Abstract
Activation-induced cytidine deaminase (AID) is shown to be essential and sufficient to induce two genetic alterations in the Ig loci: class switch recombination (CSR) and somatic hypermutation (SHM). However, it is still unknown how a single-molecule AID differentially regulates CSR and SHM. Here we identified Spt6 as an AID-interacting protein by yeast two-hybrid screening and immunoprecipitation followed by mass spectrometry. Knockdown of Spt6 resulted in severe reduction of CSR in both the endogenous Ig locus in B cells and an artificial substrate in fibroblast cells. Conversely, knockdown of Spt6 did not reduce but slightly enhanced SHM in an artificial substrate in B cells, indicating that Spt6 is required for AID to induce CSR but not SHM. These results suggest that Spt6 is involved in differential regulation of CSR and SHM by AID.
The Ig genes in antigen-stimulated B lymphocytes are diversified by two distinct genetic alteration mechanisms, namely somatic hypermutation (SHM) and class switch recombination (CSR) (1, 2). SHM causes the accumulation of point mutations in the rearranged variable (V) region genes, leading to generation of antibodies with higher affinity after cellular selection by a limited amount of antigen (1). CSR replaces the heavy chain constant region (CH) gene proximal to the VH gene, namely Cμ with one of the downstream CH genes by recombination between the switch (S) regions located 5′ to each CH gene, thereby producing antibodies with diverse effector functions without changing their antigen specificity (2).
Both SHM and CSR require activation-induced cytidine deaminase (AID), which is specifically expressed in activated B cells (3). It is well accepted that AID initiates single-strand DNA breaks essential for SHM and CSR through its cytidine deaminase activity. AID can also introduce mutations in such non-Ig loci as c-myc, Pim1, Pax5, bcl-6, and RhoH (4, 5). The number of target loci of AID seems to be larger than expected but still limited (5). Although extensive analyses have been done to uncover the exact molecular mechanism how AID induces DNA strand breaks at restricted loci, it is unknown how a single-molecule AID differentially regulates CSR and SHM or how the Ig genes and other target loci are preferentially targeted in the whole genome. To answer these questions, extensive studies were carried out to identify cofactor(s) that may account for the target specificity of the AID function. Several AID-interacting proteins have been reported, including RNA polymerase II (6), replication protein A (7), protein kinase A (8, 9), DNA-PKcs (10), MDM2 (11), CTNNBL1 (12), Spt5 (13), and PTBP2 (14). Unfortunately, however, none of these proteins could show any functional correlation to support the target specificity of AID. There is no clear mechanism to limit the number of target loci. Most of the proteins like RNA polymerase II, protein kinase A, Spt5, and PTBP2 are rather ubiquitous and interact with many proteins other than AID. PTBP2 is a splicing factor, and Spt5 is one of the transcription elongation factors that associate with RNA polymerase II. Replication protein A, DNA-PKcs, and MDM2 are proteins involved in general DNA repair. CTNNBL1 was later shown to be dispensable for CSR (15).
AID has been shown to have the nuclear localization signal and nuclear export signal in its N terminus and C terminus, respectively (16, 17). The deletion of the nuclear localization signal region of AID results in loss of the AID functions for both SHM and CSR (16). A series of mutations at the N teminus of AID also causes defects in CSR as well as SHM (18). Although no mutations at the N terminus of AID have been shown to cause CSR-specific loss of the AID function, some AID mutants with point mutations in the N-terminal region retain substantial CSR activity but severely damage SHM activity, which is most likely due to a combination of partial loss of DNA cleavage activity and less efficient cleavage of the V region compared with S regions (18–20). Conversely, a S3A mutaiton augments both CSR and SHM (21). On the other hand, the deletions and/or mutations in the nuclear export signal region (residues 183–198) result in loss of the AID function for CSR but not SHM, probably because AID with the C-terminal deletion has normal DNA cleavage activity (19, 22). The results suggest that AID has at least two functions: DNA cleavage of V and S region associated with the N-terminal region, and CSR-specific activity associated with the C-terminal region. In addition, the C-terminal region was shown to be responsible for interaction with poly (A)+ RNA (23). We proposed that the C-terminal region of AID might be responsible for generation of recombination synapsis factor (19). Therefore, we assumed that cofactors interacting with the C-terminal region of AID might be responsible for CSR-specific activity rather than DNA cleavage, whereas cofactors interacting with the N-terminal region of AID might be responsible for DNA cleavage of both V and S regions.
In the present study, we screened AID association molecules by yeast two-hybrid screening and coimmunoprecipitation. We then assessed their functional involvement in CSR and SHM. Because AID associates with a numerous molecules, we used AID mutants as negative controls and chose the association molecules specific to wild-type AID. Among these molecules we identified Spt6, whose interaction was blocked by AID mutations at the N terminus. Knockdown of Spt6 resulted in great reduction of CSR in both the endogenous Ig locus in B cells and an artificial substrate in fibroblasts. Surprisingly, however, knockdown of Spt6 did not reduce but slightly enhanced SHM in an artificial substrate in B cells. These results indicate that Spt6 is involved in differential regulation of CSR and SHM by AID.
Results and Discussion
Proteins Physically Interacting with AID.
To identify AID-interacting proteins that may account for the target specificity of AID function in CSR and SHM, we overexpressed mouse (m) and human (h) AID in a mouse B-cell line, CH12F3-2A, which can switch from IgM to IgA upon stimulation. We used AID tagged with GFP-FLAG (GF) at its C terminus to avoid masking the FLAG epitope by putative large AID association proteins. The addition of GF to the C terminus of AID had little effect on the function of AID to induce both CSR and SHM. Cytoplasmic extracts of CH12F3-2A with AID-GF were fractionated by centrifugation through a glycerol density gradient (10−50%, vol/vol). Each fraction collected was analyzed for the presence of AID-GF by GFP fluorescence (Fig. S1A). The distribution of AID-GF was broad. Similar broad distribution of endogenous AID was observed in CH12F3-2A extracts using an anti-AID antibody (Fig. S1B). However, the increase of the NaCl concentration from 150 mM to 500 mM reduced the overall size distribution and sharpened the distribution profile (Fig. S1C). The results indicate that AID interacts with a large number of cytoplasmic proteins, some of which can be removed at 500 mM NaCl. The complex formation was not due to GFP because GF alone formed a small and sharp peak. RNase A treatment reduced the size of the peak only slightly at 150 mM NaCl but hardly at 500 mM NaCl, indicating that there are some AID protein complexes containing RNA but the majority of the AID complex is formed through the protein–protein interaction.
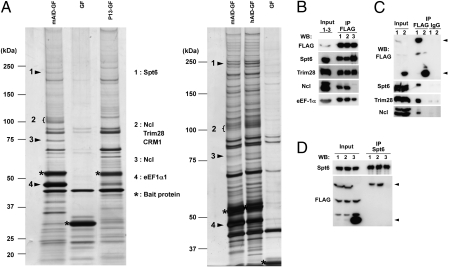
We then immunoprecipitated AID-GF interacting molecules with the anti-FLAG antibody from the cytoplasmic extracts and fractionated by SDS/PAGE, followed by MS. As shown in Fig. 1A, a huge variety of proteins were coimmunoprecipitated with AID-GF compared with GF. Therefore, we decided to compare coimmunoprecipitates between AID and its loss-of-function mutant. We used the human AID mutant P13 (M139V) defective in both SHM and CSR activities (22). Similarly diverse proteins with similar intensity were coimmunoprecipitated with the P13 mutant when we used an equal number of cells for wild-type and mutant AID (Fig.1A).
Fig. 1.
Identification of AID-interacting proteins. (A) Silver staining of proteins coimmunoprecipitated with mouse AID-GFP-FLAG (mAID-GF), human AID-GF (hAID-GF), GF, and P13-GF from cytosolic extracts prepared from CH12F3-2A cells expressing either of the AID-GFs. Bands were excised and analyzed by MS. Proteins that appeared to be obviously more abundant in mAID-GF and hAID-GF are indicated. Nucleolin appears at two positions owing to possible modification. (B) Western blot analyses of immunoprecipitates with anti-FLAG M2 antibody from mAID-GF–expressing CH12F3-2A extracts treated with DNase I (lane 2) or RNase A (lane 3). Immunoprecipitates from untreated extracts are shown in lane 1. (C) Western blot analyses of immunoprecipitates either with anti-FLAG M2 antibody or with mouse IgG from cytosolic extracts prepared from CH12F3-2A cells expressing mAID-GF (lane 1) or GF (lane 2). Arrows indicate mAID-GF or GF. (D) Western blot analyses of immunoprecipitates with anti-Spt6 mAb from cytosolic extracts prepared from CH12F3-2A cells expressing mAID-GF (lane 1), hAID-GF (lane 2), or GF (lane 3). Two nonspecific bands appeared in input by FLAG Western blot.
Among coimmunoprecipitates, Spt6, Trim28, Nucleolin, Skiv2l2, Zfp84, CRM1, and eEF1α were clearly more abundant in wild-type AID-GF (Fig. 1A). We also obtained the following two groups of coimmunoprecipitates from CH12F3-2A cells: (i) proteins involved in the nuclear–cytoplasmic transport, including importin 4 and importin β3 (Ranbp 5); and (ii) proteins involved in the translation or degradation of proteins, and chaperones, including eEF1α, Hsp70, Stip1, TCP1, CCTq, and KIAA1967. Identification of proteins in the group (i) and CRM1 indicates that the coimmunoprecipitations in the current condition is suitable to detect expected functional partners of AID because AID has been reported to be actively exported from nucleus to cytoplasm in a CRM1-dependent manner (16, 17). The degradation-related molecules and chaperons in group (ii) were coimmunoprecipitated probably because overexpressed wild-type and mutant AID-GF were targets of the degradation or inactivation machinery. Although eEF1α also showed a striking difference between wild-type AID and P13 mutant, we suspected that eEF1α was directly associated with mRNA to which AID interacts as reported previously (23). In fact the treatment of cell extracts with RNase A before immunoprecipitation significantly reduced eEF1α from the coimmunoprecipitates with AID-GF (Fig. 1B).
In a separate series of experiments, we expressed hAID-FLAG (F) and hAID mutants including L172A-F, ΔN10-hAID-F (N-terminal 10 residue truncation), and JP8Bdel-F (Table S1) in 293T cells and compared proteins immunoprecipitated with the anti-FLAG antibody by MS. We picked up seven proteins that were specifically coimmunoprecipitated with hAID-F in all four repeated experiments. The list of such proteins is shown in Table S2. Surprisingly, none of them overlapped with the above experiments in CH12F3-2A cells. The discrepancy could be at least in part due to the difference in tags to AID and cells used.
Spt6 Is Required for CSR in B Cells and Fibroblasts.
We first focused on Spt6, Trim28, and Nucleolin because they are involved in nucleic acid metabolism and most distinctly associated with wild-type AID. We confirmed that Spt6, Trim28, and Nucleolin were coimmunoprecipitated with AID-GF but not GF by Western blotting (Fig. 1B). The control IgG did not precipitate any of them (Fig. 1C). The association of Spt6 and Trim28 with AID-GF was not reduced by either RNase A or DNase I treatment of the cell extracts before immunoprecipitation, whereas the association of Nucleolin with AID-GF was almost completely abolished by RNase A treatment, suggesting that RNA bridges AID with Nucleolin (Fig. 1B). In addition, the association between Spt6 and AID was further confirmed by detection of AID-GF in immunoprecipitates with an anti-Spt6 antibody (Fig. 1D).
Interaction of AID with Spt6 was also supported by yeast two-hybrid screening. We screened a human lymph node cDNA library fused to the GAL4 activation domain (AD), with hAID fused to the GAL4 DNA-binding domain (BD) as bait, and a mouse pre B-cell cDNA library fused to the GAL4 AD, with mAID fused to the GAL4 DNA-BD as bait. The C-terminal fragments of hSPT6 (3817–5178 nt) and mSpt6 (4050–5178 nt), which contain Src homology 2 domain, were isolated from the human and mouse libraries, respectively (Fig. S2). This interaction was further confirmed by coimmunoprecipitation of hAID with hSPT6 fragments fused with GST.
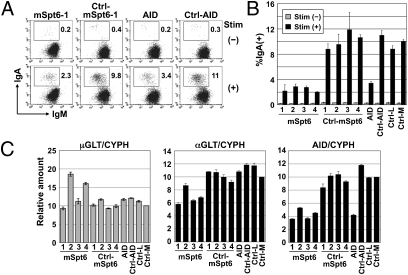
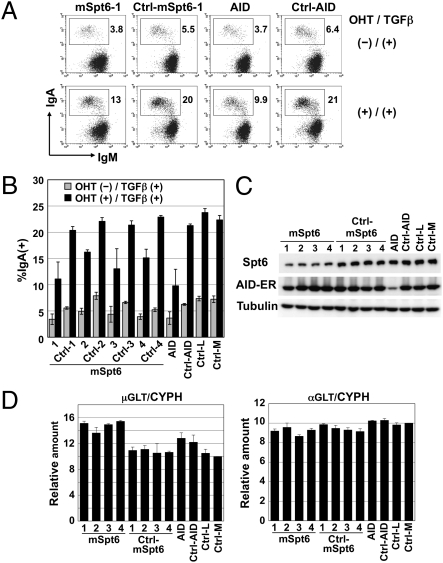
We therefore examined the functional involvement of Spt6 in CSR by knocking down its expression in CH12F3-2A cells. Knockdown of Spt6 or AID significantly reduced CSR efficiency (Fig. 2 A and B). However, 1.5 μg of Spt6 siRNA decreased AID mRNA and germline transcript (GLT) of Cα significantly, although GLT of Cμ was intact (Fig. 2C). Therefore, we further examined the effects of Spt6 knockdown using the AID-ER (fusion protein of AID and the hormone-binding domain of the estrogen receptor) system, in which CSR can be induced rapidly upon 4-hydroxy tamoxifen (OHT) addition without de novo transcription and translation of AID. Knockdown of Spt6 with 0.6 μg of siRNA reduced the efficiency of CSR almost in parallel with the degree of Spt6 protein reduction (Fig. 3 A–C). However, Stp6 knockdown did not affect the amounts of AID-ER protein and GLTs (Fig. 3 C and D). The results indicate that Spt6 is required for CSR.
Fig. 2.
CSR is inhibited by Spt6 knockdown in CH12F3-2A cells. (A and B) Spt6 knockdown severely reduced CSR efficiency. CH12F3-2A cells (1.5 × 106) were introduced with 1.5 μg of siRNAs against mSpt6, scrambled siRNA for them, an siRNA against mAID, a scrambled siRNA for it, or negative control siRNAs with low (36%) and medium (48%) GC contents (Ctrl-L and Ctrl-M, respectively). The GC contents of oligos mSpt6-1, -2, and -4 are medium (45–55%), and those of oligos mSpt6-3 and AID are low (35–45%). Twenty-four hours after siRNA introduction, cells were stimulated with CD40L, IL-4, and TGF-β for 24 h. The percentages of IgA+ cells in the live population are indicated. Representative FACS profiles are shown (A). The mean ± SD values were obtained from triplicate experiments (B). (C) siRNAs against Spt6 reduced the amount of αGLT and AID transcripts. Quantitative PCR analyses for μGLT, αGLT, and AID transcripts in Spt6-knockdown cells. Values were normalized by cyclophilin (CYPH). Unstimulated and stimulated cells were analyzed for μGLT and for αGLT and AID transcripts, respectively.
Fig. 3.
CSR is inhibited by Spt6 knockdown in mAID-ER–expressing CH12F3-2A cells without affecting the amount of AID. (A and B) Spt6 knockdown reduced CSR efficiency. CH12F3-2A cells expressing mAID-ER (1.5 × 106) were introduced with 0.6 μg of siRNAs against mSpt6, scrambled siRNA for them, an siRNA against mAID, a scrambled siRNA for it, or negative control siRNAs with low and medium GC contents (Ctrl-L and Ctrl-M, respectively). Twenty-four hours after siRNA introduction, cells were stimulated with OHT and TGF-β for 24 h. The percentages of IgA+ cells in the live population are indicated. Representative FACS profiles are shown (A). Mean ± SD values were obtained from triplicate experiments (B). (C) siRNAs against Spt6 efficiently reduced the amount of Spt6 protein but did not affect the amount of AID-ER protein. (D) Quantitative PCR analyses for μGLT and αGLT in Spt6-knockdown cells. Stimulated cells were analyzed. CYPH, cyclophilin.
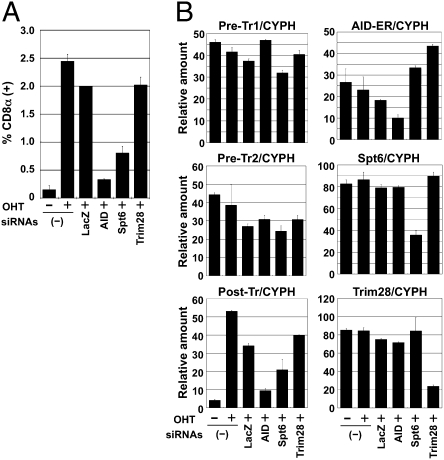
To further confirm the involvement of Spt6 in CSR, we used the artificial switch substrate in a mouse fibroblast cell line, NIH 3T3 (24). We knocked down Spt6 using a mixture of siRNAs in NIH 3T3 cells expressing the artificial switch substrate of CSR and AID-ER. siRNAs against Spt6 significantly reduced CSR efficiency in the artificial switch substrate compared with control siRNAs against LacZ (Fig. 4A). Consistently, postswitch transcripts were decreased by Spt6 knockdown, although it did not reduce the amounts of AID-ER mRNA and preswitch transcripts Pre-Tr1 and Pre-Tr2 (equivalent to GLTs) (Fig. 4B). These results further confirmed that Spt6 is required for CSR.
Fig. 4.
Spt6 knockdown inhibited CSR in the artificial switch substrate SCI (μ, α) in NIH 3T3 cells. (A) NIH 3T3 cells expressing mAID-ER and SCI (μ, α) were introduced with d-siRNAs against mSpt6, mTrim28, mAID, and LacZ together with a DsRed-expressing plasmid as a transfection indicator. Twenty-four hours after transfection, cells were stimulated with OHT for 36 h. The percentages of switched CD8α+ cells in the live and DsRed+ population are indicated. Mean ± SD values were obtained from triplicate experiments. (B) Quantitative PCR analyses for pre- (Pre-Tr1 and Pre-Tr2) and postswitch transcripts (Post-Tr), AID-ER, Spt6, and Trim28 transcripts. CYPH, cyclophilin.
Other Candidates Are Not Required for CSR.
We then examined involvement of other AID-binding proteins in CSR by knockdown assay. Knockdown of Trim28 also reduced CSR but simultaneously decreased AID mRNA in CH12F3-2A cells (Fig. S3 A and B). We thus examined the effect of Trim28 knockdown in the AID-ER system. Trim28 knockdown in the AID-ER system did not affect CSR, suggesting that Trim28 blocked CSR by inhibiting AID transcription (Fig. S3 C and D). Involvement of Trim28 in the transcriptional regulation of AID was further confirmed by the fact that both AID transcription and CSR are drastically reduced in Trim28-deficient B cells (Fig. S3 E and F). We concluded that the association of Trim28 with AID is not related to AID function. Knockdown of Trim28 did not show any significant effects on CSR in NIH 3T3 cells either (Fig. 4 A and B).
Knockdown of Nucleolin did not significantly affect CSR efficiency, either in CH12F3-2A cells or NIH 3T3 cells, although Nucleolin protein was dramatically reduced by knockdown. Knockdown of Skiv212 only slightly reduced CSR efficiency, both in CH12F3-2A cells and NIH 3T3 cells, whereas knockdown of Zfp84 did not significantly affect CSR in CH12F3-2A cells. Therefore, we concluded that Nucleolin, Skiv2l2, and Zfp84 do not play major roles in CSR, although we could not exclude the possibility that residual amounts of target proteins were still sufficient to support CSR. We then examined whether candidates identified from 293T cells by specific coimmunoprecipitation with hAID-F are involved in CSR (Table S2). Knockdown of these candidates was carried out in CH12F3-2A cells, but none of them affected CSR significantly except for hnRNPA1, which we could not knockdown. We could therefore identify only Spt6 that has functional relevance for the CSR activity of AID among all candidates detected by physical association with AID.
Spt6 Is Dispensable for SHM in B Cells.
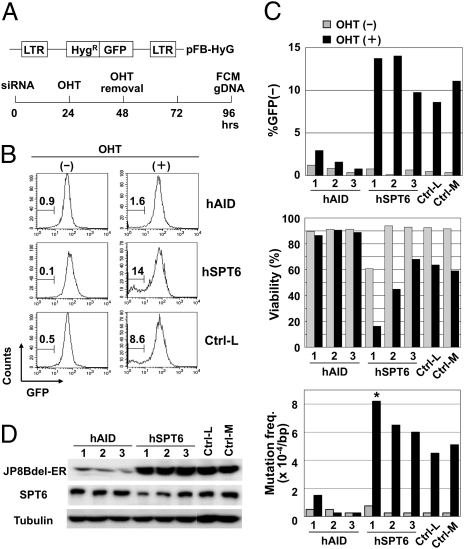
We next examined the effect of Spt6 knockdown on SHM in a human B-cell line BL2. To assess SHM efficiency sensitively and quickly, we took advantages of a modified GFP substrate of SHM (Fig. 5A). In addition, we used a C-terminal truncation mutant of AID, JP8Bdel, which has stronger SHM activity but marginal CSR activity (19, 22). In this system, OHT-activated JP8Bdel-ER protein caused loss of GFP fluorescence due to the accumulation of deleterious mutations in the GFP gene. Excessive mutations induced by JP8Bdel caused cell death. AID knockdown by three different siRNA oligos inhibited loss of GFP fluorescence, as well as the accumulation of point mutations in the GFP gene and cell death, confirming that these events were dependent on AID function and thus useful indicators for SHM (Fig. 5 B and C and Table S3).
Fig. 5.
Spt6 knockdown augmented SHM in BL2 cells. (A) Schematic representation of the artificial SHM substrate and the SHM assay procedure. BL2 cells expressing JP8Bdel-ER and the artificial SHM assay substrate were introduced with siRNAs, stimulated for 24 h with OHT, and incubated for an additional 48 h in the absence of OHT. Then cells were harvested for flow cytometry (FCM) and genomic DNA extraction. (B and C) SPT6 knockdown augmented the SHM efficiency in the artificial substrate. BL2 cells expressing JP8Bdel-ER and the artificial SHM assay substrate (1.5 × 106) were introduced with 3.0 μg of siRNAs against hSPT6, siRNAs against hAID, or negative control siRNAs with low (36%) and medium (48%) GC contents (Ctrl-L and Ctrl-M, respectively). The GC contents of oligos hAID-1, -2, -3, and hSPT6-3 are medium (45–55%) and those of oligos hSPT6-1 and -2 are low (35–45%). The percentages of GFP− cells are indicated (B). Graphical summary of the percentages of GFP− cells, viability, and mutation frequencies in the GFP sequence (C). Statistical significance was evaluated against the corresponding control oligo by χ2 test. *P < 0.05. Data are representative of three independent experiments. (D) Two siRNA oligos against SPT6 (1 and 2) reduced the amount of SPT6 protein but did not affect the amount of JP8Bdel-ER protein. Note that the other siRNA oligos against SPT6 (3) did not substantially reduce the amount of SPT6 protein.
Surprisingly, SPT6 knockdown did not inhibit but rather slightly augmented the frequency of GFP-negative cells, as well as actual mutation frequency in the GFP gene and cell death, without affecting the amount of JP8Bdel-ER protein (Fig. 5 C and D and Table S3). Although the difference was modest, the relative increase of mutation frequencies correlated well with the knockdown efficiency of each oligo against SPT6. It should be noted that both fluorescence and point mutations in the GFP gene were not reduced by SPT6 knockdown in the absence of OHT, indicating that SPT6 knockdown did not affect transcription of the SHM target. These results clearly showed that Spt6 is not required for SHM but rather inhibitory to SHM.
Spt6 Interacts with AID Through Its N Terminus.
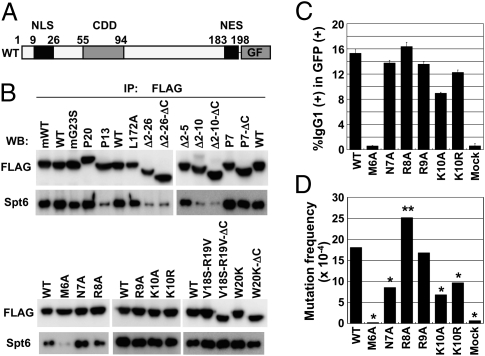
Because the C-terminal region of Spt6 interacts with AID, we then examined whether a specific region of AID is responsible for the association with Spt6. Deletion of the N-terminal residues 2–26 of AID abolished the association with Spt6, suggesting that this region contains residues involved in the interaction with Spt6 (Fig. 6 A and B and Table S1). We further tested AID mutants that carry deletion or mutation(s) in the N-terminal region and found that the deletion of residues 2–10 but not residues 2–5 of AID abolished the association with Spt6, suggesting that residues 6–10 may be responsible for the association with Spt6. Amino acid substitution experiments showed that only M6 was critical within residues 6–10 to the association with Spt6. Other mutations in the N-terminal region [G23S, V18S-R19V, W20K, and P7 (R24W)], which have been shown to reduce SHM more drastically than CSR or to abolish both, did not affect the association with Spt6 (Fig. 6B and Table S1). In agreement with Fig. 1, P13-GF actually lost the binding capacity with Spt6.
Fig. 6.
AID interacts with Spt6 through its N terminus. (A) Schematic representation of wild-type and mutant AID-GF constructs. (B) Western blot analyses of immunoprecipitates with anti-FLAG from cytosolic extracts of CH12F3-2A cells expressing wild-type AID-GF or mutant AID-GFs. All AID constructs are of human origin except for mWT and mG23S. An equal amount of wild-type and mutant AID-GF proteins was analyzed by adjusting the loading amounts of immunoprecipitates. (C) AID-deficient splenocytes were stimulated with LPS for 48 h and infected with retroviruses expressing mutant AID-GFs. Cells were stimulated for additional 48 h in the presence of LPS and IL-4. The percentages of IgG1+ cells in the GFP+ population are indicated. Mean ± SD values were obtained from triplicate experiments. (D) NIH 3T3 cells harboring an SHM substrate pI were infected with retroviruses expressing wild-type and mutant AID-GFs and cultured for 7 d. Genomic DNA was extracted for sequencing analysis. GF was used as mock. Statistical significance was evaluated against WT by χ2 test. *P < 0.001, **P < 0.05.
Next, we tried to examine the involvement of the C-terminal region (residues 183–198) of AID in the association with Spt6. To avoid insolubility of the C-terminally deleted (ΔC) AID by the spontaneous accumulation in the nucleus, we combined the deletion or mutation(s) in the N-terminal region with the deletion of the C-terminal region because most N-terminal mutants (P7, V18S-R19V, and W20K) did not accumulate in the nucleus even when nuclear export was blocked by leptomycin B (16, 18). Such N-terminal mutants that additionally lacked the C-terminal region (P7-ΔC, V18S-R19V-ΔC, and W20K-ΔC) still associated with Spt6, indicating that the C-terminal region of AID is not involved in the association with Spt6. A human AID mutant P20 carrying a 34-aa insertion after residue 182 did not show any reduction in the association with Spt6, although P20 has severe defect in CSR but little effect on SHM (22).
Dissociation of AID Mutant Activities from Their Spt6 Binding.
Studies on series of AID mutants clearly demonstrated that the C-terminal region of AID is required for CSR-specific function other than DNA cleavage. This function is assumed to be related to synapsis formation of cleaved ends (19). On the other hand, the N-terminal region of AID is required for DNA cleavage of both V and S regions. Because Spt6 is required only for CSR, the interaction of Spt6 with N-terminal residues 6–10 of AID was puzzling. To monitor the function of the mutants at residues 6–10, each was fused with GFP in the retroviral expression vector and introduced to AID-deficient spleen cells (Fig. 6C). We also tested the SHM activities of these mutants in a GFP substrate expressed in NIH 3T3 cells (Fig. 6D and Table S4). The point mutation at the residue 6 (M6A) was totally defective for both CSR and SHM. The mutations at residues 7, 9, and 10 reduced SHM as well as CSR, albeit to a less extent. By contrast, the R8A mutant rather augmented CSR and SHM activities.
Although all of R8A, R9A, and K10R mutants had significant modification of their activities, none of them changed the interaction with Spt6 (Fig. 6B). Conversely, N7A augmented interaction with Spt6, although N7A reduced both CSR and SHM activities. M6A that abolished Spt6 interaction lost both SHM and CSR, although Spt6 is involved in only CSR. Human AID mutation M6T also lost both CSR and SHM (25). It is possible that M6A mutation altered the gross structure of AID to abolish the DNA cleavage function, resulting in the loss of both CSR and SHM. Although the N-terminal region of AID seems to be responsible for its interaction with the C-terminal region of Spt6, it is not clear whether this interaction is essential for the DNA cleavage function of AID.
How Does Spt6 Differentially Regulate CSR and SHM?
The target specificity of known specific recombination is determined by combination of cis elements (the DNA sequence/structure) and trans elements (DNA binding proteins and the chromatin modification mark of the target locus). In VDJ recombination in the Ig genes, the recombination signal sequence is widely distributed in the genome, but the chromatin modification [i.e., histone3 lysine4 trimethylation (H3K4me3)] recognized by RAG2 is essential to cleave the accurate target (26, 27). In meiotic recombination, Spo11 (topoisomerase II) cleaves at loosely conserved DNA target sequences that are also recognized by zinc finger-histone methyltransferase (PRDM9) to generate H3K4me3 at the target chromatin (28–31). Without PRDM9, meiotic recombination is abortive. We have also shown that H3K4me3 at the target S region is essential for CSR (32). The FACT complex composed of SSRP1 and Spt16 is a histone chaperone and modulates the histone transmodification cascade. We have shown that the FACT complex is essential for CSR (32). In the absence of FACT, H3K4 trimethyl modifications are reduced at the Sμ and Sα regions, which is associated with S region cleavage defect.
From these studies, it is likely that Spt6 can determine the target specificity of CSR at least by two strategies: (i) recognition of DNA sequence or (ii) modification of chromatin. Because Spt6 does not bind DNA directly, it is unlikely that Spt6 directly recruits a DNA cleaving enzyme to any DNA region. In addition, Spt6 associates with RNA polymerase II, which binds both V and S regions. Spt6 is thus unlikely to guide AID specifically to S regions. Because Spt6 is another histone chaperone protein, it is important to examine whether Spt6 also affects the histone modification cascade and thus causes defect in CSR. It is also interesting to analyze why Spt6 is slightly inhibitory to SHM. The histone modification cascade in the S region and V region may be different, which triggers interesting possibilities for differential regulation of SHM vs. CSR.
There are several other possible mechanisms whereby Spt6 differentially regulates CSR and SHM. Because Spt6 has also been reported to direct Iws1-dependent mRNA splicing and export (33, 34), CSR-related function of Spt6 could involve mRNA splicing and export. Stp6 is also involved in transcriptional regulation of a large number of genes, some of which may be responsible only for CSR. Further analyses are required to uncover the precise role of Spt6 in regulation of CSR but not SHM.
Materials and Methods
RNA Interference.
A diced siRNA (d-siRNA) pool was prepared using BLOCK-iT Complete Dicer RNAi Kit according to the manufacture's instructions (Invitrogen). Primers used to amplify template cDNAs for AID, Trim28, and Spt6 of mouse origin were as follows: mAID-F: 5′-CAA GGG ACG GCA TGA GAC CTA CCT-3′; mAID-R: 5′-TCT CGC AAG TCA TCG ACT TCG TAC-3′; mTrim28-F: 5′-CCA AGG AGG TTC GAA GCT CGA TCC-3′; mTrim28-R: 5′-GGA CCT TCA GTC AGA GGC ATC AAC-3′; mSpt6-F: 5′-CAG CAG TTC CTC TAC GTG CAA ATG-3′; and mSpt6-R: 5′-ACT GGA TCA AGG CCT GGC TGT AAG-3′. Stealth siRNAs were introduced into CH12F3-2A or BL2 cells using Amaxa Nucleofector (Amaxa Biosystems). Stealth siRNAs were purchased from Invitrogen: mSpt6-1, -2, -3, and -4 (MSS209819, 209820, 209821, NM_009297_stealth_3806), AID (MSS235859), hSPT6-1, -2, and -3 (HSS110374, 110375, 110376), hAID-1, -2, and -3 (HSS126211, 126212, 126213), mTrim28-1, -2, -3, and -4 (MSS211796, 211797 211798, NM_011588_stealth_1165), and hTRIM28-1, -2, -3, and -4 (HSS115468, 115470, NM_005762_stealth_883, NM_005762_stealth_2386). The efficiency of nucleofection was confirmed to be more than 90% by introducing fluorescein-labeled siRAN oligo.
CSR Assay.
CH12F3-2A cells were stimulated for 24 h with CD40L, TGF-β, and IL-4 24 h after introducing siRNA. The surface expression of IgM and IgA was analyzed by staining cells with FITC-conjugated anti-mouse IgM (Southern Biotechnology Associates) and PE-conjugated anti-mouse IgA (Southern Biotechnology Associates). Flow cytometric analyses were performed with a FACSCalibur, and data were analyzed by CellQuest software (BD Biosciences). Live cells were selected for the analyses by forward- and side-scatter intensity and propidium iodide (PI) gatings. CH12F3-2A cells expressing AID-ER were stimulated with OHT and TGF-β for 24 h after introducing siRNA, and the surface expression of IgM and IgA was analyzed by flow cytometry as described above. NIH 3T3 cells expressing the artificial switch substrate SCI(μ, α) and mAID-ER were introduced with d-siRNA and a DsRed-expressing plasmid as a transfection indicator using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were stimulated with OHT 24 for 36 h and stained with allophyco-cyanin–conjugated anti-mouse CD8α (eBioscience). The amounts of μGLT and αGLT were evaluated by quantitative PCR as described previously (32).
SHM Assay.
The hygromycin phosphotransferase and EGFP cDNA were fused in-frame in pFB to generate an artificial SHM substrate, pFB-HyGFP. BL2 cells were introduced with AID JP8Bdel-ER and pFB-HyGFP by retroviral infection. A clone expressing AID JP8Bdel and pFB-HyGFP was chosen after selection with puromycin and hygromycin. The clone was stimulated for 24 h with OHT 24 h after introducing siRNA and incubated for an additional 48 h in the absence of OHT. Expression of GFP and survival were evaluated by flow cytometry. Live cells were selected for the analyses by forward- and side-scatter intensity and PI gatings. Genomic DNA was extracted, and GFP sequence was amplified and analyzed. NIH 3T3 cells harboring a SHM substrate pI were infected with retroviruses expressing wild-type and mutant AID-GFs and cultured for 7 d. Genomic DNA was extracted and GFP sequence was amplified and analyzed.
Supplementary Material
Acknowledgments
We thank Dr. H. Handa for anti-Spt6 mAb, Dr. Watanabe for pACT2 mouse pre-B-cell cDNA library, Dr. P. Chambon for Trim28f/f mice, and Ms. T. Toyoshima for her excellent technical assistance. This work was supported by Grant-in-Aid 17002015 for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104423108/-/DCSupplemental.
References
- 1.Kinoshita K, Honjo T. Linking class-switch recombination with somatic hypermutation. Nat Rev Mol Cell Biol. 2001;2:493–503. doi: 10.1038/35080033. [DOI] [PubMed] [Google Scholar]
- 2.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 4.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 5.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 6.Nambu Y, et al. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 8.Basu U, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Geraldes P, Platt JL, Cascalho M. The double-edged sword of activation-induced cytidine deaminase. J Immunol. 2005;174:934–941. doi: 10.4049/jimmunol.174.2.934. [DOI] [PubMed] [Google Scholar]
- 11.MacDuff DA, Neuberger MS, Harris RS. MDM2 can interact with the C-terminus of AID but it is inessential for antibody diversification in DT40 B cells. Mol Immunol. 2006;43:1099–1108. doi: 10.1016/j.molimm.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Conticello SG, et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han L, Masani S, Yu K. Cutting edge: CTNNBL1 is dispensable for Ig class switch recombination. J Immunol. 2010;185:1379–1381. doi: 10.4049/jimmunol.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinkura R, et al. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 19.Doi T, et al. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2009;106:2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei M, et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 21.Gazumyan A, et al. Amino-terminal phosphorylation of activation-induced cytidine deaminase suppresses c-myc/IgH translocation. Mol Cell Biol. 2011;31:442–449. doi: 10.1128/MCB.00349-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ta VT, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 23.Nonaka T, et al. Carboxy-terminal domain of AID required for its mRNA complex formation in vivo. Proc Natl Acad Sci USA. 2009;106:2747–2751. doi: 10.1073/pnas.0812957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. The AID enzyme induces class switch recombination in fibroblasts. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 25.Durandy A, Peron S, Taubenheim N, Fischer A. Activation-induced cytidine deaminase: structure-function relationship as based on the study of mutants. Hum Mutat. 2006;27:1185–1191. doi: 10.1002/humu.20414. [DOI] [PubMed] [Google Scholar]
- 26.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahls WP, Davidson MK. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 2010;26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2010;107:22190–22195. doi: 10.1073/pnas.1016923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.