and the amino acid composition was determined using HPLC as described above. The determined relative composition (n = 4) was: aspartic acid/asparagine, 9.25 ± 0.6%; glutamic acid/glutamine, 12.83 ± 0.48%; serine, 6.19 ± 0.12%; histidine, 2.18 ± 0.12%; glycine, 9.45 ± 0.67%; threonine, 4.94 ± 0.1%; arginine, 5.19 ± 0.23%; alanine, 8.06 ± 0.09%; tyrosine, 1.9 ± 0.04%; valine, 6.14 ± 0.13%; phenylalanine, 3.56 ± 0.08%; isoleucine, 4.49 ± 0.09%; leucine, 8.71 ± 0.1%; lysine, 7.5 ± 0.17%; proline, 3.99 ± 0.12%. Methionine, tryptophan and cysteine are degraded by the applied hydrolytic method. Relative amounts for these amino acids were taken from literature [37]: methionine, 1.7%; tryptophan, 1.1%; cysteine, 2.8%.
Cell dry weight estimation
The cell dry weight of mammalian cells in culture is not constant and increases with increasing growth rate [38, 39]. It was shown, that the volume of cells is proportional to their dry weight and can be used as an indirect measure of the biomass concentration in the culture [38]. In this study, cell size was determined using an automated cell culture analyzer (Cedex AS20, Innovatis, Bielefeld, Germany) providing an accurate determination of the cell diameter. Assuming, that the cells are spherical, the respective cell volume was calculated. The correlation between dry cell weight, DCW (pg), and cell volume, CV (μm3), determined in a control experiment using the procedure described previously [22], was DCW = 0.25 × CV. This relationship was used for calculation of biomass concentrations. The anabolic demand of the cells is shown in Table 1.
Table 1.
Anabolic demand of AGE1.HN
| Macromolecule | Anabolic demand (μmol g−1 biomass) |
|---|---|
| Carbohydrates | 438.3 |
| DNA | 45.3 |
| RNA | 180.3 |
| Lipids | 128.1 |
| Proteins | 6,433.0 |
Metabolic flux analysis
The two different procedures applied for calculation of metabolic fluxes are depicted in Fig. 2. For the calculation of metabolic fluxes over time (dynamic method), splines were fitted to the measured data for biomass and extracellular metabolites (Supplementary Fig. S1) using Matlab 7.5 (The Mathworks, Natick, MA, USA). These were used to calculate specific rates (v) for each metabolite concentration M i over time using
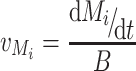 |
3 |
where B represents the respective biomass in the culture. Intracellular fluxes were calculated using
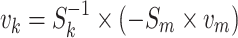 |
4 |
where v m and v k represent the measured and the calculated rate vectors. S k and S m are corresponding stoichiometric matrices.
Fig. 2.
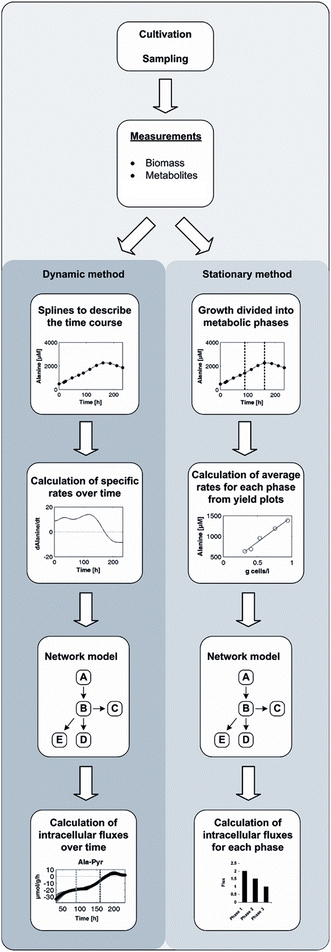
Methods applied for metabolic flux analysis. Dynamic method for calculation of the time course of metabolic fluxes and stationary method for calculation of mean fluxes for different metabolic phases
For calculating stationary fluxes, growth was divided into different metabolic phases (Fig. 2). Average rates for each metabolite M i were calculated assuming metabolic steady state for each phase using the estimated specific growth rate μ and metabolite yields:
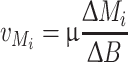 |
5 |
B represents the total biomass in the culture. The data was implemented into the metabolic network model and intracellular fluxes for each phase were subsequently calculated using Eq. 4.
Results
Growth profile of AGE1.HN during batch cultivation
During the initial growth period lasting up to 160 h the viability remained high at about 95% (Fig. 3). At 160 h, the viability started decreasing and the growth rate slowed down. The cell density at the end of the batch cultivation was about 7.5 × 106 cells ml−1 (Fig. 3). By plotting the logarithm of the cell density against time (not shown), distinct growth phases could be distinguished. The first lasted from 0 to 90 h, the second from 90 to 160 h, the third from 160 to 230 h, having growth rates of 0.351 ± 0.055, 0.279 ± 0.021 and 0.056 ± 0.011 d−1.
Fig. 3.
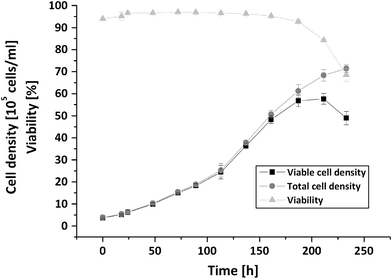
Growth of AGE1.HN. Viable cell density, total cell density and viability during batch cultivation. Mean values and standard deviations of four parallel cultivations
Exometabolome analysis and extracellular fluxes
The observed growth behavior is strongly linked to metabolite profiles (Fig. 4, Fig. S1). Pyruvate was consumed during the first 90 h of the cultivation (phase 1), and lactate, glutamate, alanine and glycine were produced. The uptake rates of glucose and pyruvate as well as the production rate of lactate were highest in the beginning and dropped until 90 h (Fig. 5). The lactate production stopped with pyruvate depletion at about 90 h indicating a coupling of these reactions (Fig. 5). The glutamate production rate was decreasing during the first phase whereas the glutamine consumption rate remained at a constant high value in this phase (Fig. 5). For most other amino acids the uptake or production rates were highest in phase 1 (Fig. S2, Table S2 and Table S3).
Fig. 4.
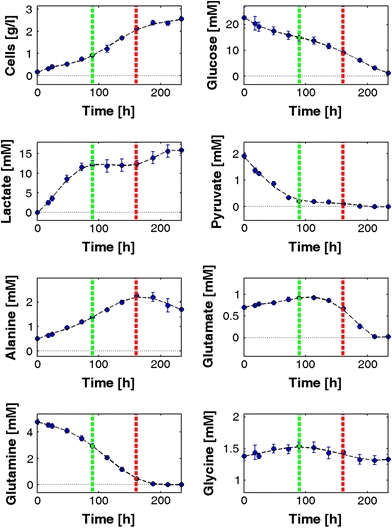
Cultivation profile of AGE1.HN. Mean values and standard deviations for cell mass and selected metabolites of four parallel cultivations are depicted. Further metabolites are depicted in Fig. S1 of the supplementary material. The first line (90 h) indicates the end of the first metabolic phase when pyruvate is depleted, the second line (160 h) the end of the second metabolic phase when glutamine is depleted
Fig. 5.
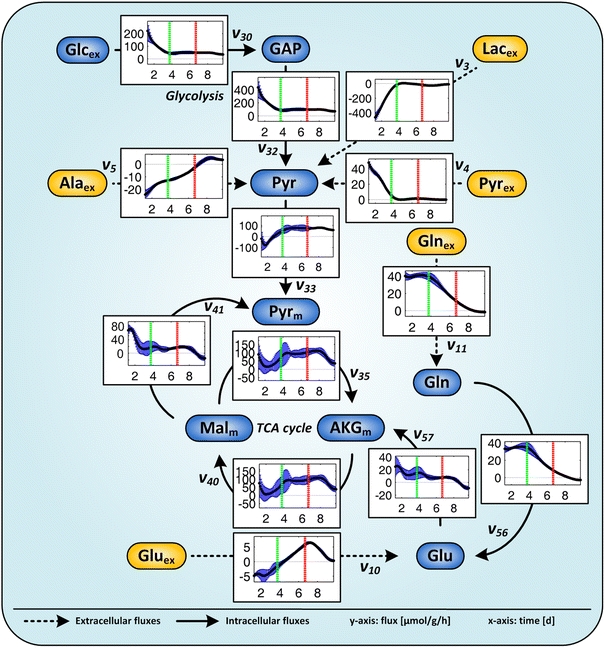
Selected metabolic fluxes of AGE1.HN over time related to biomass. Mean values and confidence intervals (90%) of four parallel cultivations. The fluxes (y-axis) are given in μmol g−1 h−1 and the time (x-axis) in days. The first line (90 h) indicates the end of the first metabolic phase when pyruvate is consumed, the second line (160 h) the end of the second metabolic phase when glutamine is consumed. Negative values indicate fluxes in the opposite direction of the arrow. Further extracellular and intracellular fluxes are depicted in Fig. S2 and Fig. S3 of the supplementary material
After 90 h (phase 2), when pyruvate was used up, lactate production stopped immediately and the cells started to consume glutamate as well as glycine (Fig. 4). The extracellular fluxes for most metabolites were significantly lower than in phase 1 (Fig. 6; Table S3). The glucose uptake rate that is almost identical to the rate of glycolysis was constant (Fig. 5) and the lactate production rate was very low (Figs. 5, 6). The glutamine uptake rate was decreasing (Fig. 5, Table S2). At the end of phase 2, glutamine was nearly completely consumed and the cells started to consume glutamate (Fig. 5; Table S2). The production rate of alanine was decreasing (Fig. 5; Fig. S2; Table S2). The rates for other amino acids remained nearly constant or were very low (Fig. S2).
Fig. 6.
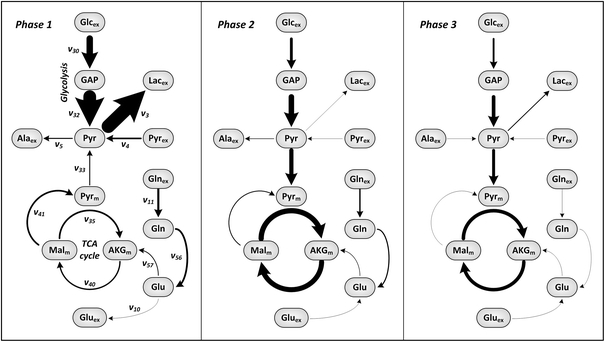
Average metabolic flux distribution of AGE1.HN in each metabolic phase. Flux symbols selected for this figure are shown in the flux map of phase 1 and are defined in the metabolic network (Fig. 1). Same experiment as shown in Fig. 5. Further data are provided in Table S3 of the supplementary material
After 160 h (phase 3), glutamine was used up which forced the cells to adapt their metabolism again. Glutamate consumption increased initially in this phase. Lactate production again increased slightly but was nevertheless very low compared to phase 1 (Fig. 5; Table S3). Alanine, which was constantly produced during phases 1 and 2, was now consumed. In addition, the cells stopped exponential growth and the viability dropped directly after glutamine depletion which indicates the important role of glutamine as substrate for AGE1.HN. Almost all of the calculated mean extracellular fluxes (Fig. 6; Table S3) were significantly different as compared to phase 1 and most of them were also significantly different as compared to phase 2. Glutamate was completely exhausted at about 215 h (Fig. 4). The glucose consumption rate was constantly decreasing until glucose was completely depleted at the end of the cultivation at 230 h (Figs. 4, 5).
In general, the standard errors for extracellular fluxes were high in the beginning and decreased with time (Fig. 5). This is due to the small differences in substrate concentrations in the beginning of the cultivation. The integral yield of cells per glucose was around 340 cells nmol−1 which is a favorable feature of AGE1.HN since it is higher compared to the corresponding yield in CHO cells which is typically ranging between 60 and 100 cells nmol−1 of glucose consumed [25, 40].
Metabolic flux analysis—intracellular fluxes
Intracellular fluxes were calculated using the stoichiometric network model depicted in Fig. 1.
Glycolysis
The highest flux through the glycolytic pathway was observed in phase 1 (Fig. 6; Table S3). Then it was steadily decreasing until 90 h (Fig. 5; Fig. S2). In phase 2, glycolytic activity was significantly lower and remained nearly constant. In phase 3, glycolytic flux was slightly decreasing until the end of the cultivation (Fig. 5; Fig. S2) but was not significantly different from phase 2 (Fig. 6; Table S3).
Pyruvate dehydrogenase activity
The calculated fluxes through the pyruvate carrier (Pyr-Pyrm, Figs. 5, 6) and through the pyruvate dehydrogenase complex (Pyrm-AcCm) were low in the beginning, i.e., not significantly different from zero (Table S3) and increased in phase 1. In phase 2, pyruvate dehydrogenase activity remained constant and was significantly higher compared to phase 1 (Figs. 5, 6; Table S3). Since lactate production was approximately 0 in phase 2, most of the pyruvate was entering the TCA cycle via pyruvate dehydrogenase. This flux was still high in phase 3 but slightly decreasing (Fig. 5).
TCA cycle and anaplerosis
The activity of the TCA cycle in phase 1 was significantly lower than in phases 2 and 3 (Figs. 5, 6; Table S3). No lactate was produced from pyruvate in phase 2. Therefore, a large fraction of pyruvate entered the TCA cycle and fluxes through the upper part as well as the lower part of the TCA cycle were very high in this phase (Figs. 5, 6; Table S3). The anaplerotic flux through the enzyme glutamate dehydrogenase (Glu-AKGm) was decreasing during the cultivation matching the decrease in available extracellular glutamine (Fig. 5). The aspartate aminotransferase flux (Asp-OAAm) was highest in phase 1 and decreasing. During phase 2, this flux remained constant until it decreased again in phase 3 (Fig. S3). Fluxes through the TCA cycle in phase 3 were similar to those in phase 2 but decreasing towards the end (Figs. 5, 6; Table S3). Glutamine was depleted and glutamate was taken up instead (Fig. 4). However, the flux through glutamate dehydrogenase was very low in phase 3 which shows that acetyl coenzyme A derived from pyruvate was by far the most important fuel for TCA cycle after glutamine depletion. The calculated flux from malate to pyruvate (Malm-Pyrm) was very high in the beginning of phase 1. In the following phases, this flux dropped and was significantly lower (Figs. 5, 6; Table S3). Interestingly during phase 1, the TCA cycle was additionally fed by the degradation of the branched chain amino acids isoleucin, leucin and valine. In the other phases, degradation of these amino acids to form intermediates of TCA cycle was not significantly different from zero.
Electron transport chain/oxidative phosphorylation
In accordance to the time courses for FADH2 and NAD(P)H production, the activity of the oxidative phosphorylation (ATPOP) was low in phase 1 and then increasing to a constant level during phase 2 until the middle of phase 3 and subsequently decreasing (Fig. S3; Table S3). The ATP-balance for all reactions included in the network without oxidative phosphorylation (ATPwOP) indicates that the ATP demand was also highest in phases 1 and 2 and lowest in phase 3.
Amino acid metabolism
The fluxes through glutamine synthetase (Gln-Glu) and glutamate dehydrogenase (Glu-AKGm) were almost constant in phase 1 and then decreasing during the cultivation, since glutamine and glutamate were successively depleted (Figs. 5, 6; Table S3). The glutamate pool was additionally fed by degradation of arginine (Arg-Glu, Fig. S3; Table S3). In phases 1 and 2, less asparagine was taken up than needed for protein synthesis which was then synthesized through the asparagine synthase from aspartate (Asn-Asp, Fig. S3; Table S3). Aspartate was feeding into the TCA cycle during the whole cultivation through the aspartate aminotransferase catalyzed reaction leading to oxaloacetate (Asp-OAAm). This flux was decreasing over time (Fig. S3). Since glycine was produced in phase 1 and then only slightly taken up (Fig. 4), it had to be produced via glycine hydroxymethyltransferase indicated by Gly-Ser in Fig. S3 and Table S3. This flux was significantly decreasing with time, because glycine was taken up in phases 2 and 3 where, additionally, the anabolic demand was decreasing because of slower growth. The flux through serine dehydratase (Ser-Pyr) was also significantly decreasing (Fig. S3; Table S3). In accordance with the time course of proline production (Fig. 5), the flux from glutamate to proline (Pro-Glu, Fig. S3; Table S3) was highest in phase 1 and then decreasing. Other degradation rates of amino acids were very low indicating that the uptake of these amino acids was just matching the anabolic demand of the cells.
Discussion
Metabolism in AGE1.HN
Metabolism and metabolic shifts of the new human cell line AGE1.HN were quantitatively analyzed in this study using time resolved metabolic flux analysis and stationary metabolic flux analysis in different phases during batch cultivation. Our results indicate that pyruvate uptake during phase 1 in combination with high levels of other substrates resulted in an inefficient metabolic phenotype characterized by overflow metabolism leading to waste product formation. In phase 2, the metabolism was switching to a most efficient state characterized by low substrate uptake rates and no lactate formation. The data indicate that a decrease in substrate levels occurring during the cultivation resulted in a slowdown of glycolysis during phase 1. Additional experiments using different pyruvate concentrations in the medium support the conclusion that there is a strong link between pyruvate in the medium and lactate production in AGE1.HN (data will be presented elsewhere). Another possibility would be that the cells could secrete lactate only until a certain concentration is reached and would then stop the production. However, in other experiments that were performed using high lactate start concentrations (about 8 mM) it was observed, that the cells were producing almost the same amount of lactate as in experiments where lactate start concentration was 0. This indicates that pyruvate depletion seems to be an important factor for cessation of lactate production. This particular phenotype was so far not observed in other mammalian cells [22, 23, 26, 41].
During exponential growth of mammalian cells under non-limiting substrate levels inefficient substrate utilization was reported for several other mammalian cell lines [22, 23, 28, 42]. Usually only a small amount of intracellular pyruvate is entering the TCA cycle under these conditions which is considered very unfavorable. Increasing flux through pyruvate dehydrogenase represents an optimization target for the cultivation of many mammalian cells, but the mechanisms and molecular reasons are still poorly understood [43, 44]. In a recent study on HEK-293, cells pyruvate dehydrogenase activity was reported to be around 22% of glycolytic activity [42]. Very low or even no activity of the pyruvate dehydrogenase complex was reported in other studies on different mammalian cells [23, 45]. In AGE1.HN cells, this was observed only during the first phase. During phases 2 and 3, pyruvate dehydrogenase activity was high. In MDCK cells it was found that an addition of pyruvate in a glutamine depleted medium resulted even in an increase in the flux from pyruvate into the TCA cycle [23]. In AGE1.HN, it seems that available pyruvate has an opposite effect triggering the conversion of pyruvate to lactate. High uptake rates of glucose and pyruvate as observed in phase 1 might lead to an increase of the intracellular pyruvate pool triggering overflow metabolism into lactate/alanine eventually resulting in an increased secretion of these waste metabolites. This might be changed by increasing reactions connecting TCA cycle and glycolysis, e.g., by stimulating pyruvate dehydrogenase and pyruvate carboxylase fluxes [46, 47]. However, the success of this strategy can not be predicted a priori. Increase of fluxes into the TCA cycle must be accompanied by an increase in oxidative phosphorylation activity which might not always be possible. Another strategy that is working in the opposite direction would be decreasing the substrate uptake rates, e.g., by knock down of transporters [48] or decreasing the lactate production, e.g., by knock down of lactate dehydrogenase [40].
Comparison of selected metabolic fluxes in AGE1.HN with those in other cells
Selected metabolic fluxes in AGE1.HN that were calculated by the stationary method for different phases are compared to fluxes published for other mammalian cells (Table 2). Glucose uptake and lactate production rates were generally lower in AGE1.HN than in other cells. Pyruvate represents an extracellular metabolite that was often neglected in other studies [26, 28]. However, as shown in this study and in MDCK cells [23, 49] or CHO cells [50], it is a very interesting metabolite that can induce huge changes in the metabolism. In AGE1.HN cells pyruvate uptake rate was by far lower than in MDCK cells that were grown in high pyruvate medium (Table 2, M2). However, the ratio of pyruvate uptake per glucose uptake was similar. The second phase in AGE1.HN showed a very efficient metabolism concerning substrate usage as already discussed before. The cells were proliferating without lactate production. Pyruvate dehydrogenase (PDH) activity was high. In all other cell lines, high lactate production was observed during the phases that were analyzed by MFA except for CHO cells in presence of galactose after glucose depletion. In order to reach higher cell densities in cultivations of AGE.HN, the second phase could be prolonged by feeding of glutamine. PDH activity in AGE1.HN was low in phase 1 and high in phases 2 and 3. A similar situation was observed in CHO cells [26] which exhibited low PDH activity and overflow metabolism during growth on glucose. After glucose depletion, these cells were consuming lactate in the presence of galactose having high PDH activity. No PDH activity was reported for MDCK cells in glutamine containing medium. In medium without glutamine and high pyruvate concentration low PDH activity was found in these cells. The absolute flux through PDH was highest in continuously cultured hybridoma cells [28], but the metabolism was by far not as efficient as in phase 2 of AGE1.HN since there was high glucose consumption and waste product formation. Absolute glutamate dehydrogenase (GDH) activity of AGE1.HN during phases 1 and 2 was lower than in other cell lines, but the relative activity compared to the glucose uptake rate was quite similar in MDCK and CHO cultures. Upper and lower TCA cycle activity in phases 2 and 3 was similar to the activity in CHO cells with glucose medium (S1 in Table 2) and in MDCK cells (M2 in Table 2) cultured without glutamine. For hybridoma cells, higher TCA cycle activity was reported.
Table 2.
Comparison of selected metabolic fluxes in AGE1.HN during different metabolic phases (P1–3) with fluxes published for other mammalian cell lines
| Flux [μmol g−1 h−1] | ||||||||
|---|---|---|---|---|---|---|---|---|
| AGE1.HN | MDCKa | CHOb | Hybridomac | |||||
| P1 | P2 | P3 | M1 | M2 | S1 | S2 | ||
| Glc uptake | 123 | 56.6 | 33.7 | 1,108.7 | 476.6 | 322.5 | 43.8d | 674.6 |
| Pyr uptake | 28.7 | 0.9 | 0.4 | 0 | 111 | nd | nd | nd |
| Lac production | 236.3 | 4.5 | 14.3 | 2,188 | 965 | 612.5 | −68.8 | 1,030 |
| Gln uptake | 37.1 | 23.5 | 2.1 | 70.9 | 0.9 | 0 | 0 | 146.9 |
| Glu production | 4 | −2.4 | −2.9 | −42.1 | −22.1 | −56.3 | −56.8 | 1.2 |
| PDH | 12.2 | 88.1 | 56.8 | 0 | 33.1 | 35.5 | 174.6 | 355.7 |
| GDH | 13.2 | 8.3 | 1.7 | 64.7 | 42.8 | 28.1 | 14 | 110.1 |
| Upper TCA | −4 | 66.4 | 57.7 | −27.3 | 43.5 | 35.5 | 148.8 | 312.5 |
| Lower TCA | 9.2 | 74.7 | 59.3 | 54.3 | 63 | 78.9 | 195.8 | 441.1 |
Fluxes are given in μmol g−1 h−1. Literature fluxes that were given per cell were normalized to biomass by assuming a cell dry weight of 800 pg cell−1 for MDCK cells [24] and 400 pg cell−1 for CHO and hybridoma cells [21, 60]
MDCK murine darby canine kidney, M1 glutamine containing medium, M2 medium with high pyruvate concentration and without glutamine, CHO chinese hamster ovary, S1 stage 1 of cultivation, S2 stage 2 of cultivation when glucose is exhausted, Glc glucose, Pyr pyruvate, Lac lactate, Gln glutamine, Glu glutamate, PDH pyruvate dehydrogenase, GDH glutamate dehydrogenase, upper TCA activity of the upper tricarboxylic acid cycle represented by the flux from citrate to α-ketoglutarate, lower TCA activity of the lower TCA cycle represented by the α-ketoglutarate dehydrogenase flux (glutamate to α-ketoglutarate), nd not determined
aMDCK cell culture [23]
bCHO cell culture [26]
cContinuous culture of hybridoma cells [28]
dGalactose uptake
The main differences between the metabolic phases of AGE1.HN are summarized in Fig. 7. The results indicate further targets for improving the metabolic phenotype of these cells to reach higher cell densities as well as to obtain a more efficient utilization of the nutrients. The high overflow metabolism in the beginning of the cultivation with channeling of pyruvate to lactate could be decreased. This could be accomplished by genetic engineering, e.g., deletion of lactate dehydrogenase [51] or introduction of pyruvate carboxylase [46], further medium optimization, e.g., reduced concentrations of pyruvate, or application of new feeding strategies [50]. The observed metabolism in phase 2 indicates the potential of this cell line to grow very efficiently having minimum formation of waste products and minimum energy spilling [52]. Therefore, it seems promising to change environmental conditions, i.e., media, substrate feeding as well as enzyme expression such as to approach the efficient metabolic state of phase 2 during a process. Studies on enzyme expression and detailed labelling experiments in combination with 13C metabolic flux analysis would provide additional hints for the best way to further improve the cell line.
Fig. 7.
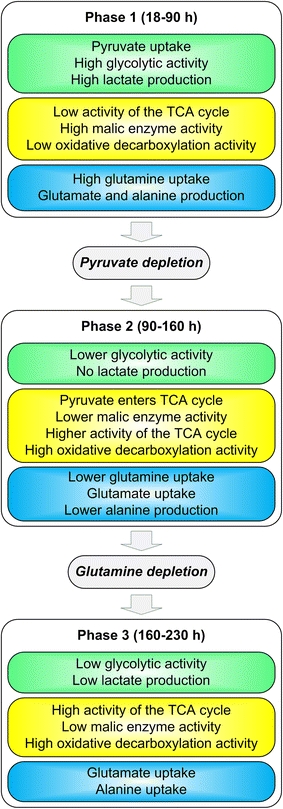
Summary of metabolic shifts during batch cultivation of AGE1.HN
Time resolved versus stationary flux analysis
The applied dynamic method is well suited to analyze and monitor metabolic shifts during the cultivation. Cell growth, cell size, extracellular as well as intracellular fluxes of mammalian cells in a cell culture process are highly dynamic which is caused by environmental changes. Therefore, the presented method that is considering the dynamics of all included metabolites and biomass is best suited to understand and finally model the process. Stationary metabolic flux analysis that was applied extensively in the past [23, 53] is only suited to describe the mean metabolism during a certain phase which is a significant disadvantage since changes during the phase considered are neglected [54]. However, if the pseudo steady state assumption during a phase is justified, it can give a useful overview of its average metabolism [55]. The information obtained by flux balance analysis can be validated [56] or further enriched by specific labelling information [57] to resolve reversible, cyclic or parallel fluxes as for example pentose phosphate pathway split [58, 59].
Concluding remarks
The application of a dynamic metabolic flux analysis method as presented in this study is well suited to describe the dynamic and adaptive behavior of the metabolism of mammalian cells during cultivation in industrially relevant media at any scale. Thus, it supports the identification of targets for improved growth and substrate utilization based on derived metabolic flux distribution. Further improvement of the method could be achieved by including labelling information as for example to monitor pentose phosphate pathway split. The presented dynamic method can be applied to almost any cultivation of a mammalian cell and could replace the widely applied but inflexible stationary MFA. In this study, metabolic shifts caused by changing environmental conditions during cultivation of AGE1.HN cells were described in detail (Fig. 7). This knowledge about the reaction of the cells to changes in the environment is a valuable basis for designing efficient feeding strategies but also providing hints for genetic modifications. In the AGE1.HN cell line major targets identified for improvement are as follows: (i) overflow metabolism during phase 1 should be reduced, e.g., by reduction of pyruvate content in the medium, (ii) phase 2 should be prolonged by specific feeding strategies, e.g., of glutamine. The dynamic metabolic flux data presented in this study contribute to the attempted systems level of understanding of this cell line that is expected to eventually improve heterologous protein production in a cell producing a heterologous protein. Generally, the method represents a valuable option to support today and future improvement of cell culture media and processes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Metabolic profiles of four cultivations (a–d) and fitted splines (TIFF 738 kb)
Extracellular fluxes of AGE1.HN over time related to biomass. Mean values and confidence intervals (90%) of four parallel cultivations. The green dotted line indicates the end of the first metabolic phase where pyruvate is consumed, the red dashed line indicates the end of the second metabolic phase where glutamine is consumed. Negative values indicate production, positive values consumption of a metabolite (TIFF 2641 kb)
Intracellular fluxes of AGE1.HN over time related to biomass. Mean values and confidence intervals (90%) of four parallel cultivations. The green dotted line indicates the end of the first metabolic phase where pyruvate is consumed, the red dashed line indicates the end of the second metabolic phase where glutamine is consumed. Abbreviations and reactions are explained in Fig. 1 and Table S5 (TIFF 3805 kb)
Main components of the applied 42-Max-UB medium (PDF 13 kb)
Time course of metabolic fluxes. Mean values (p.1 and 2) and confidence intervals (90%, p.3 and 4) of four parallel cultivations (PDF 123 kb)
Average metabolic flux distribution of AGE1.HN in each metabolic phase. Mean values and confidence intervals (90%, CI) of four parallel cultivations. The probabilities of a paired t test between the different phases are also depicted. The null hypothesis (no difference) was rejected at the 5% significance level. P1/P2, phase 1 compared with phase 2; P1/P3, phase 1 compared with phase 3; P2/P3, phase 2 compared with phase 3. Network model and abbreviations are shown in Fig. 1 (PDF 15 kb)
Enzymes considered in the metabolic network (PDF 21 kb)
Stoichiometric matrix of the metabolic network (Fig. 1) (PDF 62 kb)
Acknowledgments
This work has been financially supported by the BMBF project SysLogics—Systems biology of cell culture for biologics. We thank Michel Fritz for valuable support for the HPLC analysis as well as Christian Priesnitz, Averina Nicolae and Judith Wahrheit for their supporting work and valuable suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.O’Callaghan PM, James DC. Systems biotechnology of mammalian cell factories. Brief Funct Genomic Proteomic. 2008;7(2):95–110. doi: 10.1093/bfgp/eln012. [DOI] [PubMed] [Google Scholar]
- 2.Ferrer-Miralles N, Domingo-Espin J, Corchero JL, Vazquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb Cell Fact. 2009;8:17. doi: 10.1186/1475-2859-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat Biotechnol. 2004;22(11):1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 4.Pavlou AK, Reichert JM. Recombinant protein therapeutics—success rates, market trends, values to 2010. Nat Biotechnol. 2004;22(12):1513–1519. doi: 10.1038/nbt1204-1513. [DOI] [PubMed] [Google Scholar]
- 5.Walsh G. Biopharmaceutical benchmarks 2006. Nat Biotechnol. 2006;24(7):769–776. doi: 10.1038/nbt0706-769. [DOI] [PubMed] [Google Scholar]
- 6.Dinnis DM, James DC. Engineering mammalian cell factories for improved recombinant monoclonal antibody production: lessons from nature? Biotechnol Bioeng. 2005;91(2):180–189. doi: 10.1002/bit.20499. [DOI] [PubMed] [Google Scholar]
- 7.Rose T, Winkler K, Brundke E, Jordan I, Sandig V (2005) Alternative strategies and new cell lines for high-level production of biopharmaceuticals. In: Modern biopharmaceuticals, Wiley-VCH, Germany, pp 761–777
- 8.Sandig V, Rose T, Winkler K, Brecht R (2005) Mammalian cells. In: Production of recombinant proteins. Wiley-VCH, Germany, pp 233–252
- 9.Kumar N, Gammell P, Clynes M. Proliferation control strategies to improve productivity and survival during CHO based production culture : a summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology. 2007;53(1–3):33–46. doi: 10.1007/s10616-007-9047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.al-Rubeai M, Singh RP. Apoptosis in cell culture. Curr Opin Biotechnol. 1998;9(2):152–156. doi: 10.1016/S0958-1669(98)80108-0. [DOI] [PubMed] [Google Scholar]
- 11.Fogolin MB, Wagner R, Etcheverrigaray M, Kratje R. Impact of temperature reduction and expression of yeast pyruvate carboxylase on hGM-CSF-producing CHO cells. J Biotechnol. 2004;109(1–2):179–191. doi: 10.1016/j.jbiotec.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 12.Trummer E, Fauland K, Seidinger S, Schriebl K, Lattenmayer C, Kunert R, Vorauer-Uhl K, Weik R, Borth N, Katinger H, Muller D. Process parameter shifting: Part I. Effect of DOT, pH, and temperature on the performance of Epo-Fc expressing CHO cells cultivated in controlled batch bioreactors. Biotechnol Bioeng. 2006;94(6):1033–1044. doi: 10.1002/bit.21013. [DOI] [PubMed] [Google Scholar]
- 13.Yoon SK, Song JY, Lee GM. Effect of low culture temperature on specific productivity, transcription level, and heterogeneity of erythropoietin in Chinese hamster ovary cells. Biotechnol Bioeng. 2003;82(3):289–298. doi: 10.1002/bit.10566. [DOI] [PubMed] [Google Scholar]
- 14.Oh HK, So MK, Yang J, Yoon HC, Ahn JS, Lee JM, Kim JT, Yoo JU, Byun TH. Effect of N-acetylcystein on butyrate-treated Chinese hamster ovary cells to improve the production of recombinant human interferon-β-1a. Biotechnol Prog. 2005;21(4):1154–1164. doi: 10.1021/bp050057v. [DOI] [PubMed] [Google Scholar]
- 15.Sung YH, Song YJ, Lim SW, Chung JY, Lee GM. Effect of sodium butyrate on the production, heterogeneity and biological activity of human thrombopoietin by recombinant Chinese hamster ovary cells. J Biotechnol. 2004;112(3):323–335. doi: 10.1016/j.jbiotec.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Eyer K, Oeggerli A, Heinzle E. On-line gas analysis in animal cell cultivation: II. Methods for oxygen uptake rate estimation and its application to controlled feeding of glutamine. Biotechnol Bioeng. 1995;45(1):54–62. doi: 10.1002/bit.260450108. [DOI] [PubMed] [Google Scholar]
- 17.Jordan I, Vos A, Beilfuss S, Neubert A, Breul S, Sandig V. An avian cell line designed for production of highly attenuated viruses. Vaccine. 2009;27(5):748–756. doi: 10.1016/j.vaccine.2008.11.066. [DOI] [PubMed] [Google Scholar]
- 18.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen J. It is all about metabolic fluxes. J Bacteriol. 2003;185(24):7031–7035. doi: 10.1128/JB.185.24.7031-7035.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balcarcel RR, Clark LM. Metabolic screening of mammalian cell cultures using well-plates. Biotechnol Prog. 2003;19(1):98–108. doi: 10.1021/bp025617u. [DOI] [PubMed] [Google Scholar]
- 21.Bonarius HP, Hatzimanikatis V, Meesters KP, de Gooijer CD, Schmid G, Tramper J. Metabolic flux analysis of hybridoma cells in different culture media using mass balances. Biotechnol Bioeng. 1996;50(3):299–318. doi: 10.1002/(SICI)1097-0290(19960505)50:3<299::AID-BIT9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 22.Niklas J, Noor F, Heinzle E. Effects of drugs in subtoxic concentrations on the metabolic fluxes in human hepatoma cell line Hep G2. Toxicol Appl Pharmacol. 2009;240(3):327–336. doi: 10.1016/j.taap.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Sidorenko Y, Wahl A, Dauner M, Genzel Y, Reichl U. Comparison of metabolic flux distributions for MDCK cell growth in glutamine- and pyruvate-containing media. Biotechnol Prog. 2008;24(2):311–320. doi: 10.1021/bp0702673. [DOI] [PubMed] [Google Scholar]
- 24.Wahl A, Sidorenko Y, Dauner M, Genzel Y, Reichl U. Metabolic flux model for an anchorage-dependent MDCK cell line: characteristic growth phases and minimum substrate consumption flux distribution. Biotechnol Bioeng. 2008;101(1):135–152. doi: 10.1002/bit.21873. [DOI] [PubMed] [Google Scholar]
- 25.Deshpande R, Yang TH, Heinzle E. Towards a metabolic and isotopic steady state in CHO batch cultures for reliable isotope-based metabolic profiling. Biotechnol J. 2009;4(2):247–263. doi: 10.1002/biot.200800143. [DOI] [PubMed] [Google Scholar]
- 26.Altamirano C, Illanes A, Becerra S, Cairo JJ, Godia F. Considerations on the lactate consumption by CHO cells in the presence of galactose. J Biotechnol. 2006;125(4):547–556. doi: 10.1016/j.jbiotec.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 27.Llaneras F, Pico J. A procedure for the estimation over time of metabolic fluxes in scenarios where measurements are uncertain and/or insufficient. BMC Bioinformatics. 2007;8:421. doi: 10.1186/1471-2105-8-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonarius HP, Ozemre A, Timmerarends B, Skrabal P, Tramper J, Schmid G, Heinzle E. Metabolic-flux analysis of continuously cultured hybridoma cells using (13)CO(2) mass spectrometry in combination with (13)C-lactate nuclear magnetic resonance spectroscopy and metabolite balancing. Biotechnol Bioeng. 2001;74(6):528–538. doi: 10.1002/bit.1145. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann U, Maier K, Niebel A, Vacun G, Reuss M, Mauch K. Identification of metabolic fluxes in hepatic cells from transient 13C-labeling experiments: part I experimental observations. Biotechnol Bioeng. 2008;100(2):344–354. doi: 10.1002/bit.21747. [DOI] [PubMed] [Google Scholar]
- 30.Maier K, Hofmann U, Reuss M, Mauch K. Identification of metabolic fluxes in hepatic cells from transient 13C-labeling experiments: part II. Flux estimation. Biotechnol Bioeng. 2008;100(2):355–370. doi: 10.1002/bit.21746. [DOI] [PubMed] [Google Scholar]
- 31.Deshpande RR, Heinzle E. Online monitoring of oxygen in spinner flasks. Biotechnol Lett. 2009;31(5):665–669. doi: 10.1007/s10529-009-9919-2. [DOI] [PubMed] [Google Scholar]
- 32.Kromer JO, Fritz M, Heinzle E, Wittmann C. In vivo quantification of intracellular amino acids and intermediates of the methionine pathway in Corynebacterium glutamicum. Anal Biochem. 2005;340(1):171–173. doi: 10.1016/j.ab.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BO. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104(6):1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyde R, Taylor PM, Hundal HS. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem J. 2003;373(Pt 1):1–18. doi: 10.1042/BJ20030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg JM, Tymoczko JL, Stryer L. Biochemie. 5th. Berlin: Spektrum Akademischer Verlag; 2003. [Google Scholar]
- 36.Kaplan RS. Structure and function of mitochondrial anion transport proteins. J Membr Biol. 2001;179(3):165–183. doi: 10.1007/s002320010046. [DOI] [PubMed] [Google Scholar]
- 37.Savinell JM, Palsson BO. Network analysis of intermediary metabolism using linear optimization I. Development of mathematical formalism. J Theor Biol. 1992;154(4):421–454. doi: 10.1016/S0022-5193(05)80161-4. [DOI] [PubMed] [Google Scholar]
- 38.Frame KK, Hu WS. Cell volume measurement as an estimation of mammalian cell biomass. Biotechnol Bioeng. 1990;36(2):191–197. doi: 10.1002/bit.260360211. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen LK, Reid S, Greenfield PF. Cell cycle model to describe animal cell size variation and lag between cell number and biomass dynamics. Biotechnol Bioeng. 1997;56(4):372–379. doi: 10.1002/(SICI)1097-0290(19971120)56:4<372::AID-BIT3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 40.Kim SH, Lee GM. Down-regulation of lactate dehydrogenase-A by siRNAs for reduced lactic acid formation of Chinese hamster ovary cells producing thrombopoietin. Appl Microbiol Biotechnol. 2007;74(1):152–159. doi: 10.1007/s00253-006-0654-5. [DOI] [PubMed] [Google Scholar]
- 41.Gambhir A, Korke R, Lee J, Fu PC, Europa A, Hu WS. Analysis of cellular metabolism of hybridoma cells at distinct physiological states. J Biosci Bioeng. 2003;95(4):317–327. doi: 10.1016/s1389-1723(03)80062-2. [DOI] [PubMed] [Google Scholar]
- 42.Henry O, Jolicoeur M, Kamen A (2010) Unraveling the metabolism of HEK-293 cells using lactate isotopomer analysis. Bioprocess Biosyst Eng [DOI] [PubMed]
- 43.Modak J, Deckwer WD, Zeng AP. Metabolic control analysis of eucaryotic pyruvate dehydrogenase multienzyme complex. Biotechnol Prog. 2002;18(6):1157–1169. doi: 10.1021/bp025613p. [DOI] [PubMed] [Google Scholar]
- 44.Zeng AP, Modak J, Deckwer WD. Nonlinear dynamics of eucaryotic pyruvate dehydrogenase multienzyme complex: decarboxylation rate, oscillations, and multiplicity. Biotechnol Prog. 2002;18(6):1265–1276. doi: 10.1021/bp020097o. [DOI] [PubMed] [Google Scholar]
- 45.Neermann J, Wagner R. Comparative analysis of glucose and glutamine metabolism in transformed mammalian cell lines, insect and primary liver cells. J Cell Physiol. 1996;166(1):152–169. doi: 10.1002/(SICI)1097-4652(199601)166:1<152::AID-JCP18>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 46.Elias CB, Carpentier E, Durocher Y, Bisson L, Wagner R, Kamen A. Improving glucose and glutamine metabolism of human HEK 293 and Trichoplusia ni insect cells engineered to express a cytosolic pyruvate carboxylase enzyme. Biotechnol Prog. 2003;19(1):90–97. doi: 10.1021/bp025572x. [DOI] [PubMed] [Google Scholar]
- 47.Irani N, Wirth M, van Den Heuvel J, Wagner R. Improvement of the primary metabolism of cell cultures by introducing a new cytoplasmic pyruvate carboxylase reaction. Biotechnol Bioeng. 1999;66(4):238–246. doi: 10.1002/(SICI)1097-0290(1999)66:4<238::AID-BIT5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 48.Paredes C, Prats E, Cairo JJ, Azorin F, Cornudella L, Godia F. Modification of glucose and glutamine metabolism in hybridoma cells through metabolic engineering. Cytotechnology. 1999;30(1–3):85–93. doi: 10.1023/A:1008012518961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genzel Y, Ritter JB, Konig S, Alt R, Reichl U. Substitution of glutamine by pyruvate to reduce ammonia formation and growth inhibition of mammalian cells. Biotechnol Prog. 2005;21(1):58–69. doi: 10.1021/bp049827d. [DOI] [PubMed] [Google Scholar]
- 50.Omasa T, Furuichi K, Iemura T, Katakura Y, Kishimoto M, Suga K. Enhanced antibody production following intermediate addition based on flux analysis in mammalian cell continuous culture. Bioprocess Biosyst Eng. 2010;33(1):117–125. doi: 10.1007/s00449-009-0351-8. [DOI] [PubMed] [Google Scholar]
- 51.Chen K, Liu Q, Xie L, Sharp PA, Wang DI. Engineering of a mammalian cell line for reduction of lactate formation and high monoclonal antibody production. Biotechnol Bioeng. 2001;72(1):55–61. doi: 10.1002/1097-0290(20010105)72:1<55::AID-BIT8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Russell JB. The energy spilling reactions of bacteria and other organisms. J Mol Microbiol Biotechnol. 2007;13(1–3):1–11. doi: 10.1159/000103591. [DOI] [PubMed] [Google Scholar]
- 53.Xie L, Wang DI. Material balance studies on animal cell metabolism using a stoichiometrically based reaction network. Biotechnol Bioeng. 1996;52(5):579–590. doi: 10.1002/(SICI)1097-0290(19961205)52:5<579::AID-BIT5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 54.Niklas J, Schneider K, Heinzle E. Metabolic flux analysis in eukaryotes. Curr Opin Biotechnol. 2010;21(1):63–69. doi: 10.1016/j.copbio.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Quek LE, Dietmair S, Kromer JO, Nielsen LK. Metabolic flux analysis in mammalian cell culture. Metab Eng. 2010;12(2):161–171. doi: 10.1016/j.ymben.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Zupke C, Stephanopoulos G. Intracellular flux analysis in hybridomas using mass balances and in vitro (13)C nmr. Biotechnol Bioeng. 1995;45(4):292–303. doi: 10.1002/bit.260450403. [DOI] [PubMed] [Google Scholar]
- 57.Bonarius HP, Timmerarends B, de Gooijer CD, Tramper J. Metabolite-balancing techniques versus 13C tracer experiments to determine metabolic fluxes in hybridoma cells. Biotechnol Bioeng. 1998;58(2–3):258–262. doi: 10.1002/(SICI)1097-0290(19980420)58:2/3<258::AID-BIT20>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 58.Lee WN, Boros LG, Puigjaner J, Bassilian S, Lim S, Cascante M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1, 2–13C2]glucose. Am J Physiol. 1998;274(5 pt 1):E843–E851. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- 59.Velagapudi VR, Wittmann C, Schneider K, Heinzle E. Metabolic flux screening of Saccharomyces cerevisiae single knockout strains on glucose and galactose supports elucidation of gene function. J Biotechnol. 2007;132(4):395–404. doi: 10.1016/j.jbiotec.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 60.Xie L, Wang DI. Applications of improved stoichiometric model in medium design and fed-batch cultivation of animal cells in bioreactor. Cytotechnology. 1994;15(1–3):17–29. doi: 10.1007/BF00762376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolic profiles of four cultivations (a–d) and fitted splines (TIFF 738 kb)
Extracellular fluxes of AGE1.HN over time related to biomass. Mean values and confidence intervals (90%) of four parallel cultivations. The green dotted line indicates the end of the first metabolic phase where pyruvate is consumed, the red dashed line indicates the end of the second metabolic phase where glutamine is consumed. Negative values indicate production, positive values consumption of a metabolite (TIFF 2641 kb)
Intracellular fluxes of AGE1.HN over time related to biomass. Mean values and confidence intervals (90%) of four parallel cultivations. The green dotted line indicates the end of the first metabolic phase where pyruvate is consumed, the red dashed line indicates the end of the second metabolic phase where glutamine is consumed. Abbreviations and reactions are explained in Fig. 1 and Table S5 (TIFF 3805 kb)
Main components of the applied 42-Max-UB medium (PDF 13 kb)
Time course of metabolic fluxes. Mean values (p.1 and 2) and confidence intervals (90%, p.3 and 4) of four parallel cultivations (PDF 123 kb)
Average metabolic flux distribution of AGE1.HN in each metabolic phase. Mean values and confidence intervals (90%, CI) of four parallel cultivations. The probabilities of a paired t test between the different phases are also depicted. The null hypothesis (no difference) was rejected at the 5% significance level. P1/P2, phase 1 compared with phase 2; P1/P3, phase 1 compared with phase 3; P2/P3, phase 2 compared with phase 3. Network model and abbreviations are shown in Fig. 1 (PDF 15 kb)
Enzymes considered in the metabolic network (PDF 21 kb)
Stoichiometric matrix of the metabolic network (Fig. 1) (PDF 62 kb)


