Summary
Human exonuclease 1 (hExo1) plays important roles in DNA repair and recombination processes that maintain genomic integrity. It is a member of the 5′ structure-specific nuclease family of exonucleases and endonucleases that includes FEN-1, XPG, and GEN1. We present structures of hExo1 in complex with a DNA substrate, followed by mutagenesis studies, and propose a common mechanism by which this nuclease family recognizes and processes diverse DNA structures. hExo1 induces a sharp bend in the DNA at nicks or gaps. Frayed 5′ ends of nicked duplexes resemble flap junctions, unifying the mechanisms of endo- and exo-nucleolytic processing. Conformational control of a mobile region in the catalytic site suggests a mechanism for allosteric regulation by binding to protein partners. The relative arrangement of substrate binding sites in these enzymes provides an elegant solution to a complex geometrical puzzle of substrate recognition and processing.
Introduction
Human Exonuclease 1 (hExo1) is essential for maintaining genomic stability by nucleolytic processing of DNA intermediates involved in repair and recombination. hExo1 functions in several DNA repair pathways: it confers the primary exonuclease activity employed in mammalian mismatch repair (MMR) (Genschel et al., 2002; Wei et al., 2003); it is involved in DNA resection during double strand break repair (DSBR) (Zhu et al., 2008); it is important for telomere maintenance through promotion of recombination at transcription-induced telomeric structures (Vallur and Maizels, 2010b). Deficiency of mismatch repair can have profound deleterious effects on human health, such as spontaneous mutability, hereditary nonpolyposis colorectal cancer (HNPCC), and the development of 15–25% of sporadic tumors (Kolodner, 1995; Peltomaki, 2003). Failure to repair double strand breaks can result in chromosomal rearrangements or deletions, leading to carcinogenesis and premature aging (Hartlerode and Scully, 2009).
The first characterized role of hExo1 was its exonuclease function in human mismatch repair. hExo1 excises mismatches in this repair pathway, and requires a nick 5′ to the excision region to perform 5′-3′ hydrolysis on double-stranded DNA (Dzantiev et al., 2004; Genschel et al., 2002; Genschel and Modrich, 2003; Zhang et al., 2005). hExo1 interacts with a number of MMR proteins, including MutLα and the DNA lesion recognition proteins MutSα and MutSβ (Nielsen et al., 2004; Schmutte et al., 1998; Schmutte et al., 2001); these interactions directly modulate exonucleolytic activity (Genschel and Modrich, 2003, 2009). Binding of hExo1 to MutSα in a mismatch- and ATP-dependent manner is required for processive 5′-3′ hydrolysis (Genschel and Modrich, 2003). Additionally, studies in yeast suggest that Exo1 may also play a structural role in mismatch repair through stabilization of complexes containing multiple MMR proteins (Amin et al., 2001). In DBSR, hExo1 interacts with a different assembly of protein partners during homologous recombination (Mimitou and Symington, 2008; Zhu et al., 2008). Depletion of hExo1 results in an increase in the development of double strand breaks (Gravel et al., 2008; Nimonkar et al., 2008).
hExo1 is a member of the 5′ structure-specific nuclease family of metalloenzymes that are involved in multiple DNA repair pathways. This family includes FEN-1 (flap endonuclease 1), that participates in processing of Okazaki fragments; GEN1 (gap endonuclease 1), involved in Holliday junction (HJ) resolution, and XPG (xeroderma pigmentosum complementation group G) that processes DNA bubble structures (Tomlinson et al., 2010). These proteins share a conserved N-terminal catalytic core nuclease region, but exhibit individual preferences for structurally distinct DNA substrates. The C-terminal regions of these proteins are divergent in sequence. Although structures of human FEN-1 and FEN-1 homologs have been determined (Ceska et al., 1996; Chapados et al., 2004; Devos et al., 2007; Dore et al., 2006; Feng et al., 2004; Hosfield et al., 1998; Hwang et al., 1998; Matsui et al., 2002; Mueser et al., 1996; Sakurai et al., 2005), these lack either the assembled two-metal active site required for catalysis, or a DNA substrate. Consequently, many questions concerning catalytic mechanism and substrate recognition have remained unanswered.
Here we present the structure of the hExo1 N-terminal catalytic domain (residues 1–352) in complex with a 10-bp duplex with a three-base 3′ single-strand extension. This structure mimics a gapped duplex and represents a model intermediate structure in mismatch repair (Genschel and Modrich, 2003), and is likely to correspond to a substrate in double-strand break repair (Nimonkar et al., 2008). hExo1 recognizes nicked, gapped, or blunt DNA in vitro (Genschel and Modrich, 2003; Lee and Wilson, 1999). The active site accommodates both 5′ ends and 5′ flaps, which undergo exo- and endonucleolytic cleavage respectively (Lee and Wilson, 1999). Elucidating the structural features by which the active site can accommodate such different substrates and activities is therefore essential for understanding this enzyme and other members in the FEN family. We find that hExo1 binds at the junction of single- and double-stranded DNA by stabilizing a sharp bend that can be accommodated only in nicked or gapped DNA. A metal center cleaves the nicked strand immediately adjacent to this junction. The scissile bond is placed at the metal center, if the last two bases at the 5′ end of the double-stranded region fray. Such fraying converts a double-stranded region into a short single-stranded segment. Consequently the 5′ end of a nick could resemble the basal region of a flap, making endo- and exonucleolytic activities equivalent. Although we do not observe a 5′ flap directly, the structure of the complex indicates a probable path whereby the flap strand exits out of the active site and binds along the protein surface. The enzyme therefore solves the problem of diverse substrate recognition and processing by setting up a series of surface sites that each bind and process the various segments of structured DNA substrates.
The hExo1 catalytic domain shares ~20% sequence identity with other FEN-1 family members. Much of the domain structure is structurally homologous with FENs, but we also identify important structural differences, which encode the functional specializations that differentiate these enzymes from each other. Analysis of these similarities and differences has enabled us to propose a unified model for substrate recognition and processing for the FEN enzyme family, based on the insight that a common relative geometrical arrangement of the active site, 5′- and 3′-binding sites can accommodate and distinguish different structured DNA substrates. All family members induce a sharp bend in the substrate for nick or gap recognition; fraying at the 5′ end of the duplex removes apparent differences between nick and flap processing; (non)accommodation of 5′ or 3′ flaps in surface binding sites further encodes substrate specificity. The structure-based hypotheses generated by our observations and models are open to testing in future experiments.
Results
Overview of the hExo1 structure
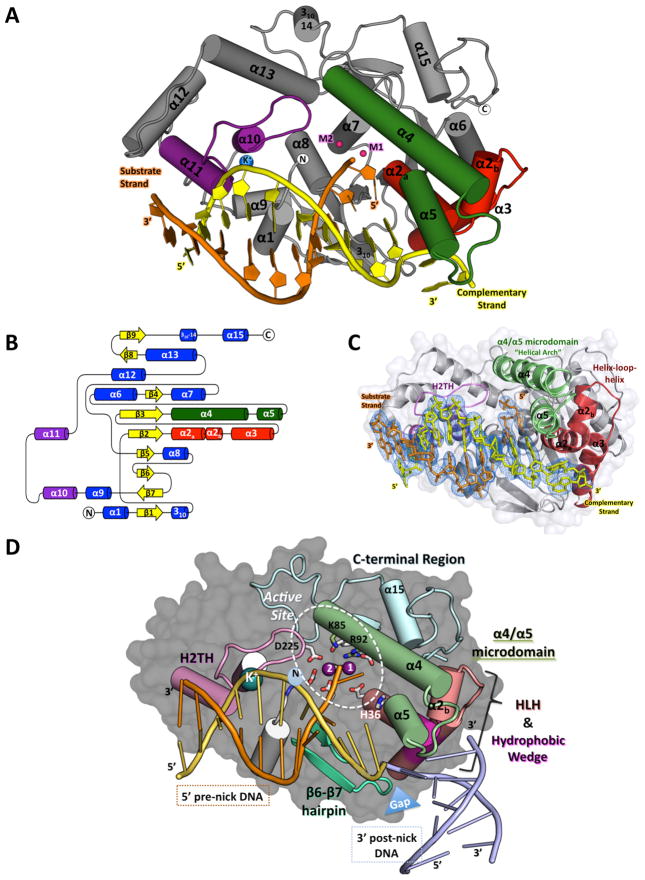
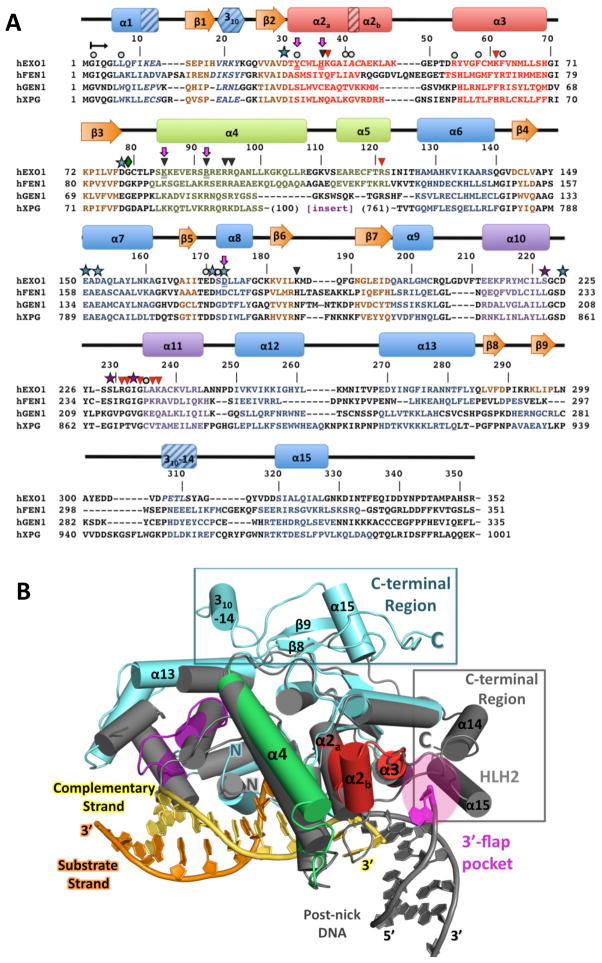
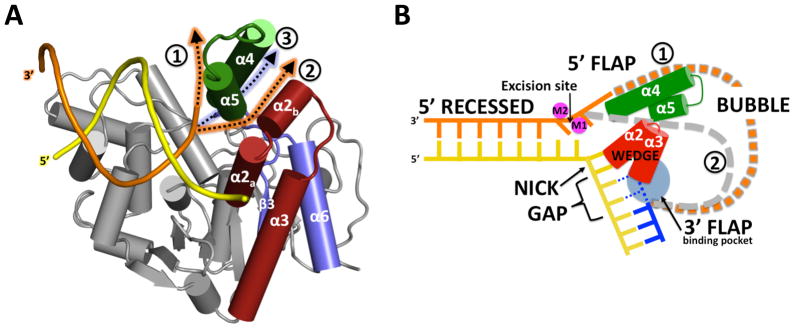
The N-terminal domain of hExo1 forms a bean-shaped core with a helical protrusion, and a number of surface grooves (Figure 1A–D). The structure can be divided into four regions (defined in reference to the nick at which the enzyme binds): the pre-nick DNA binding region which holds the double-stranded segment with the substrate strand that is cleaved, the post-nick binding region which in our complexes binds the single-stranded gap, the active site region containing a metal center at which DNA cleavage takes place, and a C-terminal segment (Figure 1D). The DNA substrate is sharply bent at the dsDNA-ssDNA junction (Figure 1C) by a protrusion of the protein (α2, α3), the hydrophobic wedge, also observed in FEN-1 family members (Chapados et al., 2004), accounting for the specificity of this enzyme family for nicked or gapped DNA. The regions that are preserved and differ in the enzyme family are shown in a structure-based sequence alignment (Figure 2A) of FEN-1 family members with hExo1 and a structural alignment (Figure 2B) of Archaeoglobus fulgidus FEN-1 with hExo1. Particularly noteworthy are the divergences in the post-nick binding region which accommodates 3′ nick termini or 3′ flaps, and the portion of the C-terminal domain of hExo1.
Figure 1. Structure of the hExo1 catalytic domain in complex with a 5′-recessed end substrate.
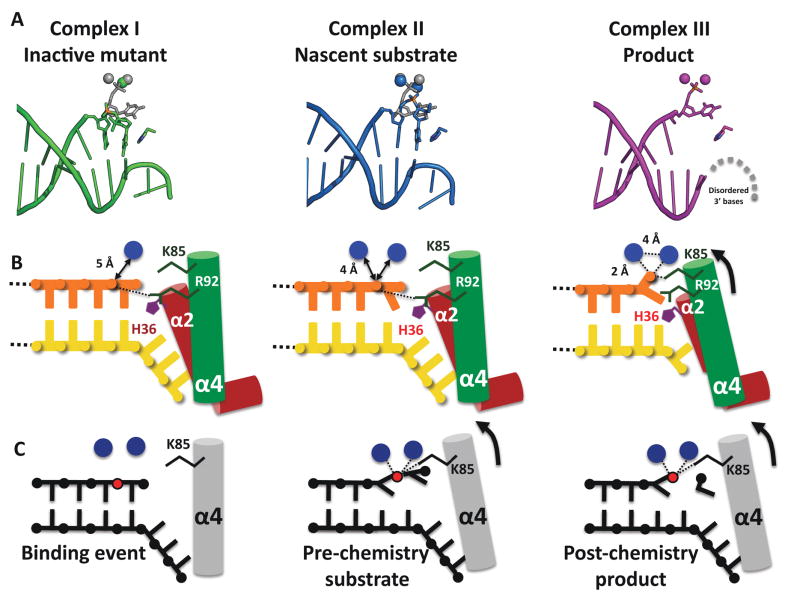
(A) Structure of the hExo1 catalytic domain bound to a gapped DNA substrate mimic. Conserved DNA binding motifs are indicated: α4-α5 micro-domain (“helical arch”), green; α2-α3 helix-loop-helix, red; K+ ion-binding site (blue) in helix-two-turn-helix (H2TH), purple. Two active-site metal ions are shown (red).
(B) Topology map of secondary structure elements in the hExo1 catalytic domain.
(C) Simulated-annealing omit electron density map calculated to 2.5 Å resolution contoured at 1σ around DNA of the hExo1 D173A complex. Note the ~90° bend of the 3′ complementary DNA strand. DNA binding motifs are colored as in A.
(D) Key functional features of the of hExo1-DNA complex. DNA binding motifs are colored as in B. C-terminal region is blue; hydrophobic wedge region of α2-α3 is shown in magenta; β6-β7 hairpin is teal. hExo1 DNA substrate is orange and yellow; modeled post-nick DNA from A. fulgidus FEN-1 structure (PDB 1RXW) is blue. A gap between the pre- and post- nick region is highlighted with a blue triangle. Active site residues are shown as sticks. Mn2+ (magenta) and K+ ions (blue) are highlighted. See also Figure S1.
Figure 2. Structure and sequence alignments of hExo1 with other human 5′ structure-specific nuclease superfamily members.
(A) Structure-based sequence alignment of human 5′ structure-specific nuclease family members. Residues with assigned secondary structure are colored as in Figure 1. 310-helices are shown with diagonal lines. Note that the sequence of the large spacer region (insert) in hXPG is not shown. hExo1 residues mutated in this study are underlined and highlighted with purple arrows. Functionally important residues are indicated: conserved acidic residues in the active site, blue stars; K+-chelating residues, purple stars; interactions with complementary (non-substrate) DNA strand, black triangles; interactions with DNA substrate strand, red triangles; van der Waals contacts with DNA, grey circles. (B) Comparison of hExo1 with FEN-1. Alignment of structures of hExo1 (teal; motifs colored as in 2A) and A. fulgidus FEN-1 (grey; PDB ID 1RXW) Structures reveal significant divergence in the C-terminal region (boxed), and the 3′ flap binding motif of FEN-1 (magenta).
DNA bending at nicks or gaps
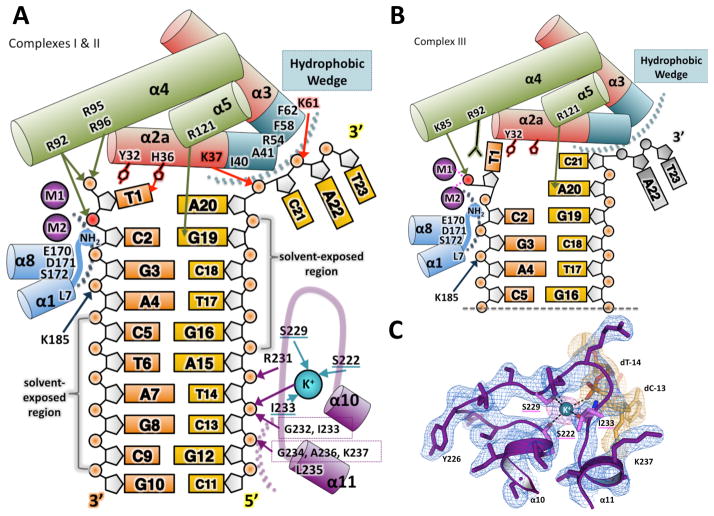
Helices α2 and α3 are located at the boundary between the pre- and post-nick regions of the DNA (Figure 1). In this region the DNA is sharply bent (~90°), and binding of intact duplex DNA is blocked by a number of hydrophobic residues that create a “wedge” motif. We observe hydrophobic interactions between residues I40, A41 (α2), F58 and F62 (α3) and the C21 and A22 bases at the 3′ terminus (Figure 3). A comparable interaction has been observed in A. fugidus FEN-1 DNA complex which has duplex instead of single-stranded DNA bound to the post-nick binding region (Chapados et al., 2004).
Figure 3. Interactions between the hExo1 protein and DNA substrate.
(A) Schematic of interactions between hExo1 and DNA in the complex I and II (nascent substrate) structures, colored as in Figure 1A. The hydrophobic wedge is at the ends of α2 and α3 (teal). The scissile phosphodiester bond is red. Hydrogen bonds between DNA substrate and protein side chains are indicated with arrows: main-chain hydrogen bonding residues are boxed. Residues coordinating K+ are underlined in teal; van der Waals contacts are indicated by dotted arcs.
(B) Schematic of interactions between hExo1 and DNA in product structure (complex III). Two active site metals and K85 coordinate the terminal phosphate. The terminal base is flipped. Disordered nucleotides A22 and T23 on the complementary strand are modeled in grey.
(C) The K+-binding site in the H2TH domain. The composite omit 2Fo-Fc electron density for the protein (blue) and the DNA density (orange) is contoured at 1σ. An anomalous difference Fourier map (magenta) of the Ba2+-substituted complex is superimposed (5σ contour). See also Figure S2.
The pre-nick DNA-binding region
The pre-nick DNA-binding region is formed by a shallow surface groove that contains discrete patches of positive charge (Figure S1). The DNA and protein make strikingly few contacts in this pre-nick region (Figure 3). The cleaved DNA strand (“substrate”) makes interactions only in the vicinity of the active site, and is completely solvated at 4th base from the 5′ terminus and beyond. The complementary, uncleaved strand forms contacts with a helix-two-turn-helix (H2TH) motif (α10-α11), a modified version of the canonical helix-hairpin-helix nucleotide binding motif also observed in FEN-1 structures (Ceska et al., 1996; Chapados et al., 2004; Feng et al., 2004; Hosfield et al., 1998; Tomlinson et al., 2010). This H2TH domain forms hydrogen bonds with the phosphate oxygens of nucleotides C13 and T14 via the main-chain amides of residues 232–237, located in the turn region and at the beginning of α11. The T14 phosphate is also coordinated by a potassium ion bound in the turn region of the H2TH motif. The K+-binding site (Figure 3A, 3C) is formed by interactions with two main-chain carbonyl groups (S222, I233), the hydroxyl of S229, the DNA (T14 phosphate oxygen), and solvent (two water molecules). This K+ coordination has not been observed in other 5′ nuclease structures, determined in the absence of DNA. It may therefore assemble only in the presence of DNA, suggesting interactions that can be readily formed and re-formed upon sliding in a processive processing mode of the enzyme. This site is almost identical to one observed elsewhere in DNA Polymerase β (Figure S2) (Pelletier et al., 1994). In Pol β, it has been suggested that this site facilitates movement of the protein along the DNA substrate backbone (Pelletier et al., 1996).
The post-nick DNA-binding region
In the post-nick DNA-binding region the ssDNA segment of the complementary (non-substrate) strand is contacted by a helix-loop helix (HLH) motif (α2-α3) observed also in FEN-1. However, a second HLH motif (HLH2) present in a number of FEN-1 structures is absent in hExo1 (Figure 2B).
In the gap-mimic substrate presented here, the uncleaved DNA strand is present, but its partner DNA strand 3′ to a nick is absent. Nevertheless, it is clear that several elements in the structure could interact with the latter, including a hairpin loop between strands β6 and β7 (Figure 1D). The 3′ end of the missing strand is predicted to interact with the α2-α3 HLH region. In hExo1 the α2 helix makes a ~90° bend at residue A41 not seen in FEN-1. In A. fulgidus FEN-1 the HLH2 region, which is absent in hExo1, interacts with phosphate groups of the 3′ flap in a double-flap substrate (Chapados et al., 2004). It has not yet been determined whether such a double-flap substrate can be processed by hExo1. We postulate that these differences in interactions in the vicinity of the 3′ nicked DNA strand play key roles in DNA substrate selection by preventing, accommodating, or stabilizing binding of 3′ flaps.
The active site
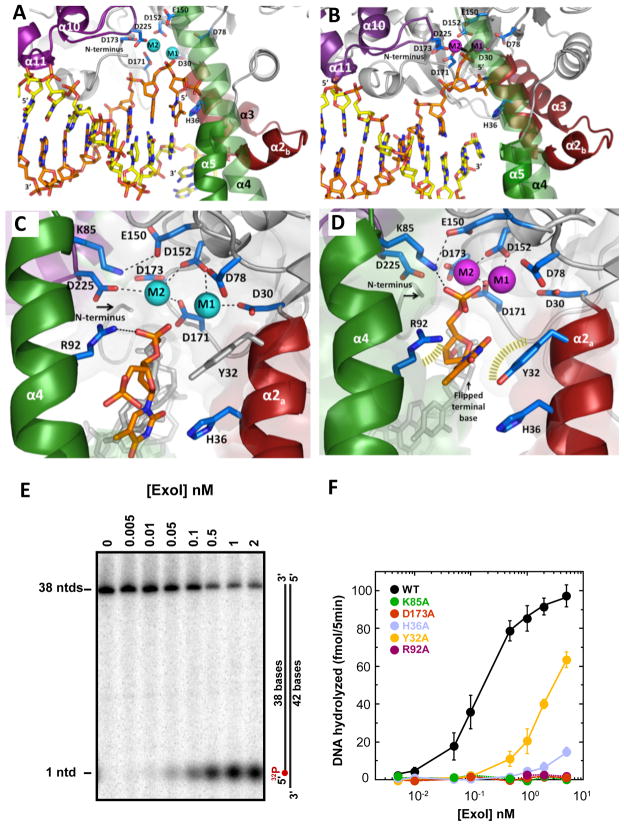
Our analysis of the active site based on three structures of enzyme-DNA complexes: an inactive mutant (D173A) with Ca2+ (Complex I), and two wild-type enzyme structures with Ba2+ (Complex II) or Mn2+ (Complex III) (see Figure S5 for metal dependence and enzyme activity). The active site is located at the boundary between the pre- and post-nick DNA-binding regions (Figure 1D). The FEN-1 enzyme family members are metalloenzymes, involving at least two Mg2+ cations in the active site (Feng et al., 2004; Hwang et al., 1998; Mueser et al., 1996; Sakurai et al., 2005). Although one of the two metals in the active site is absent in the mutant Ca2+ complex (Figure S3), the DNA is bound similarly to the other structures, showing that the enzyme binds at nicks in the absence of catalytic activity or a complete metal center.
The metal positions in both the Ba2+ (complex II), and the Mn2+ (complex III) complexes are well defined by anomalous scattering (Figure S4). The metals are coordinated by five acidic residues, which interact with site M1 (D30), or site M2 (D171, D173, D225), or both (D152). Alanine mutations in D78, D173 and D225 abolish activity, and D78A or D225A mutants alter substrate affinity (Lee et al., 2002) (Figure 4E–F). This loss of function has been observed also for all the conserved active site acidic residues of human FEN-1 (Shen et al., 1996, 1997).
Figure 4. Interactions in the hExo1 active site.
(A) Nascent substrate structure (Complex II) with two Ba2+ ions (blue) coordinated by conserved carboxylates. Substrate-binding structural elements, metal ions, and N-terminus are indicated. Structural elements are colored according to scheme in Fig 1.
(B) Product structure (Complex III) with two Mn2+ ions (magenta) bound at the active site.
(C) Close-up of complex II active site (rotated 90° from A). Two Ba2+ ions (cyan) are coordinated by conserved carboxylate residues; the scissile phosphate is coordinated by R92. Given the limits of resolution of these structures, interactions (dashed lines) are approximate.
(D) Close-up of complex III active site (rotated 90° from B. Two Mn2+ ions (magenta) are coordinated by conserved acidic residues and phosphate; the phosphate also interacts with K85. Approximate interactions within 2.8 Å are shown by dashed lines.
(E) Activity of wild-type hExo1 catalytic domain determined by titration (concentrations indicated) against a fixed concentration (25nM) of a 5′-recessed substrate (37 °C for 5 minutes), followed by separation using denaturing polyacrylamide gel electrophoresis and visualized by autoradiography (See Supplementary Material).
(F) Activity of alanine mutants. Amount of DNA hydrolyzed was plotted as a function of protein concentration for Exo1 catalytic domain (WT) (black), K85A (green), D173A (red), H36A (light blue), R92A (magenta), and Y32A (orange). Error bars represent standard deviation from at least three replicates of the experiment. We cannot rule out contribution to residual activity by a contaminating E. coli nuclease that could have copurified with the protein, but consider this unlikely because all mutants were expressed at similar level and displayed nearly identical chromatographic properties. See also Figures S3–S6 and Movie S1.
The hydrolytic center is postulated to be similar to the classic two-metal mechanism originally described for the Klenow exonuclease activity (Beese and Steitz, 1991; Steitz and Steitz, 1993)), although this has not yet been established definitively (Syson et al., 2008). If so, the scissile bond is expected to be located in-between the two metal centers. In the Ba2+ complex II, the scissile bond is close to but not in direct contact with the metal center (Figure 4A, 4C, S3). We postulate that this complex corresponds to a “nascent substrate” in which an increase in ionic radius of Ba2+ relative to Mg2+ and slight distortions due to the phosphorothioate backbone prevent the phosphate oxygen from being placed in between the two metals. In Klenow fragment, a similar situation has been interpreted as the consequence of differences between phosphodiester and phosphorothioate coordination geometries (Brautigam and Steitz, 1998; Brautigam et al., 1999). In the Mn2+ complex III, the 5′ phosphate of the nicked strand interacts directly with the two metals (Figure 4B, 4D, S4). This complex is therefore likely to correspond to a product conformation.
Mutagenesis shows that, in addition to the metal centers, a number of other interactions are important for catalysis. R92 on α4 interacts with the scissile bond in complexes I and II (Figure 4C, S3). The R92A mutant results in significant loss of activity (Figure 4E–F). A similar result was observed for FEN-1 from Pyrococcus furiosis and P. horikoshii (Allawi et al., 2003; Matsui et al., 2002). K85 on α4 interacts with the terminal phosphate in the product, but not the nascent substrate complex (Figure 4C, 4D). In this study, we find that K85A also results in a dramatic loss of activity (Figure 4F). A similar observation has been reported for the corresponding FEN-1 mutant (Sengerova et al., 2010).
Duplex fraying at the 5′ nick terminus
In complex II, the terminal base no longer forms a canonical base-pair with its partner on the template strand seen in complex I. The terminal phosphate points towards R95 and R96 on the α4 helix (Figure S3). In the product complex (complex III), the terminal base has flipped out of the duplex. This conformation is stabilized by van der Waals interactions with R92 on α4, and a π-stacking interaction with Y32 on α2 (Figure 4D). The displaced base observed in Complex III corresponds to the penultimate base in a pre-chemistry substrate which has the scissile bond positioned adjacent to the metal center (Figure 5). Accordingly, these observations suggest that the last two bases in the exonucleolytic substrate are displaced out of their duplex interactions (“frayed”). This fraying has two consequences: it positions the scissile bond in the vicinity of the metal center; it unifies the mechanisms of exo- and endo-nucleolytic cleavage of nicks and flaps respectively (see Movie S1).
Figure 5. Assignment of structures to steps in the hExo1 reaction cycle.
(A) Comparison of active site in complex I (mutant), complex II (Ba2+ “nascent substrate”), and complex III (Mn2+ “product”) showing positions of DNA, metals, and H36.
(B) Schematic representation of some mechanistically relevant interactions. Complex I contains one Ca+ ion at some distance away from the phosphate in the scissile bond. Complex II contains two Ba2+ ions in the active site and exhibits 5′ basepair fraying. In both pre-chemistry structures the scissile 5′ phosphate interacts with R92. We propose that in a catalytically active metal complex, the scissile phosphate is bound by K85.
(C) Schematic representation of proposed hExo1 reaction cycle. After binding of the DNA and assembly of the metal sites (left), the scissile bond moves into the active site via phosphate coordination with two metal ions as well as K85 (center). Both the base poised for cleavage and the penultimate base are unpaired (frayed) from the duplex into the active site. Post-chemistry the 5′ phosphate of the product strand is coordinated by two metal ions (right).
In all three structures, H36 interacts to a greater or lesser extent with the terminal base, and may therefore play a critical part in the fraying process. The H36A mutant reduces activity 150-fold, whereas the Y32A mutant results in a 20-fold drop in activity (Figure 4F), indicating that both residues participate, with H36 fulfilling a central role.
A mobile α4-α5 microdomain
As in FEN-1, a two-helix structure α4-α5 (also known as the helical arch, helical clamp, or I domain) forms one side of the active site (Ceska et al., 1996; Chapados et al., 2004; Devos et al., 2007; Hosfield et al., 1998; Mueser et al., 1996; Sakurai et al., 2005). These helices contain a large number of positively charged residues: in hExo1, α4 and α5 contain eleven positively charged residues (Figure S1). This segment binds the terminal 5′ phosphate of the substrate, and provides residues that contribute to the fraying of the two end bases of the duplex.
The mobility and ordering of the α4 and α5 helices in the 5′ nucleases varies greatly; they are disordered in a number of homolog structures, and a disorder-to-order transition has been invoked upon flap substrate binding in FEN-1 (Devos et al., 2007; Dore et al., 2006; Hosfield et al., 1998; Hwang et al., 1998; Matsui et al., 2002; Sakurai et al., 2005). In the hExo1 structures presented here α4, α5, and part of β3 form a mobile microdomain that adopts different conformations. Alignment of the three structures by their β-sheet core reveals movements >3 Å between equivalent Cα atoms of the mutant and product complexes (Figure S6).
The C-terminal region
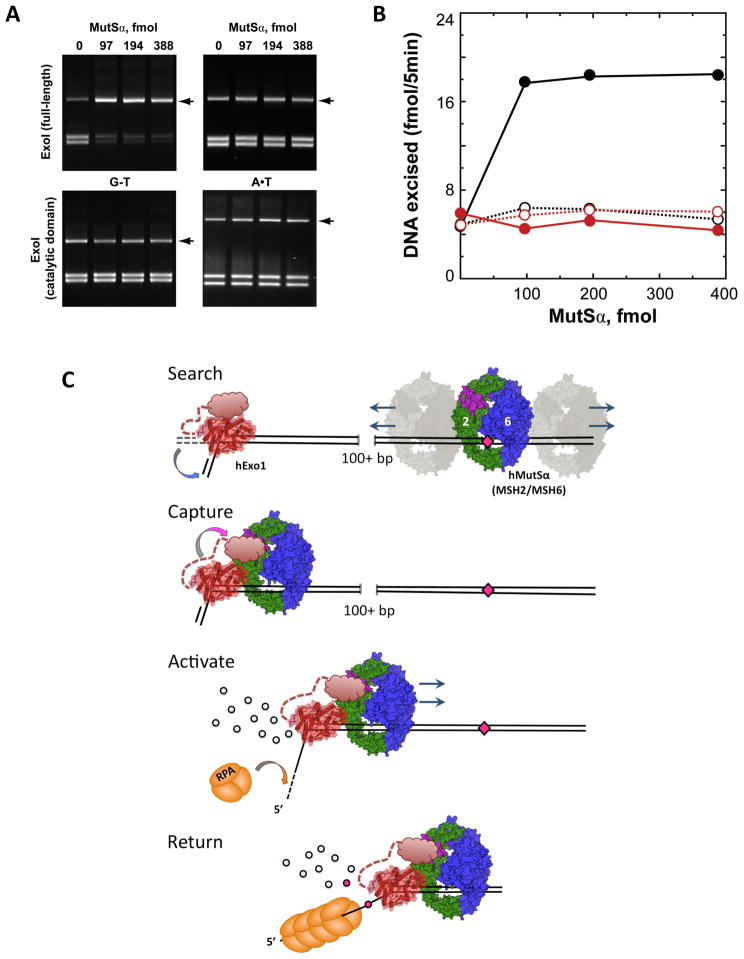
The C-terminal region (residues 285–345) (Figure 1B, 1D) consists of strands β8 and β9, helices α14 and α15, and loop regions that are stabilized by a network of H-bonds and van der Waals interactions. This region adopts a structure distinct in sequence, fold, and location (on the opposite face of the molecule) from the corresponding region of FEN1 (Figure 2B). The C-terminal region interacts with the active site through interactions at the base of α4 (L82, P83, S84) and α9 (E150, Y149, Y157, E154). In particular, E150 on α9 links C-terminal Q285 with Lys 85 on α4, which participates in catalysis (Figure S7). The α4 and α9 residues are conserved among Exo1 subclass of 5′ nucleases, but diverge from FEN-1 subclass nucleases (Figure 2A). We postulate that these interactions couple catalysis to binding events that modulate the conformation of the entire C-terminal domain (which is absent in this structure) through binding interactions with other proteins such as the MMR protein MutSα (Genschel and Modrich, 2003; Schmutte et al., 1998; Schmutte et al., 2001). In a reconstituted reaction MutSα stimulates mismatch excision by full-length hExoI, but not the C-terminal deletion, consistent with the proposed function of the C-terminal domain (Figure 6A, B).
Figure 6. Model of 5′ exonuclease activity in human DNA mismatch repair.
(A) MutSα-stimulated mismatch provoked excision. Activation of hExoI by MutSα was scored as a function of MutSα concentration in the presence of 280 fmol MutLα, 900 fmol replication protein A (RPA), and 27 fmol of either full length hExoI (Top) or hExoI catalytic domain (bottom) with heteroduplex (left) or homoduplex (right) DNA substrates. After incubation for 5 min at 37 °C, reaction products were digested with NheI and ClaI, and separated by electrophoresis on a 1% agarose gel (See Supplementary Material). Excised molecules are resistant to cleavage by NheI, and are indicated by arrows. (B) Extents of excision on G–T (closed circles) and A–T (open circles) substrates are plotted for full-length hExoI (black) and hExoI catalytic domain (red). Quantitation is based on a single representative experiment, depicted in (A). (C) A model for the interaction between MutSα and hExoI. MutSα is bound to a mismatch (pink diamond); hExo1 (red) is pre-bound at a nick and bends the DNA. The hExo1 C-terminal domain is depicted as an unstructured ensemble connected by a flexible linker. MutSα is shown in green (MSH2) and blue (MSH6) with its putative hExo1 binding region (Schmutte et al., 2001) in purple. Following mismatch recognition, first MutSα searches for hExo1 via bidirectional, one-dimensional diffusion along the DNA (search). Next, MutSα encounters hExo1 pre-bound at a nick and interacts with the C-terminal domain (capture) thereby alleviating the autoinhibition of the Exo1 (activate). Excised nucleotides are shown as black circles. Finally, the entire complex travels back towards the site of the mismatch, driven by the unidirectional 5′ to 3′ exonucleolytic cleavage of DNA (return). RPA (orange) binds to single-stranded regions of DNA uncovered by hExo1 activity. See also Figure S7.
Discussion
Here we report the structure of human Exonuclease 1 (hExo1) in complex with a DNA substrate that represents a gapped substrate, with the 5′ end of the nicked strand placed within the active site where exonucleolytic cleavage occurs. These observations enable us to propose a model for the interactions between hExo1 and MutSα in mismatch repair, and to propose a unified mechanism for the endonucleolytic and exonucleolytic cleavage activities and substrate recognition for this protein family.
Control of catalytic activity by conformational coupling
In the hExo1 structures presented here α4, α5, and part of β3 form a mobile microdomain that adopts different conformations (Figure S6) and contributes to substrate recognition and cleavage (Figure 4). In other FEN family member structures, determined in the absence of a DNA substrate, this region is disordered (Devos et al., 2007; Dore et al., 2006; Hwang et al., 1998; Matsui et al., 2002; Sakurai et al., 2005). Binding interactions that stabilize one conformation over another, or cause order/disorder transitions, clearly have the potential to exercise considerable control over nuclease activity.
Parts of the α4/α5 helices are exposed and could form binding sites with protein partners, extended parts of DNA substrates. Other parts pack against the α2/α3 helices, which also form an extensive exposed surface that potentially can interact with partners. Binding to either of these helical regions is likely to affect the mobility and conformation of the α4 helix. The C-terminal domain also could interact with α4/α5 microdomain, mediated through contacts with the C-terminal region present in the structure determined here (Figure S7). MutSα and PCNA interact with this domain (Liberti et al., 2010; Schmutte et al., 1998; Schmutte et al., 2001); similar interactions with other proteins could couple hExo1 activity to other pathways such as double-stranded break repair (Doherty et al., 2005; Gravel et al., 2008; Nimonkar et al., 2008; Sharma et al., 2003).
A model for the interaction between hExo1 and MutSα in mismatch repair
The interaction between hExo1 and MutSα is critical for DNA mismatch repair (Amin et al., 2001; Genschel et al., 2002; Nielsen et al., 2004; Schmutte et al., 2001; Wei et al., 2003). It has been well established that repair requires a nick at which hExo1 is recruited by MutSα, initially bound at a mismatch as far as 1000 base pairs away (Fang and Modrich, 1993; Genschel et al., 2002; Genschel and Modrich, 2003). Furthermore, the C-terminal (but not N-terminal) hExo1 domain interacts with MSH2 (Schmutte et al., 1998; Schmutte et al., 2001). Based on the structural information of the hExo1 DNA complexes and our observation that deletion of the C-terminal hExo1 domain abolishes MutSα-provoked excision, we propose a model for this recruitment process (Figure 6C).
In the absence of stimulatory factors, the full-length hExo1 is not very active, but still binds to nicks or gaps (Genschel et al., 2002; Genschel and Modrich, 2003; Lee and Wilson, 1999). We postulate that the C-terminal fragment which is partially absent in our structure functions as an auto-inhibitory domain by modulating the conformation of the α4/α5 microdomain. Upon binding to a mismatch, MutSα clamps to the dsDNA duplex, and carries out a search via bidirectional one-dimensional diffusion along the DNA (Gradia et al., 1997; Gradia et al., 1999). Upon encountering the pre-bound hExo1 at a nick, MutSα captures the C-terminal domain, relinquishing its auto-inhibitory interactions with the catalytic domain, and activating exonucleolytic activity by enabling the α4/α5 microdomain to adopt a conformation that positions the scissile bond over the metal center (see above). The activated hExo1-MutSα complex now returns to the mismatch site, driven by the unidirectional 5′-3′ hExo1 exonuclease activity.
A model for 5′ flap interactions
Although hExo1 is predominantly exonucleolytic, it also can process 5′ flaps endonucleolytically. We do not observe a 5′ flap directly, but our structure indicates that such a single-stranded segment needs to be guided out of the crowded active site. Conflicting models have been proposed for the recognition and processing of 5′ flap structures invoking either “tracking” of the nuclease along the single-stranded flap or “threading” of the flap through an aperture in the protein (Barnes et al., 1996; Ceska et al., 1996; Dervan et al., 2002; Gloor et al., 2010a, b). Our structures indicate that the protein must bind first at nicks, ruling out the tracking model, consistent with other observations (Gloor et al., 2010b; Hohl et al., 2007).
The threading model posits that the 5′ flap passes through the covalently closed arch formed by α4, α5 and their connecting loop. Although a model for such a path can be constructed by linear extension of the DNA in the currently observed complexes (Figure 7A, path 3), such a model raises a number of questions concerning the energy source for this process (how the single-stranded segment is pulled through the aperture in the absence of a driving force such as ATP hydrolysis), accommodation of the flap prior to threading (as the enzyme binds to the nick prior to threading a flap), and potential divergence in the evolution of substrate recognition and processing mechanisms in the superfamily (XPG and GEN1 cut bubbles and HJs, respectively, which do not have free ends and cannot thread). A non-threading path involving binding to a surface groove or cleft (which may subsequently close through a conformational change) could provide a mechanistically simpler alternative. A path can be constructed by extending a single-stranded 5′ end of the substrate strand to follow approximately the natural curvature of the DNA backbone set up in the double-stranded segment in the 5′-binding region, allowing the DNA to exit the active site by passing in front of α4 (Figure 7A, path 1). We propose that slight re-arrangements in the highly mobile α4 helix could position the scissile bond of a flap over the metal center. In path 2 the mobility of the α4/α5 microdomain opens a cleft between it and the α2 region, thereby providing flap exit path (Figure 7A, path 2). Neither proposed path for 5′ flaps invokes threading through the protein; instead, flap interactions take on the characteristic of a conventional binding event, consistent with recent biochemical experiments (Gloor et al., 2010a), and enabling development of a unified mechanism for the superfamily (see below).
Figure 7. A unified model for substrate recognition in the 5′ structure-specific nuclease family.
(A) Models of interactions between hExo1 and a 5′-flap DNA substrate. The observed 5′ recessed (pre-nick) DNA complex is shown in yellow (complementary strand) and orange (substrate strand). Two alternate paths (orange arrows) for accommodation of a 5′ flap in hExo1 are shown; path 1 (orange arrow) shows a path that extends the 5′ DNA flap in the nascent substrate complex in front of mobile helix α4; path 2 (orange arrow) passes through a cleft between the α4/α5 microdomain and the α2 helix, which could open up if the microdomain moves. A third path (blue arrow) threads the DNA through the protein, below the arch formed by the loop connecting α4 and α5, which cannot accommodate bubbles or HJs in a unified model.
(B) Schematic representation of the unified substrate-binding model. Selected structural elements that define alternate possible paths 1 or 2 are colored.
A unified model for substrate discrimination and nuclease activities in the FEN family
FEN family members recognize or distinguish between five DNA structures: simple nicks, gaps and 3′-overhangs (hExo1, FEN), 5′ flaps (FEN, hExo1, GEN1), 5′ and 3′ double flaps (FEN), bubbles (XPG), and HJs (GEN). Bubbles and HJs can be thought of as logical extensions of the combined 5′, 3′ double flap substrates: a bubble is covalently connected to a 5′, 3′ double flap; a HJ is a fused double flap (Figure 7B). The similarities in the hydrolytic mechanism and diversities in substrate recognition are reflected in the patterns of structure (Figure 1) and sequence alignments (Figure 2).
All FEN family members recognize nicks or gaps. Consequently, the hydrophobic wedge which induces a sharp bend in the DNA at such sites is structurally conserved, although its sequence diverges. The nucleolytic mechanism is similarly conserved among the family members, as indicated by the presence of the acidic patch for binding metals in the catalytic site. This conservation extends to residues involved in fraying at the 5′ ends of nicks, which unifies the exo- and endo-nucleolytic activities.
All the DNA substrates possess a pre-nick duplex DNA region, the binding region for this element is also highly conserved. The K+-binding site is postulated to facilitate sliding of DNA in a processive mode (but not in XPG). With exception of hGEN1, these interactions are conserved across all FEN family members. GEN1 processes HJs, and may not depend on processivity.
The diversity of 5′ and 3′ substrate structures presents a geometrical puzzle for a common mechanism of DNA processing. It is impossible to thread all these varied substrates through a conserved, covalently closed aperture located within the α4-α5 helical arch. In particular, bubbles or HJs cannot be threaded without introducing a covalent break in either the DNA or the protein. However, it may be possible to pass a 5′ extension out of the active site without needing to invoke threading through the protein (see above), providing a unified model of structured DNA substrate processing: 5′, 3′ ends are all recognized by grooved surface features (Figure 7B).
The α4/α5 microdomain is highly divergent among the family members and encodes recognition of the 5′ flap or its equivalent in bubbles and HJs. Similarly, 3′ DNA substrate features are encoded by the interactions with highly divergent α2/α3 region and the post-nick segment. If the 3′ end of the nicked strand is bound in this region, then 3′ flaps could be limited in length (or not be able to bind). Conversely, surface grooves or clefts could guide flaps, bubbles, or HJs. hExo1 does not appear to have such a groove (consistent with its function), but an insertion in the α2 and α3 helices could form these. Such insertions are indicated for XPG and GEN in the structure-based sequence alignment.
For those members of the enzyme family that are regulated by interactions with protein partners, we propose that the mobile α4/α5 microdomain plays an important role in such control mechanisms either through direct interactions, or indirectly though the C-terminal domain. The concerted motions of the α4 and α5 helices together with part of β3 appear to terminate at the highly conserved G79, which may act as a part of a hinge.
The DNA substrate complexes of hExo1 presented here begin to clarify a number of long-standing questions in the mechanism of the FEN nucleases. All family members induce a sharp bend in the substrate for nick or gap recognition; fraying at the 5′ end of the duplex removes apparent differences between nick and flap processing and position the scissile phosphate; surface binding sites encode recognition of 5′ and 3′ DNA substrate structures through conventional binding interactions. Within this general framework, recognition of simple nicked (or gapped) ends, flaps, and covalently connected flaps (bubbles) or HJs can be accommodated. Accommodation of additional structures in flap regions, such as double-stranded segments, or telomeric G4 DNA (Vallur and Maizels, 2010a, b) will depend both on the distance from the nick or gap junction and potential interactions with other parts of the particular enzyme such as the highly divergent C-terminal domain. The elegant simplicity by which Nature has solved complex topological puzzles in substrate recognition and processing is remarkable.
Experimental Procedures
Cloning, expression, purification, and activity determination of hExo1 catalytic domain constructs
Wild-type, mutant, and selenomethionine-labelled Exo1 N-terminal domains (residues 1–352) were cloned, expressed and purified as described in Supplementary Information. Exonuclease assays were carried in 20 mM Tris-HCl (pH 7.6), 0.75 mM HEPES-KOH, 120 mM KCl, 5mM MgCl2, 250 μg/mL bovine serine albumin, 1.5 mM ATP, 1mM glutathione, 60 μM dithiothreitol, and 1% glycerol. Activities of wild-type and mutant Exo constructs were compared by incubating 25 nM radiolabelled 5′-recessed DNA substrate with 0.005–5 nM protein in 5 μL for 5 min at 37 °C. Reaction products were analyzed by denaturing polyacrylamide gel electrophoresis (Figure 4E).
Crystallization, data collection, and structure determination
Wild-type and mutant proteins were co-crystallized (see Supplementary Information) with a 5′-recessed-end substrate (top strand: 5′-(p)-TCGACTAGCG; bottom strand: 5′-CGCTAGTCGACAC) in the presence of Ca2+; Ba2+ and Mn2+ were introduced by exchange in crystal soaks. Diffraction data were collected at The Advanced Photon Source (APS) at Argonne National Laboratory, beamlines 22-ID and 22-BM (SER-CAT), and The Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory, beamline 12.3.1 (SIBYLS); experiments were conducted at 100 K. Native diffraction data were collected to 2.5 Å resolution and selenomethionine data were collected to 3.3 and 3.4 Å resolution at λ=0.9794 Å at the selenium K edge. Data were scaled in space group P21212 using HKL2000 (Otwinowski, 1998). The structure of the hExo1 catalytic domain (D173A) DNA complex was determined by selenium-SIRAS experimental phasing (Hendrickson et al., 1990). Two Se peak data sets and a native data set were used to determine experimental phases with SHARP (Bricogne et al., 2003). The model contained two molecules in the asymmetric unit, and five selenium sites per monomer were observed. The model was built manually in COOT (Emsley and Cowtan, 2004). Initial solvent-flattened maps lacked side chain density and connectivity and were improved using partial model phase combination and B-factor sharpening in CNS (Brünger et al., 1998). PHENIX Autobuild (Adams et al., 2002) was used to improve initial model and electron density. The structure was refined using CCP4 (Winn et al., 2003) and CNS with a maximum likelihood target and phase probability distribution.
Wild-type crystals diffracted to 3.1-Å resolution and were scaled in the P43212 space group using HKL2000. Wild-type barium derivative data were measured at λ=1.2Å, and manganese derivative data were collected at λ=1.0 Å or 1.25 Å. Molecular replacement phases were calculated using PHASER (Storoni et al., 2004) using the D173A mutant structure as a model. Initial low-resolution refinement was carried out in CNS using deformable elastic network (DEN) restraints (Schroder et al., 2007). The hExo1 D173A structure was input as a reference model. DNA was built manually in COOT using electron-density maps from this refinement. Metal ion positions were identified using phased anomalous difference Fourier maps. Structures were refined using PHENIX and CNS with a maximum likelihood target and phase probability distribution. Final model coordinates were checked with MOLPROBITY (Davis et al., 2004).
Mismatch-provoked excision assays
Mismatch-provoked excision reactions were carried out in the buffer described in Experimental Procedures, and contained 24 fmol of a 6440-bp circular G-T heteroduplex (or control A-T homoduplex) with a strand break located 128 bp 5′ to the mismatch. Reactions (20-μl) were assembled on ice by addition of 1 μl each of MutSα, MutLα, and RPA, diluted as described (Genschel and Modrich, 2003), to 16 μl of a solution containing all other components except ExoI. Reactions were initiated by addition of 1 μl of ExoI directly to the above solution on ice, and immediately transferred to a 37 °C water bath and incubated for 5 minutes. Samples were deproteinized by Proteinase K treatment followed by phenol extraction. Extent of excision was scored by NheI resistance assay as described (Genschel et al., 2002 JBC, 2002).
Supplementary Material
Acknowledgments
We thank Anita Changela, Shannon Kelly, and Miaw-Sheue Tsai for assistance with cloning and purification. This work was supported by National Institutes of Health grants R01 GM091487 to LSB, R01 GM45190 to PM, and P01 CA092584 to LSB and PM. PM is an Investigator of the Howard Hughes Medical Institute. Use of the Advanced Photon Source was supported in part by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Use of the SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory, was supported in part by the DOE program Integrated Diffraction Analysis Technologies (IDAT) and the DOE program Molecular Assemblies Genes and Genomics Integrated Efficiently (MAGGIE) under Contract Number DE-AC02-05CH11231 with the U.S. Department of Energy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Allawi HT, Kaiser MW, Onufriev AV, Ma WP, Brogaard AE, Case DA, Neri BP, Lyamichev VI. Modeling of flap endonuclease interactions with DNA substrate. J Mol Biol. 2003;328:537–554. doi: 10.1016/s0022-2836(03)00351-6. [DOI] [PubMed] [Google Scholar]
- Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CJ, Wahl AF, Shen B, Park MS, Bambara RA. Mechanism of tracking and cleavage of adduct-damaged DNA substrates by the mammalian 5′- to 3′-exonuclease/endonuclease RAD2 homologue 1 or flap endonuclease 1. J Biol Chem. 1996;271:29624–29631. doi: 10.1074/jbc.271.47.29624. [DOI] [PubMed] [Google Scholar]
- Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Steitz TA. Structural Principles for the Inhibition of the 3′-5′ Exonuclease Activity of Escherichia coli DNA Polymerase I by Phosphorothioates. Journal of Molecular Biology. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- Brautigam CA, Sun S, Piccirilli JA, Steitz TA. Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′–5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry. 1999;38:696–704. doi: 10.1021/bi981537g. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Ceska TA, Sayers JR, Stier G, Suck D. A Helical Arch Allowing Single-stranded DNA to Thread Through T5 5′-Exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan JJ, Feng M, Patel D, Grasby JA, Artymiuk PJ, Ceska TA, Sayers JR. Interactions of mutant and wild-type flap endonucleases with oligonucleotide substrates suggest an alternative model of DNA binding. Proc Natl Acad Sci U S A. 2002;99:8542–8547. doi: 10.1073/pnas.082241699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos JM, Tomanicek SJ, Jones CE, Nossal NG, Mueser TC. Crystal structure of bacteriophage T4 5′ nuclease in complex with a branched DNA reveals how flap endonuclease-1 family nucleases bind their substrates. J Biol Chem. 2007;282:31713–31724. doi: 10.1074/jbc.M703209200. [DOI] [PubMed] [Google Scholar]
- Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM., Jr RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J Biol Chem. 2005;280:28085–28094. doi: 10.1074/jbc.M500265200. [DOI] [PubMed] [Google Scholar]
- Dore AS, Kilkenny ML, Jones SA, Oliver AW, Roe SM, Bell SD, Pearl LH. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 2006;34:4515–4526. doi: 10.1093/nar/gkl623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fang W-h, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- Feng M, Patel D, Dervan JJ, Ceska T, Suck D, Haq I, Sayers JR. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat Struct Mol Biol. 2004;11:450–456. doi: 10.1038/nsmb754. [DOI] [PubMed] [Google Scholar]
- Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- Genschel J, Modrich P. Functions of MutL{alpha}, RPA, and HMGB1 in 5′-directed mismatch repair. J Biol Chem. 2009;284:21536–21544. doi: 10.1074/jbc.M109.021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor JW, Balakrishnan L, Bambara RA. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J Biol Chem. 2010a doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor JW, Balakrishnan L, Bambara RA. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J Biol Chem. 2010b;285:34922–34931. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R. The Human Mismatch Recognition Complex hMSH2-hMSH6 Functions as a Novel Molecular Switch. Cell. 1997;91 doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Dunand-Sauthier I, Staresincic L, Jaquier-Gubler P, Thorel F, Modesti M, Clarkson SG, Scharer OD. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 2007;35:3053–3063. doi: 10.1093/nar/gkm092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Hwang KY, Baek K, Kim HY, Cho Y. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat Struct Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- Kolodner RD. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- Lee B, Nguyen LH, Barsky D, Fernandes M, Wilson DM., 3rd Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 2002;30:942–949. doi: 10.1093/nar/30.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- Liberti SE, Andersen SD, Wang J, May A, Miron S, Perderiset M, Keijzers G, Nielsen FC, Charbonnier JB, Bohr VA, et al. Bi-directional routing of DNA mismatch repair protein human exonuclease 1 to replication foci and DNA double strand breaks. DNA Repair (Amst) 2010 doi: 10.1016/j.dnarep.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui E, Musti KV, Abe J, Yamasaki K, Matsui I, Harata K. Molecular structure and novel DNA binding sites located in loops of flap endonuclease-1 from Pyrococcus horikoshii. J Biol Chem. 2002;277:37840–37847. doi: 10.1074/jbc.M205235200. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–775. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueser TC, Nossal NG, Hyde CC. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 fmaily of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acd Sci USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. Denzo and Scalepackprograms: An oscillation data processing suite for macromolecular crystallography 1998 [Google Scholar]
- Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: Implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537–4542. [PubMed] [Google Scholar]
- Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- Schroder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengerova B, Tomlinson C, Atack JM, Williams R, Sayers JR, Williams NH, Grasby JA. Bronsted analysis and rate-limiting steps for the T5 flap endonuclease catalyzed hydrolysis of exonucleolytic substrates. Biochemistry. 2010;49:8085–8093. doi: 10.1021/bi100895j. [DOI] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J Biol Chem. 2003;278:23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- Shen B, Nolan JP, Sklar LA, Park MS. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J Biol Chem. 1996;271:9173–9176. doi: 10.1074/jbc.271.16.9173. [DOI] [PubMed] [Google Scholar]
- Shen B, Nolan JP, Sklar LA, Park MS. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallographica Section D-Biological Crystallography. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Syson K, Tomlinson C, Chapados BR, Sayers JR, Tainer JA, Williams NH, Grasby JA. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J Biol Chem. 2008;283:28741–28746. doi: 10.1074/jbc.M801264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson CG, Atack JM, Chapados B, Tainer JA, Grasby JA. Substrate recognition and catalysis by flap endonucleases and related enzymes. Biochem Soc Trans. 2010;38:433–437. doi: 10.1042/BST0380433. [DOI] [PubMed] [Google Scholar]
- Vallur AC, Maizels N. Complementary roles for exonuclease 1 and Flap endonuclease 1 in maintenance of triplet repeats. J Biol Chem. 2010a;285:28514–28519. doi: 10.1074/jbc.M110.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallur AC, Maizels N. Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric g4 DNA. PLoS ONE. 2010b;5:e8908. doi: 10.1371/journal.pone.0008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr, Kneitz B, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nuclease Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.