Abstract
Temozolomide (TMZ) is the preferred chemotherapeutic agent in the treatment of glioma following surgical resection and/or radiation. Resistance to TMZ is attributed to efficient repair and/or tolerance of TMZ-induced DNA lesions. The majority of the TMZ-induced DNA base adducts are repaired by the base excision repair (BER) pathway and therefore modulation of this pathway can enhance drug sensitivity. N-methylpurine DNA glycosylase (MPG) initiates BER by removing TMZ-induced N3-methyladenine and N7-methylguanine base lesions, leaving abasic sites (AP sites) in DNA for further processing by BER. Using the human glioma cell lines LN428 and T98G, we report here that potentiation of TMZ via BER inhibition [methoxyamine (MX), the PARP inhibitors PJ34 and ABT-888 or depletion (knockdown) of PARG] is greatly enhanced by over-expression of the BER initiating enzyme MPG. We also show that methoxyamine-induced potentiation of TMZ in MPG expressing glioma cells is abrogated by elevated-expression of the rate-limiting BER enzyme DNA polymerase β (Polβ), suggesting that cells proficient for BER readily repair AP sites in the presence of MX. Further, depletion of Polβ increases PARP inhibitor-induced potentiation in the MPG over-expressing glioma cells, suggesting that expression of Polβ modulates the cytotoxic effect of combining increased repair initiation and BER inhibition. This study demonstrates that MPG overexpression, together with inhibition of BER, sensitizes glioma cells to the alkylating agent TMZ in a Polβ-dependent manner, suggesting that the expression level of both MPG and Polβ might be used to predict the effectiveness of MX and PARP-mediated potentiation of TMZ in cancer treatment.
Keywords: base excision repair, methoxyamine, N-methylpurine DNA glycosylase, poly(ADP-ribose) polymerase, temozolomide
Temozolomide (TMZ) is an oral chemotherapeutic agent approved for the treatment of anaplastic astrocytoma and newly diagnosed glioblastoma.1 TMZ has also demonstrated clinical activity in metastatic melanoma and is under clinical evaluation for use in other cancers, including leukemia, lymphoma, aerodigestive tract, pancreatic, and neuroendocrine tumors, as well as cancers that have metastasized to the brain.2 TMZ causes cancer cell cytotoxicity by methylating genomic DNA, producing cytotoxic and/or mutagenic abnormal DNA bases.3,4 The major site of methylation is at the N7 position of guanine (>70%) followed by the N3 position of adenine (9.2%) and the O6 atom of guanine (5%).3 However, the ability of cancer cells to recognize and repair those DNA lesions confers chemotherapeutic resistance and limits therapeutic efficacy.4,5 The majority of TMZ-induced DNA lesions, including N7-methyl guanine and N3-methyl adenine, are repaired by the base excision repair (BER) pathway,3 while the O6-methyl adduct of guanine is directly removed by O6-methylguanine-DNA methyltransferase (MGMT).6,7 Although O6-methylguanine constitutes only a small proportion of the base lesions produced by TMZ, it is the most cytotoxic of all the lesions induced by TMZ and constitutes a significant fraction of TMZ-induced cytotoxicity.2 Since O6-methylguanine–induced cytotoxicity is mediated through the mismatch repair (MMR) pathway, sensitivity to TMZ requires both low MGMT repair activity and functional MMR.2 A significant percentage of gliomas lack expression of MGMT due to hypermethylation of the MGMT promoter, whereas at least half of glioblastoma multiforme (GBM) express MGMT, and the expression is associated with resistance to chemotherapy and poor prognosis.8,9 Loss of function of the MMR protein MSH6, due to somatic mutations, has also been shown to be associated with glioblastoma recurrence post irradiation and TMZ treatment.10 Therefore, it is important to either overcome resistance resulting from MGMT activity or find an alternative that improve the efficacy of TMZ in the presence of MGMT activity. However, MGMT inhibitors (e.g., Patrin)11 have not shown clinical efficacy.2,12 A viable option may be to target the BER pathway. Pharmacological inhibition of the BER pathway, which repairs the N7-methylguanine and N3-methyladenine lesions induced by TMZ, has been shown to enhance TMZ-induced cytotoxicity independent of MGMT status.13
The repair of TMZ-induced base damage by the BER pathway starts with the recognition and removal of the damaged bases by N-methylpurine DNA glycosylase (MPG), also known as alkyladenine DNA glycosylase (AAG).7 The abasic site (AP site) produced following the action of MPG is then hydrolyzed by AP endonuclease 1 (APE1), resulting in the incision of the damaged DNA strand and formation of a 3′OH group and 5′deoxy-ribose phosphate (5′dRP) group in the repair gap.14 Poly(ADP-ribose) polymerase 1 (PARP1) together with PARP2 and poly(ADP-ribose) glycohydrolase (PARG) recognizes the DNA strand interruption and facilitates the recruitment of subsequent BER proteins, including the BER scaffold protein XRCC1 and DNA polymerase β (Polβ).14 Polβ subsequently hydrolyzes the 5′dRP moiety and inserts a single nucleotide, preparing the strand for ligation by a complex of DNA ligase IIIα and XRCC1 to complete the repair process.15
Enhanced sensitivity to alkylating agents has been observed by modulating the BER pathway in preclinical studies, suggesting BER modulation is an attractive target for chemotherapy potentiation.16 Currently, several BER proteins are under active investigation as potential targets for chemotherapy sensitization, including APE1,17 PARP1,18 PARG,19 and Polβ.20–24 Methoxyamine (MX) is a small molecule that specifically inhibits BER25 and is currently being evaluated in phase I clinical trials (TRC102; ClinicalTrials.gov Identifier: NCT00692159). Methoxyamine inhibits the repair of AP sites by binding to and modifying the AP site, rather than directly inhibiting the enzyme APE1. AP sites modified by MX are refractory to APE1, preventing its processing by the ensuing steps of BER, and the MX-modified AP site is highly cytotoxic.26 Methoxyamine potentiates a wide range of DNA damaging agents that produce AP sites regardless of the status of MMR, MGMT, and p53.17
PARP1 (ARTD1) is the founding member of a large family of poly(ADP-ribose) polymerases.27–29 It is the primary enzyme catalyzing the transfer of ADP-ribose units from NAD+ to target proteins including PARP1 itself. Under normal physiologic conditions, PARP1 facilitates the repair of DNA base lesions by helping recruit the BER proteins XRCC1 and Polβ.30 Inhibition of PARP1 results in decreased repair of DNA base damage and increased sensitivity of cells to alkylating agents, which makes it an attractive and effective target for chemotherapy sensitization.31 Many PARP inhibitors have been developed and tested in several tumor types.32 They have been shown to enhance the cytotoxic effect of TMZ against glioma,33–35 leukemia,36 lung,37,38 and colon38–40 carcinoma cells. Further, it has been shown recently that a PARP inhibitor (ABT-888) + TMZ has broad activity in multiple histologic types in subcutaneous, orthotopic, or metastatic tumor models.41 PARG is the main enzyme responsible for the degradation of poly ADP-ribose (PAR) in vivo via endo- and exoglycosidic cleavage.28 Although complete ablation of PARG activity leads to early embryonic lethality, embryonic stem cells derived from a PARG null mouse42 and cells from PARG110 (one of three isoforms of PARG)-deficient mice43 have been shown to be sensitive to alkylating agents and ionizing radiation. In addition, inhibition of PARG activity was demonstrated to sensitize malignant melanoma to TMZ in mouse models.19
Overexpression of MPG has been reported to sensitize human breast cancer cells,24 osteosarcoma cells,44 and ovarian cancer cells45 to the chemotherapeutic agent TMZ. The increased sensitivity has been shown to be the result of increased repair initiation of the nontoxic N7-methylguanine lesion,46 saturating the rating-limiting enzyme Polβ and resulting in accumulation of cytotoxic 5′dRP repair intermediates.23 Since most BER inhibitors (e.g., AP site repair inhibition by MX or PARP and PARG inhibition) inhibit the steps following glycosylase-mediated repair initiation, we hypothesize that MPG overexpression might increase BER inhibitor-induced sensitization of glioma cells to the alkylating agent TMZ. In this study, we show that overexpression of MPG sensitizes glioma cells (LN428 and T98G) to MX, the PARP inhibitors PJ34 and ABT-888, or PARG inhibition (knockdown) following exposure to TMZ, demonstrating that increased initiation of BER combined with inhibition of the ensuing repair steps provides enhanced sensitization of glioma cells to TMZ. Further, we show that depletion of Polβ enhances the sensitization induced by the combination of increased repair initiation and BER inhibition, whereas elevated expression of Polβ abrogates the sensitization. Further, we observed wide variability in mRNA expression for MPG, Polβ, and PARP1 in GBM tumors, as compared with normal brain tissue. As our functional analyses suggest that the expression status of both MPG and Polβ might be used to predict the effectiveness of TMZ plus BER inhibitors in the treatment of glioma, we propose that future analyses include protein expression evaluation of key BER proteins and/or measurement of key BER enzyme activities from tumor biopsies to aid in treatment optimization.
Materials and Methods
Chemicals and reagents
Alpha Eagle's minimal essential medium (EMEM) was from Mediatech or InVitrogen. Fetal bovine serum (FBS), heat inactivated FBS, Pen/Strep/Ampho, glutamine, and antibiotic/antimycotic were from InVitrogen. TMZ was obtained from the National Cancer Institute Developmental Therapeutics Program. A TMZ stock solution was prepared in dimethyl sulfoxide (DMSO) at 100 mM. Puromycin, gentamicin, and neomycin were purchased from Clontech Laboratories, Irvine Scientific, and InVitrogen, respectively. PJ34 and methoxyamine hydrochloride were purchased from Calbiochem and Sigma, respectively. ABT-888 was kindly provided by Abbott Laboratories. The plasmid pSV2MGMT was kindly provided by B. Kaina.
Plasmid expression and RNAi vectors
Human WT and mutant (N169D) MPG were expressed using the plasmid pRS1422 or pIRES-neo-MPG(N169D), respectively, as described previously.22 The construction of mammalian expression plasmids of Flag-tagged human WT and mutant Polβ (K72A) was described previously.22 The shuttle vectors (control: pLKO.1-puro-turbo green fluorescent protein [GFP]; PARG small hairpin RNA [shRNA]: pCMV-tGFP-PARG) of the HIV-based lentiviral shRNA expression system were from Sigma. Lentiviruses expressing PARG-specific or control shRNA were prepared by the University of Pittsburgh Cancer Institute (UPCI) lentiviral facility. The shRNA target sequences for PARG are described in detail in Supplementary Table S1.
Cell culture and cell line development
The glioblastoma cell line LN428 (kindly provided by Ian Pollack; University of Pittsburgh, PA) was cultured in Alpha EMEM supplemented with 10% heat inactivated FBS, glutamine, antibiotic/antimycotic, and gentamicin, as we have described previously.22 LN428 is an established glioblastoma-derived cell line with mutations in p53 and deletions in p14ARF and p16 and is WT for PTEN.47,48 Additional glioma cell lines used herein are detailed in Table 1. Briefly, T98G, A-172, DBTRG-05MG, M059K, M059J, and U87MG cells we obtained from ATCC. The LN215, LN235, LN319, and LN444 cell lines were obtained from Dr S.-Y. Cheng (UPCI) and Dr E. Van Meir (Emory University). Cells were maintained at 37°C with 5% CO2. Human MPG (WT), mutant MPG (N169D), Flag-Polβ (WT), and Flag-Polβ (K72A) expressing cell lines were developed as described previously.22 Lentiviral particles were generated by cotransfection of 4 plasmids (pLKO.1-puro-TurboGFP [control, expresses GFP] or pCMV-tGFP-PARG [PARG shRNA: coexpresses PARG shRNA and GFP] together with pMD2.g[VSVG], pVSV-REV and PMDLg/pRRE) into 293-FT cells49,50 using FuGene 6 Transfection Reagent, as described previously.22 Forty-eight hours after transfection, lentivirus-containing supernatant was collected and passed through 0.45-µM filters to isolate the viral particles. Lentiviral transduction was performed as described earlier.22 Briefly, 6.0 × 104 cells were seeded into a 6-well plate 24 h before transduction. Cells were transduced for 18 h at 32°C and then cultured for 72 h at 37°C. Cells expressing GFP or both GFP and PARG-specific shRNA were isolated by fluorescence activated cell sorting, or FACS.
Table 1.
Characteristics of the glioma cell lines used in this study
| Cell Line | ATCC # | Tissue | Other details | Citation |
|---|---|---|---|---|
| LN428 | - | Brain, glioblastoma | Mutations in p53, deletions in p14ARF and p16, WT for PTEN | 22,47,48 |
| T98G | CRL-1690 | Brain, glioblastoma | Elevated expression of MGMT | 61 |
| A-172 | CRL-1620 | Brain, glioblastoma | - | 77 |
| DBTRG-05MG | CRL-2020 | Glioblastoma; brain; glial cell | - | 78 |
| M059K | CRL-2365 | Malignant; brain; glioblastoma | Proficient for DNA-PK expression | 79 |
| M059J | CRL-2366 | Malignant; brain; glioblastoma | Deficient for DNA-PK expression | 79 |
| U87MG | HTB-14 | Brain, glioblastoma, astrocytoma | PTEN null; predicted to be null for MRE11B and XPC | 80 |
| LN215 | - | Brain, glioblastoma | - | 81,82 |
| LN235 | - | Brain, glioblastoma | - | 81,82 |
| LN319 | - | Brain, glioblastoma | - | 81,82 |
| LN444 | - | Brain, glioblastoma | - | 81,82 |
LN428 cell lines engineered to overexpress MGMT (LN428/MGMT) were developed by plasmid transfection. Briefly, 1.5 × 105 cells were seeded into 60-mm dishes and incubated for 24–30 h at 5% CO2 at 37°C. The human MGMT expression plasmid [pIRES-Puro-hMGMT] was transfected using FuGene 6 Transfection Reagent (Roche) according to the manufacturer's instructions. Stable cell lines were selected in puromycin (0.5 µg/mL) for 2 weeks, individual clones (stably expressing human MGMT) were expanded, and 30 µg of nuclear extract was analyzed by immunoblot analysis for the expression of human MGMT protein.
Cell cytotoxicity assay
Short-term cell survival assay
TMZ- or TMZ + MX–induced cytotoxicity was determined by an MTS assay, a modified MTT assay, as described previously.22 Results were calculated from the average of 3 or 4 separate experiments and are reported as the percentage of treated cells relative to the cells without treatment (% control).
Long-term cell survival assay
Cells were seeded into a 6-well plate 24 h before exposure to PJ34 (4 μM), ABT-888 (10 μM) or DMSO as control.Thirty minutes later, cells were treated with TMZ alone, TMZ plus PJ34 (2 μM), or TMZ plus ABT-888 (5 μM) for 6 h. Cells were washed with PBS, trypsinized, resuspended, and counted before being re-seeded into 3 100-mm cell culture dishes at 8000 (for LN428 cell lines) or 3000 (for T98G cell lines) cells each. Cells were incubated with or without 2 μM PJ34 or 5 μM ABT-888 for 10 days, and the cells were counted. Results were calculated from 3 independent experiments and reported as percentage relative to the control treatment (% control).
Cell extract preparation and Western blot
Nuclear extracts were prepared and protein concentrations were determined as described previously.22 Twenty µg of protein was loaded on a precast 4–20% Tris-glycine gel (Invitrogen) or 25 µg of protein was loaded onto a precast 4–20% Mini-PROTEAN TGX gel (Bio-Rad). For the whole cell extracts used in probing PAR, 3 × 106 cells were seeded into a 100 mm cell culture dish 24 h before drug treatment. Cells were then treated with TMZ (1.5 mM) or DMSO (control) for different periods of time. After treatment, the cells were washed twice with cold PBS, collected, and lysed in 400 µL of 2X Laemmli buffer (2% sodium dodecyl sulfate, 20% glycerol, 62.5 mM Tris-HCl, pH 6.8, 0.01% bromophenol blue). Samples were boiled for 8 min and extracts from approximately 1.5 × 105 cells were loaded in each lane on a 4–12% pre-cast Tris-glycine gel(Invitrogen, ) for immunoblot analysis. The following primary antibodies were used in the immunoblot assays: anti-human MPG (Mab; clone 506-3D);22 anti-Polβ (Mab clone 61; Thermo Fisher Scientific); anti-APE1 (EMD Biosciences); anti-PARP1 (BD Pharmingen); anti-PCNA (Santa Cruz, Biotechnology); anti-PAR (Clone 10H, kindly provided by Dr M. Ziegler, University of Bergen, Norway); and anti-MGMT (Novus).
Isolation and analysis of total RNA from normal brain and GBM tumor tissue
Approval by an institutional review board was obtained under the University of Pittsburgh tissue banking protocol, and all subjects provided written informed consent for participation. Formalin-fixed paraffin-embedded (FFPE) tumor and normal tissue were obtained and evaluated by a board-certified pathologist (RLH) to verify that representative sections were used. All tissue samples were obtained using an honest broker and samples were de-identified. Total cellular RNA was isolated from archival FFPE tumor (a GBM) and normal brain tissue using the RecoverAll Total Nucleic Acid isolation kit (Ambion), and the final concentration was determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific). Following isolation, cDNA was synthesized from 50 ng of RNA using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (part #4375575), essentially as we have described previously.51 Briefly, cDNA was preamplified for 10 cycles using the TaqMan™ PreAmp Master Mix (part # 4391128) and diluted 1:5. The preamplified cDNA was next analyzed using validated Applied Biosystems TaqMan Gene Expression Assays (human MPG: Hs00357983-G1; human Polβ: Hs01099715-M1; and human PARP1: Hs00911369-G1) and normalized to the expression of human β-actin (part #4333762T). Expression analysis was determined using the ▵▵CT protocol as per the manufacturer to determine the relative level of expression, as compared with human β-actin among all samples. From each tumor sample, expression was normalized to the level of expression in a normal brain sample (sample #20).
Quantitative RT-PCR analysis
Expression of MPG, Polβ, and PARP1 in the cell lines was measured by quantitative reverse transcriptase (qRT)-PCR using an Applied Biosystems StepOnePlus system as described previously.22 Briefly, 80 000 cells were lysed and reverse transcribed using the Applied Biosystems Taqman Gene Expression Cells-to-CT Kit. Each sample was analyzed in triplicate, and the results are an average of all 3 analyses. Analysis of mRNA expression was conducted as per the manufacturer (▵▵CT method). The Applied Biosystems TaqMan Gene Expression Assays used were as follows: human MPG: Hs00357983-G1; human Polβ: Hs01099715-M1; and human PARP1: Hs00911369-G1. Each were normalized to the expression of human β-actin (part #4333762T).
DNA glycosylase molecular beacon activity assay
All oligodeoxyribonucleotides were purchased from Integrated DNA Technologies, including the following: FD-Con, 6-FAM-dGCACTATTGAATTGACACGCCATGTCGATCAATTCAATAGTGC-Dabcyl, where 6-FAM is carboxyfluorescein and Dabcyl is 4-(4′-dimethylaminophenylazo) benzoic acid; FD-MPG1, 6-FAM-dGCACTXTTGAATTGACACGCCATGTCGATCAATTCAATAGTGC-Dabcyl, where X is 1,N6-ethenoadenine (ɛA). These oligodeoxyribonucleotides were designed to form a stem-loop structure with 13 nucleotides in the loop and 15 base pairs in the stem. Carboxyfluorescein (6-Fam) is a fluorescent molecule that is quenched by Dabcyl in a nonfluorescent manner via Förster Resonance Energy Transfer (FRET).52,53 Therefore, when the DNA is in a stem-loop structure, the 6-FAM at the 5′ end and the Dabcyl at the 3′ end are brought into close proximity. The close proximity of the 6-FAM to Dabcyl enables efficient quenching of 6-FAM by Dabcyl. If the ɛA is removed by MPG and the DNA backbone is hydrolyzed by APE1, the 6-FAM–containing oligonucleotide (4 bases in length) will dissociate from the hairpin at 37°C (Fig. 1C) and the 6-FAM dissociation from the DNA hairpin prevents the quenching by Dabcyl. The increase in 6-FAM–mediated fluorescence is proportional to the amount of ɛA removed. Any increase in fluorescence in control beacon with a normal adenine would be the result of nonspecific cleavage of the DNA backbone.
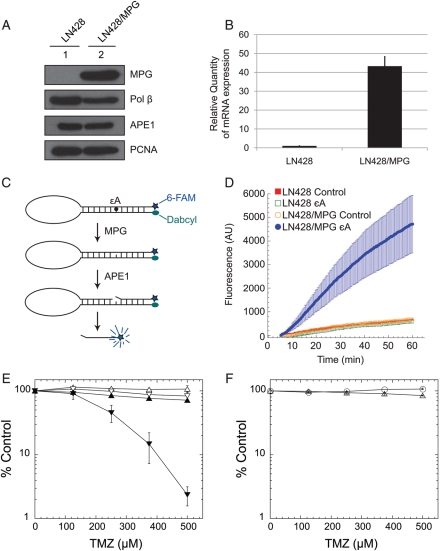
Fig. 1.
Overexpression of MPG in LN428 cells dramatically increases MX-induced potentiation of TMZ. (A) MPG overexpression as determined by immunoblot analysis of nuclear proteins isolated from the LN428 cells (lane 1) or LN428/MPG cells (lane 2). Expression levels of the BER proteins Polβ and APE1 are also shown. PCNA expression is shown as a loading control. (B) MPG overexpression as determined by qRT-PCR analysis in LN428 and LN428/MPG cells. (C) A schematic diagram showing the mechanism of the DNA Glycosylase Molecular Beacon Activity Assay that is used to measure the MPG-mediated DNA glycosylase activity in cell lysates. (D) Increased DNA glycosylase activity in MPG over-expressing LN428/MPG cells as determined by the DNA Glycosylase Molecular Beacon Activity Assay. DNA glycosylase activity specific for removal of the MPG substrate ɛA was measured in nuclear lysates from the control cell line (LN428) and the MPG over-expression cell line (LN428/MPG), as described in the Materials and Methods section. Each lysate was analyzed using either the control-beacon or the ɛA-beacon: LN428 lysates (control-beacon, red filled squares; ɛA-beacon, green open squares) and LN428/MPG lysates (control-beacon, orange open circles; ɛA-beacon, filled blue circles). Results are reported as the mean fluorescence response unit of 3 independent experiments ± S.E. (E) MPG overexpression increases the MX-induced potentiation of TMZ in LN428 cells. LN428 cells (open triangle) or LN428/MPG cells (inverted open triangle) were cultured in 96-well plates for 24 h prior to exposure to MX (filled symbols). Following exposure to MX (60 mM) for 30 min, cells were treated with TMZ together with MX (30 mM) for 48 h. Viable cells were determined using a modified MTT assay as described previously.22 Plots show the % viable cells as compared with untreated (control) cells. Means are calculated from quadruplicate values in each experiment. Results indicate the mean ± S.E. of 3 independent experiments. (F) Overexpression of the glycosylase mutant MPG (N169D) in LN428 cells does not increase the MX-induced potentiation of TMZ. 24 h after seeding into 96-well plates, LN428 cells overexpressing mutant MPG (N169D) were treated with (open triangle) or without (open circle) MX (60 mM) for 30 min. Following MX pretreatment, cells were exposed to TMZ in the presence (open triangle) or absence (open circle) of MX (30 mM) for an additional 48 h. Viable cells were counted, and results are reported as in Fig. 1E.
To ensure that the beacons correctly adapted a stem-loop structure, each was incubated at 95°C for 3 min. The beacons were removed from the heat and allowed to slowly cool overnight to room temperature in an insulated container. Once the hairpin was formed, no measurable fluorescence was detected (not shown) and the hairpin was stable at 37°C for greater than 120 min. However, when heated to 95°C, the hairpin unfolds, resulting in maximum fluorescence intensity (not shown). Nuclear protein extracts were prepared as described above (in the section “Cell extract preparation and Western blot”). Approximately 500 μL of nuclear protein extracts were dialyzed twice using the Slide-A-Lyzer Dialysis Cassette with a 7000 molecular weight cut-off. The samples were dialyzed for 90 min at 4°C in the following buffer: 50 mM Hepes, pH7.5, 100 mM KCl, 0.5 mM ethylene-diaminetetraacetric acid (EDTA), 20% glycerol, and 1 mM DTT. Reactions were performed using 10 μg of dialyzed protein extract and beacon substrate (final conc = 40 nM) in the following buffer: 25 mM HEPES-KOH pH7.8, 150 mM KCl, 0.5 mM EDTA, 1% glycerol, and 0.5 mM DTT. Fluorescence was measured every 20 s for 60 min, using a StepOnePlus real-time PCR system and expressed as arbitrary units (AU).
Molecular beacon data analysis
The fluorescence data were analyzed to enable comparisons across cell lines and for comparison of control and lesion-containing BER beacons. We eliminated the background fluorescence due to incubation of the beacon alone by subtracting the fluorescence values of a control well containing no protein extract from all wells using that molecular beacon. To enable comparisons across different cell lines, molecular beacons, and trials, we selected the fluorescence value of the 5-min time point as the zero value for each well. We subtracted this value from all other time points in that well so all graphs begin from zero AU and 5 min after initiating the reaction. Five minutes was selected as the point from which to begin comparisons, because time points earlier than 4 min contained variations in absolute fluorescence measurements independent of the molecular beacon and cell line (not shown). Five minutes was selected to eliminate the variable measurements and to facilitate valid comparisons between trials and conditions. The mean of 3 separate trials was plotted, with error bars representing the standard error of the mean.
DNA extraction and MSP assay for human MGMT promoter
DNA was purified from 5 × 106 LN428 cells and T98G cells using the DNeasy tissue kit (Qiagen) according to the manufacturer's instruction, and methylation of the MGMT promoter was determined by methylation-specific PCR (MSP), as we have described previously.54 The sense and antisense primers for the methylated human MGMT promoters were 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ and 5′-GCACTCTTCCGAAAACGAAACG-3′, respectively, and the primers used to detect the unmethylated human MGMT promoters were 5′-TTTGTGTTTTGATGTTTGTA GGTTTTTGT-3′ and 5′-AACTCCACACTCTTCCAAAAACAAAACA-3′, respectively.54 The PCR products (93 bp for the unmethylated human MGMT promoter and 81 bp for the methylated human MGMT promoter) were analyzed by 4% agarose gel electrophoresis (Invitrogen-Gibco) using Universal unmethylated DNA (Chemicon International) as a negative control DNA and Universal methylated DNA (Chemicon International) as a positive control DNA.
Cloning and expression of human MGMT
The human MGMT cDNA (pSV2MGMT) was amplified by PCR using primers hMGMT-F (CACCATGGACAAGGATTGTGAAAT) and hMGMT-R (CTAGTTTCGGCCAGCAGG CG). MGMT cDNA was then cloned via a topoisomerase cloning procedure into the pENTR-D cloning plasmid (Invitrogen), as per the manufacturer's protocol. The human MGMT open reading frame (ORF) was transferred from pENTR-hMGMT to a Gateway modified pIRES-Puro plasmid via LR recombination reaction, as per the manufacturer (Invitrogen).
Results
MX-induced potentiation of TMZ is enhanced by overexpression of MPG
To test our hypothesis that increased repair initiation by MPG will further sensitize glioma cells exposed to BER inhibitors, we stably overexpressed WT MPG in the LN428 glioma cell line. Overexpression of MPG was confirmed at the protein and mRNA levels using immunoblot (Fig. 1A) and qRT-PCR analyses, respectively (Fig. 1B), with an approximate 40-fold increase of mRNA.
To confirm the increased glycosylase activity in the MPG overexpressing cells (LN428/MPG), we developed a real-time, quantitative fluorescent MPG activity assay using a modified form of molecular beacons, similar to those previously reported for oxidative damage.55,56 However, instead of incorporating multiple base lesions into the stem,55,56 we designed a BER beacon (BER-beacon) with a single base lesion to more accurately and quantitatively determine lesion repair rates. This unique BER-beacon comprises a single DNA oligodeoxynucleotide designed to form a stem-loop structure and contains a 5′ fluorophore (6-FAM) and a 3′ quencher (Dabcyl) on either end of the oligonucleotide. A 1,N6-ethenoadenine lesion (ɛA), a substrate of MPG,57 was positioned in the stem region of the BER-beacon at base #5 from the 5′end and is used to probe for MPG activity. The same BER-beacon structure with a normal adenine was used as the control substrate. Following removal of ɛA by MPG and subsequent DNA strand excision by APE1 5′ to the AP site, the fluorophore 6-FAM is separated from the quencher (Dabcyl) and the increase in fluorescence signal (517 nm) is proportional to the level of MPG activity (Fig. 1C). The LN428 lysate incubated with the control beacon (Fig. 1D, red filled squares) had a minimal (if any) increase in fluorescence, indicating the control beacon is largely intact. The LN428 lysate had little or no endogenous MPG activity, since when incubated with the beacon containing the MPG-specific substrate ɛA, there was no observable change in fluorescence (Fig. 1D, green open squares). The LN428/MPG lysate also did not have a negligible increase in fluorescence when incubated with the control beacon (Fig. 1D, orange open circles), indicating that MPG overexpression does not increase cleavage of normal DNA. However, the LN428/MPG lysate exhibited robust MPG activity visible with a large increase in fluorescence when incubated with the molecular beacon containing the MPG substrate ɛA (Fig. 1D, filled blue circles). This corresponded to an overall 7.9-fold increase in MPG activity (measured at 60 min), as compared with the LN428 cells and an estimated rate of repair (based on the slope of the curve from 15 to 30 min) of 107.00 AU/min, whereas the background rate of repair in the LN428 cell lysate was similar to the background signal using the control beacon (14.64 AU/min). This demonstrates that the LN428/MPG cell line has increased functional MPG and does not recognize normal DNA as a substrate. These data are in line with our earlier report showing that overexpression of MPG results in an increase in DNA glycosylase activity.23
Using a short-term cell survival assay (48 h MTS assay), we next assayed the potentiation of TMZ by MX in the LN428 cells, with or without MPG overexpression. MX sensitized both cell lines to TMZ, but sensitization of the LN428 cells was minimal (Supplementary Fig. S1). In the LN428 cells, MX induced a 1.5-fold increase in sensitivity to TMZ (LN428 IC50, TMZ treatment: 1.5 mM; TMZ + MX treatment: 1.0 mM). However, the potentiation of TMZ induced by MX was significantly greater in the LN428/MPG cells, decreasing the half maximal inhibitory concentration (IC50) in the combined treatment 4-fold, as compared with the LN428 cells (LN428/MPG IC50, TMZ treatment: 0.8 mM; TMZ + MX treatment: 0.25 mM)(Fig. 1E and Supplementary Fig. S1). To confirm that MPG overexpression-induced potentiation is a result of elevated glycosylase activity, we overexpressed a mutant MPG (N169D) in the glioma cell line LN428. This active-site mutant has been shown to have 100-fold less glycosylase activity than WT MPG.58 Overexpression of the mutant MPG did not sensitize LN428 cells to a combined treatment of MX and TMZ (Fig. 1F), supporting our hypothesis that MPG overexpression-induced sensitization is due to increased DNA glycosylase activity in the cells.
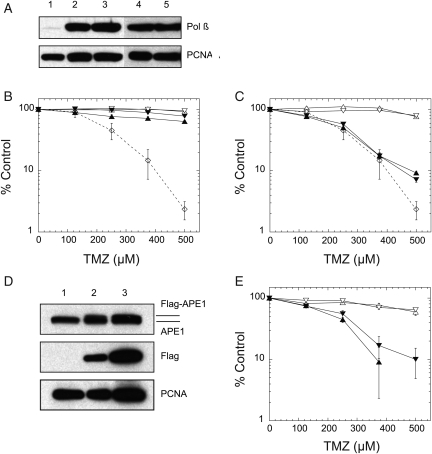
MX-induced potentiation of TMZ is regulated by the expression of Polβ
Although MX reacts efficiently with AP sites in vitro,25 it is also possible that a fraction of the AP sites produced following TMZ exposure will be processed by APE1 and subsequently repaired in vivo. To investigate the possibility that robust BER would alter the MX-induced potentiation of TMZ, we overexpressed Polβ, the rate-limiting enzyme of the BER pathway,59 and assayed MX-induced potentiation. Overexpression of WT Polβ in the LN428/MPG cells (Fig. 2A) completely abrogated the potentiation induced by MX (Fig. 2B, compare to Fig. 1E). In contrast, overexpression of a 5′dRP lyase null mutant (K72A) of Polβ15,60 (Fig. 2A) did not affect the MX-induced potentiation of TMZ (Fig. 2C). Further, to determine whether increased expression of APE1 affects MX-induced potentiation of TMZ, we overexpressed APE1 in the LN428/MPG cells (Fig. 2D and Supplementary Fig. S2B). Interestingly, increased expression of APE1 did not alter the potentiation of TMZ induced by MX (Fig. 2E). A possible explanation for this latter observation is that although overexpression of APE1 increased its mRNA level by 20-fold, its protein level was only slightly increased, which might not be sufficient to significantly increase the number of AP sites processed by APE1 (see Fig. 2D and Supplementary Fig. S2B).
Fig. 2.
Overexpression of the BER protein Polβ, but not, APE1 reverses the MX-induced potentiation of TMZ in LN428/MPG cells. (A) Overexpression of WT Polβ or the 5′dRP lyase null mutant (K72A) of Polβ in LN428/MPG cells as determined by immunoblot analysis of nuclear protein extracted from LN428/MPG/VC cells (vector control, lane 1), LN428/MPG/Flag-Polβ-WT cells (clone 1 and 6 expressing Flag-tagged WT Polβ, lanes 4 &5) and LN428/MPG/Flag-Polβ-K72A cells (clone 5 and 16 expressing Flag-tagged mutant Polβ, lanes 2 & 3). PCNA is shown as a loading control. (B) Over-expression of Polβ reverses the MX-induced potentiation of TMZ in LN428/MPG cells. Cell viability assays were performed and results are reported as in Fig. 1E. LN428/MPG/Flag-Polβ-WT clone 1 cells (triangle) and LN428/MPG/Flag-Polβ-WT clone 6 cells (inverted triangle) were treated with TMZ only (open symbols) or with TMZ and MX (filled symbols). Dotted line with diamond symbols shows LN428/MPG cells treated with MX and TMZ as shown in Fig. 1E. (C) Overexpression of mutant Polβ (K72A) does not reverse the MX-induced potentiation of TMZ in LN428/MPG cells. Cell viability assays were performed and results were reported as in Fig. 1E. LN428/MPG/Flag-Polβ-K72A clone 5 cells (triangle) and LN428/MPG/Flag-Pol β-K72A clone 16 cells (inverted triangle) were treated with TMZ only (open symbols) or with TMZ and MX (filled symbols). The Dotted line with diamond symbols shows LN428/MPG cells treated with MX and TMZ as shown in Fig. 1E. (D) Immunoblot shows overexpression of Flag-APE1 in the LN428/MPG cells. Lane 1: LN428/MPG/vector control; lane 2: LN428/MPG/Flag-APE1 clone 4 and lane 3: LN428/MPG/Flag-APE1 clone 8. PCNA was used as a loading control. (E) Overexpression of APE1 does not reverse the MX-induced potentiation of TMZ in LN428/MPG cells. Cell viability assays were performed and results are reported as in Fig. 1E. LN428/MPG/Flag-APE1 clone 4 cells (triangle) and LN428/MPG/Flag-APE1 clone 8 cells (inverted triangle) were treated with TMZ only (open symbols) or with TMZ and MX (filled symbols).
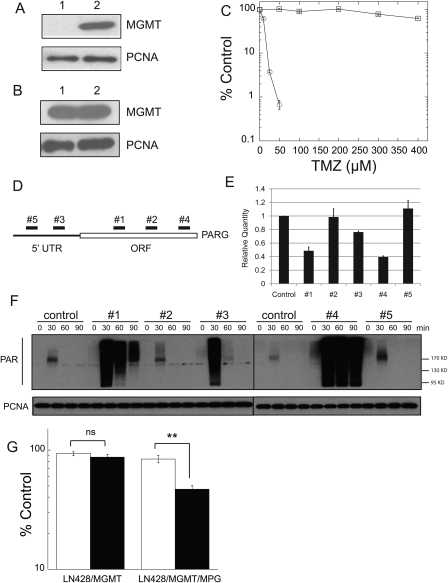
PARG deficiency-induced potentiation of TMZ is enhanced by over-expression of MPG in the presence of MGMT
Next, we addressed chemotherapy sensitization in an MGMT-positive background. The LN428 cell line used in our study has no detectable expression of MGMT (Fig. 3A; lane 1) as a result of epigenetic silencing by promoter methylation (Supplementary Fig. S2A). To study BER inhibition-induced chemotherapy potentiation in the presence of MGMT expression, we transfected the LN428 and LN428/MPG cells with a mammalian expression plasmid (pIRES-Puro-hMGMT), and cell clones stably expressing MGMT were selected for further analysis (Fig. 3B). Overexpression of MGMT yielded LN428 cells resistant to TMZ in a long-term cell survival assay (Fig. 3C).
Fig. 3.
Over-expression of MPG increases the PARG KD–induced potentiation of TMZ in LN428/MGMT cells. (A) MGMT expression, as determined by immunoblot analysis of nuclear protein extracted from LN428 cells (lane 1) and T98G cells (lane 2; used as a positive control). PCNA expression is shown as a loading control. (B) MGMT overexpression, as determined by immunoblot analysis of nuclear protein extracted from LN428/MGMT cells (lane 1) and T98G cells (lane 2; used as a positive control). PCNA expression is shown as a loading control. (C) LN428/MGMT cells are resistant to TMZ, as compared with LN428 cells. Cell viability assays were performed and results are reported as in Fig. 1E. LN428, open circle; LN428/MGMT, open rectangle. (D) A schematic diagram showing the relative target specificity of the 5 PARG shRNA constructs targeting PARG mRNA. (E) Decreased PARG mRNA expression levels induced by the 5 shRNA constructs targeting PARG. Results are reported as the mean ± SE of 3 independent qRT-PCR experiments. (F) PARG KD induces delayed degradation of PAR in LN428/MPG cells following exposure to 1.5 mM TMZ, as demonstrated by immunoblot analysis. (G) PARG KD (PARG KD, black columns; control, white columns) significantly reduced cell survival following exposure to 300 µM TMZ in cells expressing MPG (LN428/MGMT/MPG), as determined by the long-term cell survival assay. Sensitization was not statistically significant (ns) in LN428/MGMT cells with low MPG expression. However, sensitization was statistically significant (**P < 0.01) in LN428/MGMT/MPG cells.
Although poly(ADP-ribosyl)ation of PARP1 and other BER proteins facilitates the repair of base lesions, the dynamics between PAR synthesis and degradation is also important for the effectiveness of the repair process.19 Previously, it was reported that a deficiency in the degradation of PAR negatively affects the repair of base lesions and sensitizes cells to base damage.31 Since PARG is the primary enzyme responsible for degrading PAR in vivo, we investigated whether PARG-KD-induced potentiation of TMZ can be enhanced by overexpression of MPG. We first screened five different shRNA constructs targeting PARG (Fig. 3D) using an HIV-lentiviral system49,50 in the LN428/MPG cells for effective KD of the enzyme. Using RNA prepared from LN428/MPG cells expressing each of the 5 PARG-specific shRNAs, qRT-PCR results showed that the cells expressing shRNA#1 and shRNA#4 have the lowest levels of PARG mRNA (Fig. 3E). To assay the impact of PARG-KD on the ability of cells to degrade DNA damage-induced PAR formation, control cells and cells treated with 1.5 mM TMZ were lysed at different time points and the lysates were probed for PAR in immunoblot analyses. Consistent with the qRT-PCR results, expression of PARG shRNA #1 and #4 greatly decreased the degradation of PAR following exposure to TMZ (Fig. 3F). Based on these results, we decided to use shRNA #4 for effective PARG KD in the following experiments. Further, to target tumor cells that express MGMT, we assayed PARG KD–induced potentiation of TMZ in the LN428 cell lines with overexpressed MGMT. First, we generated stable PARG KD in the MGMT expressing LN428 and LN428/MPG cell lines, as determined by qRT-PCR, using the PARG shRNA #4 lentivirus (Supplementary Fig. S2C and S2D). Next, using long-term cell survival assays, we probed the PARG KD–induced potentiation of TMZ in these cell lines. The results demonstrated that a deficiency in degrading PAR as a result of PARG KD significantly (P < 0.005) sensitized cells to TMZ (300 µM) in the MPG overexpressing cells (LN428/MGMT/MPG) by decreasing the percent cell viability from 87% to 47% (Fig. 3G), while sensitization by PARG KD was not statistically significant (P > 0.1) in the parental cells that exhibit a low (almost undetectable) level of MPG expression (LN428/MGMT) (Fig. 3G).
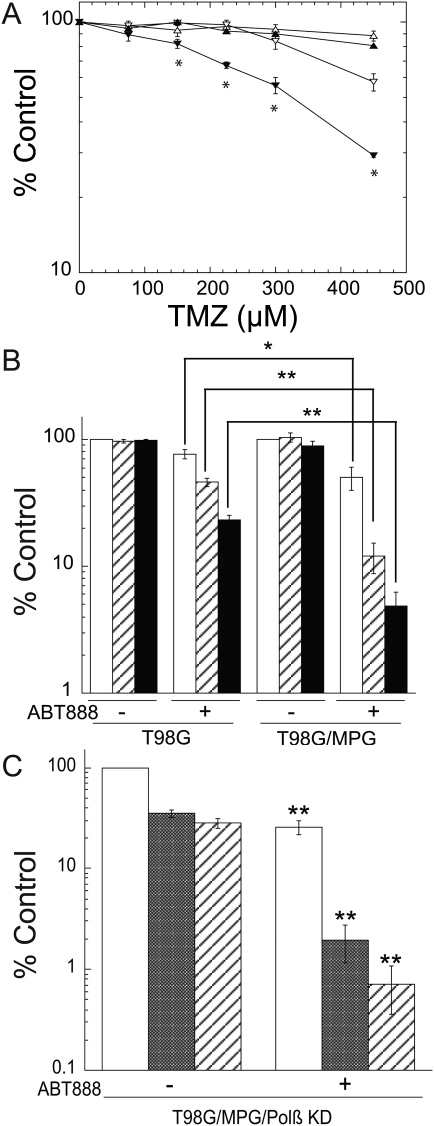
PARP inhibitor-induced potentiation of TMZ is enhanced by overexpression of MPG
Using a long-term cell survival assay, we next assessed whether the PARP inhibitor–induced potentiation of TMZ is affected by overexpression of MPG. We have previously shown that the PARP inhibitor PJ34 (2 µM) significantly reduced the level of PARP activation following exposure to TMZ.22 Here we show that pre- (4 µM) and cotreatment with PJ34 (2 µM) significantly sensitized cells to TMZ, with P < 0.01 for TMZ doses higher than 150 µM, and sensitization by PJ34 was not observed in the parental cells with a low level of MPG expression (LN428/MGMT) (Fig. 4A). To further confirm that overexpression of MPG increases the PARP inhibition–induced potentiation of TMZ in glioma cells, we used a second glioma cell line, T98G,61 which has endogenous elevated expression of MGMT (Fig. 3A and B). We inhibited BER using the clinically relevant PARP inhibitor ABT-88862 in similar experiments as those conducted in the LN428 cell lines. We first overexpressed MPG in the T98G cells using a mammalian expression plasmid (pRS1422). Overexpression of MPG in the T98G cells increased its mRNA level (10-fold) and protein level as determined by immunoblot and qRT-PCR analyses (Supplementary Fig. S3A and S3B). Consistent with previous reports that demonstrate ABT-888 potentiates TMZ in diverse tumor models,41,62 treatment with ABT-888 sensitized T98G cells to TMZ (Fig. 4B). More importantly, overexpression of MPG significantly increased the potentiation induced by ABT-888 (Fig. 4B, P < 0.05 and P < 0.01). Depletion of Polβ in the MPG-overexpressing T98G cells (T98G/MPG/Polβ KD) enhanced the ABT-888–mediated sensitization of the cells to TMZ treatment (IC50 < 25 µM). Similar to the T98G/MPG cells, ABT-888 treatment alone resulted in cell killing in the T98G/MPG/Polβ KD cells, albeit the killing effect was much stronger, as it killed ∼70% of cells as compared with 30% in the T98G/MPG cells (Fig. 4C and Fig. 4B). A combined treatment with TMZ and ABT-888 in the T98G/MPG/Polβ KD cells induced significantly increased cytotoxicity compared with TMZ treatment alone (Fig. 4C, P < 0.01), suggesting that the expression status of Polβ also plays a role in determining the ABT-888–induced potentiation of TMZ. These results demonstrate that increased BER repair initiation enhances the PARP inhibitor–induced potentiation of TMZ via a process that is also dependent on the expression of Polβ. Hence, the expression level of both MPG and Polβ in tumors might be used as a biomarker for alkylator chemotherapy potentiation by methoxyamine or PARP inhibitors.
Fig. 4.
Overexpression of MPG increases the PARP inhibitor-induced potentiation of TMZ in glioma cells with elevated levels of MGMT. (A) PJ34 significantly sensitized LN428/MGMT/MPG cells, but not the LN428/MGMT cells, to TMZ, as measured by long-term cell survival assays. LN428/MGMT cells (triangle); LN428/MGMT/MPG cells (reversed triangle); TMZ treatment only (open symbols); and PJ34 and TMZ treatment (filled symbols). Results were calculated as the percentage survival relative to non-TMZ–treated control cells (% control) and reported as the mean ± SE of 3 independent experiments (*P < 0.01, Student's t-test). (B) Overexpression of MPG in T98G cells significantly increased the ABT-888–induced potentiation of TMZ, as measured by long-term cell survival assays. No TMZ treatment controls (white bars); 50 µM TMZ treatment (lined bars) and 100 µM TMZ treatment (black bars). Results were calculated and reported as in Fig. 4A. Statistics, Student's t-test, *P < 0.05; **P < 0.01. (C) Polβ depletion by shRNA combined with overexpression of MPG in T98G cells significantly increased the ABT-888–induced potentiation of TMZ. No TMZ treatment controls (white bars); 25 µM TMZ treatment (grey bars); and 50 µM TMZ treatment (lined bars). Results were calculated and reported as in Fig. 4A. Statistics comparing between treatments, with or without ABT-888, Student's t-test, **P < 0.01.
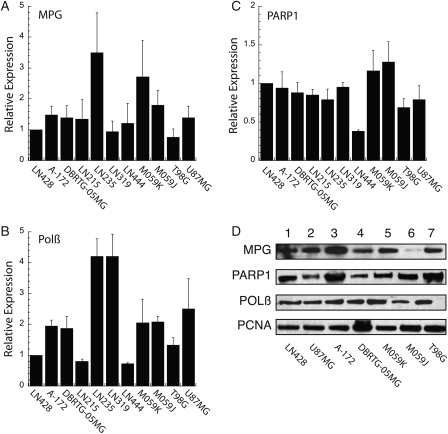
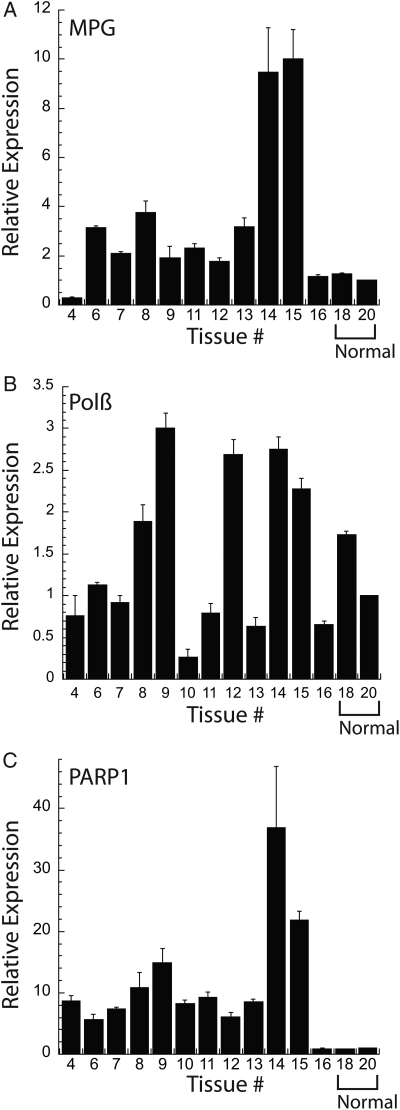
These functional and drug-induced cytotoxicity analyses prompted us to next determine if glioma cell lines and glioma tumors present with varying levels of expression for MPG, Polβ and PARP1 mRNA, and/or protein. We obtained additional established glioma cell lines and characterized the mRNA expression of MPG, Polβ, and PARP1 by qRT-PCR. As shown (Fig. 5A–C), the mRNA expression was variable across the 11 cell lines. Both MPG and Polβ mRNA expression varied as much as 4-fold compared with the LN428 cell line, whereas PARP1 mRNA expression was relatively constant. In some cases, we were also able to analyze protein expression by immunoblot. As shown in Fig. 5D, Polβ protein expression was relatively constant, whereas variations in protein expression were observed for MPG and PARP1. It should be noted that the relationship between mRNA and protein expression is not always 1:1, as suggested previously.63 Interestingly, the mRNA expression pattern in the GBM tumors was considerably more varied. In this analysis, expression was normalized to the expression of each mRNA in a normal brain tissue sample (Fig. 6). Both normal brain samples presented with relatively similar expression levels for all 3 mRNAs analyzed. However, the tumor tissue showed significant variability in the expression of these key BER genes: MPG mRNA expression varied as much as 10-fold (Fig. 6A), Polβ mRNA expression varied as much as 8-fold (Fig. 6B), and PARP1 mRNA expression varied as much as 40-fold compared with normal brain (Fig. 6C).
Fig. 5.
Expression profile of MPG, PARP1, and Polβ in established glioma cell lines. (A–C) The relative expression of mRNA for (A) MPG, (B) Polβ, and (C) PARP1 in GBM cell lines, as indicated in the figure, was measured by quantitative RT-PCR using an Applied Biosystems StepOnePlus system, as described in the Materials and Methods, normalizing to the LN428 cell line across samples. Analysis of mRNA expression was conducted as per the manufacturer (▵▵CT method) and normalized within each sample to the expression of human β-actin (part #4333762T). (D) MPG, Polβ, and PARP1 expression, as determined by immunoblot analysis of nuclear protein extracted from the indicated cells. PCNA expression is shown as a loading control.
Fig. 6.
Relative mRNA expression levels of MPG, PARP1, and Polβ in GBM tumors as compared with normal brain. (A–C) The relative expression of mRNA for (A) MPG, (B) Polβ, and (C) PARP1 in GBM tumors (tissue #4–16), as indicated in the figure, was measured by quantitative RT-PCR using an Applied Biosystems StepOnePlus system, as described in the Materials and Methods, normalizing to the normal brain sample (tissue #20) across tissue samples. Analysis of mRNA expression was conducted as per the manufacturer (▵▵CT method), normalized within each sample to the expression of human β-actin (part #4333762T).
Discussion
MPG initiates the repair of a spectrum of DNA base lesions,64 in particular the repair of alkylated bases.7 It has been demonstrated that MPG expression levels vary considerably in human breast cancer,65 astrocytic tumors,66 and glioblastoma. In addition, MPG possesses multiple post-translational modifications and interacts with many DNA repair proteins, including XRCC1 and HR23A, suggesting that the glycosylase activity of MPG may be under tight cellular regulation.14 Here, we demonstrate that BER inhibitor–mediated sensitization of glioma cells to TMZ is enhanced by overexpression of MPG. Glioma cells with elevated expression of MPG exhibited dramatically increased potentiation of TMZ via several BER inhibitors, including MX, and the PARP inhibitors PJ34 and ABT-888, or by PARG depletion (PARG KD). The enhanced potentiation of TMZ in the MPG-overexpressing glioma cell lines observed in these studies is in line with a previous report showing that MX-induced sensitization is increased by MPG overexpression in ovarian cancer cells.45 However, the expression level of MPG is not the only factor that controls the MX-induced potentiation of TMZ, as it is also related to the efficiency and expression of the BER pathway proteins that process AP sites and downstream repair intermediates. From our experiments (Fig. 2B and C), we show that overexpression of the wild-type BER rate-limiting enzyme Polβ, but not the 5′dRP lyase activity null mutant of Polβ (K72A), in the MPG-overexpressing cells abrogates the MPG-dependent potentiation. Therefore, it is the collective expression status of both MPG and Polβ that defines the sensitization induced by MX. It is possible that the presence of Polβ lyase activity modulates the binding efficiency of MX to the AP site; thus elevated expression of Polβ abrogates the MX-induced potentiation of TMZ in the MPG-overexpressing cells. This is consistent with a recently suggested BER biochemical model of substrate channeling,67 as well as the finding that PARP1 recognizes AP sites.68 However, these studies also raise the possibility that the 5′dRP lesion, the substrate of the lyase activity of Polβ, may also be recognized and bound by MX, suggesting that increased expression of Polβ competes with MX for the binding and processing of 5′dRP and leads to cytotoxic protection. APE1 is the main enzyme that directly competes with MX for the processing of AP sites in cells, yet overexpression of the enzyme did not alter the MX-induced potentiation of TMZ (Fig. 2E). A possible explanation might be that although APE1 mRNA levels were increased by more than 20-fold (Supplementary Fig. S2B), the protein level of APE1 was only slightly increased (Fig. 2D). Since APE1 is an abundant enzyme in cells, a slight increase in the level of APE1 protein may not change the ratio of AP sites processed by APE1 or MX.
As discussed in our previous report, the dynamics between PAR synthesis and degradation not only are involved in facilitating the repair of base lesions, but also act as a mediator of cell death via hyperactivation of PARP and subsequent cellular energy depletion in response to the accumulation of unrepaired BER intermediates.22 Thus, although inhibition of the hyperactivation of PARP and PAR synthesis provides a short-term cell survival advantage, damage-induced DNA lesions persist in cells due to the inhibition of the role of PARP in repair. Cells harboring the unrepaired DNA lesions will eventually die due to the accumulation of double strand breaks (DSBs), as cells go through multiple rounds of replication.69 Therefore, in the context of chemotherapy sensitization involving PARP inhibition or depletion of PARG (PARG KD), the long-term assay (10 days) for cell survival, which allows for multiple rounds of DNA replication, is more suitable than the short-term (2 days) MTS assay. For this reason, all the cell survival assays involving PARG or PARP inhibition were conducted using the long-term assay as described in “Materials and Methods.” PARG is the primary enzyme for degrading PAR in human cells. It has been reported that the PARG inhibitor GPI 16552 chemosensitizes malignant melanoma to TMZ,19 which implies that not only poly(ADP-ribosyl)ation of target proteins by PARP but also the rapid clearance of PAR by PARG is important for cell survival following DNA base damage. In line with the previous report demonstrating that PARG inhibition sensitizes melanoma to TMZ,19 we report herein that shRNA-mediated PARG downregulation sensitizes glioma cells to TMZ. More importantly, we show that the sensitization is greatly enhanced in cells with elevated expression of MPG (Fig. 3G).
PARP has recently become the focus of investigations of chemotherapy potentiation since the publication of a sensitive phenotype induced by PARP inhibitors in breast cancer cells bearing a loss of BRCA1 or BRCA2 function.70,71 Currently, PARP inhibitors are under phase 0 to phase 2 clinical trials in combination with the clinical alkylating agent TMZ.32 The rationale for combining a PARP inhibitor with TMZ is generally considered to be the inhibition of repair of TMZ-induced DNA lesions via inhibiting the role of PARP in BER. However, it is not known if the status of the BER pathway inherent in cancer cells has an impact on the potentiation induced by PARP inhibitors. In this study, using the PARP inhibitors PJ34 and ABT-888, we demonstrated that PARP inhibitor–induced potentiation of TMZ is significantly enhanced in glioma cells with elevated expression of MPG (Fig. 4A), suggesting that increased repair initiation of TMZ-induced base lesions can further sensitize cancer cells to PARP inhibition, and the expression level of MPG in cancer cells may predict clinical outcome. The functional significance of these proof-of-principle studies is enhanced by our expression analysis of 3 key BER genes in GBM tumors. We find considerable variability in the expression of the BER genes MPG, Polβ, and PARP1. These findings are in line with those reporting elevated expression of MPG65,66,72 and Polβ73 in tumors as well as the recent findings of upregulation of PARP1 in triple-negative breast cancer, medulloblastoma, and pediatric glioma.74–76
This study addresses the relationship between DNA glycosylase and Polβ expression and chemotherapy sensitization via BER inhibition (MX, the PARP inhibitors PJ34 and ABT-888, or PARG KD). We demonstrated that the BER inhibition-induced potentiation of TMZ is enhanced by over-expression of the BER initiating enzyme MPG, suggesting that combining inhibition of repair and robust initiation of the BER pathway is an effective means to improved chemotherapy efficacy. Further we suggest that the expression level of both MPG and Polβ in cancer cells might be used to predict effectiveness when combining BER inhibition and alkylating agents.
Supplementary Material
Funding
This work was supported by grants from the American Cancer Society (RSG-05-246-01), the National Institutes of Health (CA132385; GM087798 and CA148629) and the National Brain Tumor Society to RWS. Support for the UPCI Lentiviral Facility was provided by the Cancer Center Support Grant from the National Institutes of Health (P30 CA047904). Support was also provided by the University of Pittsburgh Department of Pharmacology & Chemical Biology and a John S. Lazo Cancer Pharmacology Fellowship to EMG. BM was supported as a Hampton University/UPCI joint cancer education summer program fellow (P20-CA132385).
Supplementary Material
Acknowledgments
We thank I. Pollack (UPCI) for the LN428 cells and M. Ziegler (Univ. of Bergen) for the PAR Ab (clone 10H). We would also like to thank Maureen Lyons-Weiler (UPCI) for isolating RNA from the tissue samples and Jonette Werley (Univ. of Pittsburgh) for preparation of the FFPE tumor and normal brain samples.
Conflict of interest statement. None declared.
References
- 1.Mrugala MM, Chamberlain MC. Mechanisms of disease: Temozolomide and glioblastoma–look to the future. Nat Clin Pract Oncol. 2008;5:476–486. doi: 10.1038/ncponc1155. [DOI] [PubMed] [Google Scholar]
- 2.Tentori L, Graziani G. Recent approaches to improve the antitumor efficacy of temozolomide. Curr Med Chem. 2009;16:245–257. doi: 10.2174/092986709787002718. [DOI] [PubMed] [Google Scholar]
- 3.Sobol RW. Temozolomide. In: Schwab M, editor. Encyclopedia of Cancer. Berlin, Heidelberg, New York: Springer; 2009. [Google Scholar]
- 4.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkaria JN, Kitange GJ, James CD, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Wood RD, Mitchell M, Sgouros J, et al. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 8.Hegi ME, Diserens AC, Gorlia T, et al. Mgmt gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 9.Pollack IF, Hamilton RL, Sobol RW, et al. Mgmt expression strongly correlates with outcome in childhood malignant gliomas: Results from the ccg-945 cohort. J Clin Oncol. 2006;24:3431–3437. doi: 10.1200/JCO.2006.05.7265. [DOI] [PubMed] [Google Scholar]
- 10.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein msh6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMurry TB. Mgmt inhibitors–the trinity college-paterson institute experience, a chemist's perception. DNA Repair (Amst) 2007;6:1161–1169. doi: 10.1016/j.dnarep.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Ranson M, Hersey P, Thompson D, et al. Randomized trial of the combination of lomeguatrib and temozolomide compared with temozolomide alone in chemotherapy naive patients with metastatic cutaneous melanoma. J Clin Oncol. 2007;25:2540–2545. doi: 10.1200/JCO.2007.10.8217. [DOI] [PubMed] [Google Scholar]
- 13.Adhikari S, Choudhury S, Mitra PS, et al. Targeting base excision repair for chemosensitization. Anti-cancer agents in medicinal chemistry. 2008;8:351–357. doi: 10.2174/187152008784220366. [DOI] [PubMed] [Google Scholar]
- 14.Almeida KH, Sobol RW. A unified view of base excision repair: Lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobol RW, Prasad R, Evenski A, et al. The lyase activity of the DNA repair protein ß-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 16.Kinsella TJ. Coordination of DNA mismatch repair and base excision repair processing of chemotherapy and radiation damage for targeting resistant cancers. Clin Cancer Res. 2009;15:1853–1859. doi: 10.1158/1078-0432.CCR-08-1307. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Gerson SL. Therapeutic impact of methoxyamine: Blocking repair of abasic sites in the base excision repair pathway. Current Opinion in Investigative Drugs. 2004;5:623–627. [PubMed] [Google Scholar]
- 18.Chalmers AJ. The potential role and application of parp inhibitors in cancer treatment. Br Med Bull. 2009;89:23–40. doi: 10.1093/bmb/ldp005. [DOI] [PubMed] [Google Scholar]
- 19.Tentori L, Leonetti C, Scarsella M, et al. Poly(adp-ribose) glycohydrolase inhibitor as chemosensitiser of malignant melanoma for temozolomide. Eur J Cancer. 2005;41:2948–2957. doi: 10.1016/j.ejca.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Jaiswal AS, Banerjee S, Panda H, et al. A novel inhibitor of DNA polymerase {beta} enhances the ability of temozolomide to impair the growth of colon cancer cells. Mol Cancer Res. 2009;7:1973–1983. doi: 10.1158/1541-7786.MCR-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushina Y, Manita D, Takeuchi T, et al. The inhibitory action of kohamaic acid a derivatives on mammalian DNA polymerase beta. Molecules. 2009;14:102–121. doi: 10.3390/molecules14010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang J, Goellner EM, Wang XW, et al. Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Molecular Cancer Research. 2010;8:67–79. doi: 10.1158/1541-7786.MCR-09-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trivedi RN, Almeida KH, Fornsaglio JL, et al. The role of base excision repair in the sensitivity and resistance to temozolomide mediated cell death. Cancer Res. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 24.Trivedi RN, Wang XH, Jelezcova E, et al. Human methyl purine DNA glycosylase and DNA polymerase ß expression collectively predict sensitivity to temozolomide. Molecular Pharmacology. 2008;74:505–516. doi: 10.1124/mol.108.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosa S, Fortini P, Karran P, et al. Processing in vitro of an abasic site reacted with methoxyamine: A new assay for the detection of abasic sites formed in vivo. Nucleic Acids Res. 1991;19:5569–5574. doi: 10.1093/nar/19.20.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L, Bulgar A, Miao Y, et al. Combined treatment with temozolomide and methoxyamine: Blocking apurininc/pyrimidinic site repair coupled with targeting topoisomerase iialpha. Clin Cancer Res. 2007;13:1532–1539. doi: 10.1158/1078-0432.CCR-06-1595. [DOI] [PubMed] [Google Scholar]
- 27.Hottiger MO, Hassa PO, Luscher B, et al. Toward a unified nomenclature for mammalian adp-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Hassa PO, Haenni SS, Elser M, et al. Nuclear adp-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber V, Dantzer F, Ame JC, et al. Poly(adp-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 30.Dantzer F, Ame JC, Schreiber V, et al. Poly(adp-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 31.Fisher AE, Hochegger H, Takeda S, et al. Poly(adp-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(adp-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratnam K, Low JA. Current development of clinical inhibitors of poly(adp-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 33.Cheng CL, Johnson SP, Keir ST, et al. Poly(adp-ribose) polymerase-1 inhibition reverses temozolomide resistance in a DNA mismatch repair-deficient malignant glioma xenograft. Mol Cancer Ther. 2005;4:1364–1368. doi: 10.1158/1535-7163.MCT-05-0128. [DOI] [PubMed] [Google Scholar]
- 34.Tentori L, Leonetti C, Scarsella M, et al. Systemic administration of gpi 15427, a novel poly(adp-ribose) polymerase-1 inhibitor, increases the antitumor activity of temozolomide against intracranial melanoma, glioma, lymphoma. Clinical Cancer Research. 2003;9:5370–5379. [PubMed] [Google Scholar]
- 35.Tentori L, Portarena I, Torino F, et al. Poly(adp-ribose) polymerase inhibitor increases growth inhibition and reduces g(2)/m cell accumulation induced by temozolomide in malignant glioma cells. Glia. 2002;40:44–54. doi: 10.1002/glia.10113. [DOI] [PubMed] [Google Scholar]
- 36.Tentori L, Portarena I, Vernole P, et al. Effects of single or split exposure of leukemic cells to temozolomide, combined with poly(adp-ribose) polymerase inhibitors on cell growth, chromosomal aberrations and base excision repair components. Cancer Chemotherapy & Pharmacology. 2001;47:361–369. doi: 10.1007/s002800000248. [DOI] [PubMed] [Google Scholar]
- 37.Miknyoczki SJ, Jones-Bolin S, Pritchard S, et al. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by cep-6800, a poly(adp-ribose) polymerase inhibitor. Mol Cancer Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 38.Calabrese CR, Almassy R, Barton S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(adp-ribose) polymerase-1 inhibitor ag14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 39.Calabrese CR, Batey MA, Thomas HD, et al. Identification of potent nontoxic poly(adp-ribose) polymerase-1 inhibitors: Chemopotentiation and pharmacological studies. Clin Cancer Res. 2003;9:2711–2718. [PubMed] [Google Scholar]
- 40.Curtin NJ, Wang LZ, Yiakouvaki A, et al. Novel poly(adp-ribose) polymerase-1 inhibitor, ag14361, restores sensitivity to temozolomide in mismatch repair-deficient cells. Clin Cancer Res. 2004;10:881–889. doi: 10.1158/1078-0432.ccr-1144-3. [DOI] [PubMed] [Google Scholar]
- 41.Palma JP, Wang YC, Rodriguez LE, et al. Abt-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res. 2009;15:7277–7290. doi: 10.1158/1078-0432.CCR-09-1245. [DOI] [PubMed] [Google Scholar]
- 42.Koh DW, Lawler AM, Poitras MF, et al. Failure to degrade poly(adp-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortes U, Tong WM, Coyle DL, et al. Depletion of the 110-kilodalton isoform of poly(adp-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang D, Zhong ZY, Zhang QH, et al. [effect of adenoviral n-methylpurine DNA glycosylase overexpression on chemosensitivity of human osteosarcoma cells] Zhonghua Bing Li Xue Za Zhi. 2006;35:352–356. [PubMed] [Google Scholar]
- 45.Fishel ML, He Y, Smith ML, et al. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res. 2007;13:260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 46.Rinne ML, He Y, Pachkowski BF, et al. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Res. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park MJ, Kim MS, Park IC, et al. Pten suppresses hyaluronic acid-induced matrix metalloproteinase-9 expression in u87mg glioblastoma cells through focal adhesion kinase dephosphorylation. Cancer Res. 2002;62:6318–6322. [PubMed] [Google Scholar]
- 48.Ishii N, Maier D, Merlo A, et al. Frequent co-alterations of tp53, p16/cdkn2a, p14arf, pten tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zufferey R, Dull T, Mandel RJ, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zufferey R, Nagy D, Mandel RJ, et al. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 51.Yoshizawa K, Jelezcova E, Brown AR, et al. Gastrointestinal hyperplasia with altered expression of DNA polymerase ß. PLoS ONE. 2009;4:e6493. doi: 10.1371/journal.pone.0006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 53.Yaron A, Carmel A, Katchalski-Katzir E. Intramolecularly quenched fluorogenic substrates for hydrolytic enzymes. Anal Biochem. 1979;95:228–235. doi: 10.1016/0003-2697(79)90210-0. [DOI] [PubMed] [Google Scholar]
- 54.Taioli E, Ragin C, Wang XH, et al. Recurrence in oral and pharyngeal cancer is associated with quantitative mgmt promoter methylation. BMC Cancer. 2009;9:354. doi: 10.1186/1471-2407-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maksimenko A, Ishchenko AA, Sanz G, et al. A molecular beacon assay for measuring base excision repair activities. Biochem Biophys Res Commun. 2004;319:240–246. doi: 10.1016/j.bbrc.2004.04.179. [DOI] [PubMed] [Google Scholar]
- 56.Zielinska A, Davies OT, Meldrum RA, et al. Direct visualization of repair of oxidative damage by ogg1 in the nuclei of live cells. J Biochem Mol Toxicol. 2010 doi: 10.1002/jbt.20346. (in press). [DOI] [PubMed] [Google Scholar]
- 57.Lau AY, Wyatt MD, Glassner BJ, et al. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, aag. Proceedings of the National Academy of Science. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connor EE, Wilson JJ, Wyatt MD. Effects of substrate specificity on initiating the base excision repair of n-methylpurines by variant human 3-methyladenine DNA glycosylases. Chem Res Toxicol. 2005;18:87–94. doi: 10.1021/tx049822q. [DOI] [PubMed] [Google Scholar]
- 59.Sobol RW, Kartalou M, Almeida KH, et al. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J Biol Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 60.Sobol RW, Horton JK, Kuhn R, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 61.Stein GH. T98g: An anchorage-independent human tumor cell line that exhibits stationary phase g1 arrest in vitro. Journal of cellular physiology. 1979;99:43–54. doi: 10.1002/jcp.1040990107. [DOI] [PubMed] [Google Scholar]
- 62.Donawho CK, Luo Y, Penning TD, et al. Abt-888, an orally active poly(adp-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 63.Gygi SP, Rochon Y, Franza BR, et al. Correlation between protein and mrna abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svilar D, Goellner EM, Almeida KH, et al. Base excision repair and lesion-dependent sub-pathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011 doi: 10.1089/ars.2010.3466. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerda SR, Turk PW, Thor AD, et al. Altered expression of the DNA repair protein, n-methylpurine-DNA glycosylase (mpg) in breast cancer. FEBS Lett. 1998;431:12–18. doi: 10.1016/s0014-5793(98)00697-8. [DOI] [PubMed] [Google Scholar]
- 66.Kim NK, Ahn JY, Song J, et al. Expression of the DNA repair enzyme, n-methylpurine-DNA glycosylase (mpg) in astrocytic tumors. Anticancer Res. 2003;23:1417–1423. [PubMed] [Google Scholar]
- 67.Prasad R, Shock DD, Beard WA, et al. Passing the baton: Substrate channeling in mammalian base excision repair pathways. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khodyreva SN, Prasad R, Ilina ES, et al. Apurinic/apyrimidinic (ap) site recognition by the 5′-drp/ap lyase in poly(adp-ribose) polymerase-1 (parp-1) Proc Natl Acad Sci U S A. 2010;107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Shi Y, Guan R, et al. Potentiation of temozolomide cytotoxicity by poly(adp)ribose polymerase inhibitor abt-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol Cancer Res. 2008;6:1621–1629. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- 70.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of brca2-deficient tumours with inhibitors of poly(adp-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 71.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in brca mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 72.Kim NK, An HJ, Kim HJ, et al. Altered expression of the DNA repair protein, n-methylpurine-DNA glycosylase (mpg) in human gonads. Anticancer Res. 2002;22:793–798. [PubMed] [Google Scholar]
- 73.Albertella MR, Lau A, O'Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Barton VN, Donson AM, Kleinschmidt-DeMasters BK, et al. Parp1 expression in pediatric central nervous system tumors. Pediatr Blood Cancer. 2009;53:1227–1230. doi: 10.1002/pbc.22141. [DOI] [PubMed] [Google Scholar]
- 75.Goncalves A, Finetti P, Sabatier R, et al. Poly(adp-ribose) polymerase-1 mrna expression in human breast cancer: A meta-analysis. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-010-1199-y. (in press) [DOI] [PubMed] [Google Scholar]
- 76.Pizem J, Popovic M, Cor A. Expression of gli1 and parp1 in medulloblastoma: An immunohistochemical study of 65 cases. J Neurooncol. 2010 doi: 10.1007/s11060-010-0431-2. [DOI] [PubMed] [Google Scholar]
- 77.Giard DJ, Aaronson SA, Todaro GJ, et al. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 78.Kruse CA, Mitchell DH, Kleinschmidt-DeMasters BK, et al. Characterization of a continuous human glioma cell line dbtrg-05mg: Growth kinetics, karyotype, receptor expression, and tumor suppressor gene analyses. In Vitro Cell Dev Biol. 1992;28A:609–614. doi: 10.1007/BF02631035. [DOI] [PubMed] [Google Scholar]
- 79.Allalunis-Turner MJ, Barron GM, Day RS, 3rd, et al. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat Res. 1993;134:349–354. [PubMed] [Google Scholar]
- 80.Clark MJ, Homer N, O'Connor BD, et al. U87mg decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van Meir E, Sawamura Y, Diserens AC, et al. Human glioblastoma cells release interleukin 6 in vivo and in vitro. Cancer Res. 1990;50:6683–6688. [PubMed] [Google Scholar]
- 82.Van Meir EG, Kikuchi T, Tada M, et al. Analysis of the p53 gene and its expression in human glioblastoma cells. Cancer Res. 1994;54:649–652. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.