Abstract
Protein translocation in Escherichia coli is mediated by the translocase that in its minimal form consists of the protein-conducting channel SecYEG, and the motor protein, SecA. SecYEG forms a narrow pore in the membrane that allows passage of unfolded proteins only. Molecular dynamics simulations suggest that the maximal width of the central pore of SecYEG is limited to 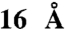 . To access the functional size of the SecYEG pore, the precursor of outer membrane protein A was modified with rigid spherical tetraarylmethane derivatives of different diameters at a unique cysteine residue. SecYEG allowed the unrestricted passage of the precursor of outer membrane protein A conjugates carrying tetraarylmethanes with diameters up to
. To access the functional size of the SecYEG pore, the precursor of outer membrane protein A was modified with rigid spherical tetraarylmethane derivatives of different diameters at a unique cysteine residue. SecYEG allowed the unrestricted passage of the precursor of outer membrane protein A conjugates carrying tetraarylmethanes with diameters up to 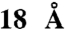 , whereas a
, whereas a 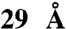 sized molecule blocked the translocation pore. Translocation of the protein-organic molecule hybrids was strictly proton motive force-dependent and occurred at a single pore. With an average diameter of an unfolded polypeptide chain of
sized molecule blocked the translocation pore. Translocation of the protein-organic molecule hybrids was strictly proton motive force-dependent and occurred at a single pore. With an average diameter of an unfolded polypeptide chain of 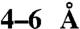 , the pore accommodates structures of at least
, the pore accommodates structures of at least 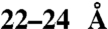 , which is vastly larger than the predicted maximal width of a single pore by molecular dynamics simulations.
, which is vastly larger than the predicted maximal width of a single pore by molecular dynamics simulations.
Keywords: secretion, Sec-system
In Escherichia coli, about 30% of the proteins synthesized in the cell accomplish their function outside the cytoplasm. Consequently, these proteins need to be translocated across or inserted into the inner membrane. The main system involved in protein translocation and membrane protein insertion is the Sec translocase with, as central component, a membrane-embedded protein-conducting pore, the SecYEG complex (also termed translocon) (1). Most membrane proteins are targeted to SecYEG as ribosome-bound nascent chains involving the signal recognition particle (SRP) and the SRP receptor (FtsY). The ribosome subsequently docks onto the SecYEG complex, and while chain elongation continues, the newly synthesized membrane protein is threaded into the membrane. The majority of the secretory proteins (preproteins) are targeted to the membrane in a posttranslational fashion. This involves the molecular chaperone SecB that transfers the preprotein to the SecYEG-bound motor protein SecA. SecA utilizes cycles of ATP binding and hydrolysis to pass the preprotein in a stepwise fashion through the translocon (2). SecYEG is the integral membrane heterotrimeric complex (3) and constitutes the translocation pore. SecY forms the core of this pore. Based on the X-ray structure of the homologous SecYEβ complex from the archaeon Methanocaldococcus jannaschii (4), SecY consists of 10 transmembrane segments (TMs) that are organized as two halves: the N-terminal TMs 1–5 and the C-terminal TMs 6–10. The two halves are hinged by a loop that connects TMs 5 and 6 giving the overall structure a clamshell-like conformation (5). The clamshell encompasses a central pore-like structure with a funnel like appearance with, in the middle, a hydrophobic constriction. At the periplasmic face of the membrane this pore is closed by a reentrance loop (“plug”) that connects TMs 1 with TMs 2. It has been proposed that the inserting signal sequence of the preprotein inserts at a lipid exposed lateral gate between TM2 and TM7 whereupon the clamshell is opened through a widening of the central constriction and a displacement of the plug. The SecYEβ complex of M. jannaschii has been crystallized in an idle state in the absence of the SecA motor domain or the ribosome, and is considered as a resting state, in which the pore is tightly sealed by the central constriction comprises six hydrophobic residues and the plug domain (6). The structure of a SecA–SecYEG complex of Thermotoga maritima suggests a preopen state of the channel with a major movement of the lateral gate helices TM7, TM8, and TM5, and a partial displacement of the plug leaving a narrow gap in the lateral gate of 5 Å (7). A recent crosslinking analysis of the lateral gate region suggested that the lateral gate needs to open up to at least 8 Å to support protein translocation (8). In membranes, SecYEG forms higher order oligomers, most notably dimers (9, 10), and this oligomerization is promoted by SecA and by the ribosome. A cryo-EM structure of the ribosome-bound E. coli SecYEG complex with an inserting membrane protein suggests that SecYEG binds the ribosome as a dimer with only one of the pores accommodating the translocating polypeptide chain (11). A crosslinking analysis of a SecA-associated preprotein translocation intermediate indicates an association with only one of the two SecYEG monomers (12). Thus far, it is unknown if the dimeric represents a functional or structural unit. In this respect, a recent cryo-EM analysis the homologous mammalian and yeast Sec61p complex indicates the presence of a single pore bound to the ribosome (13).
A central unresolved question concerns the functional width of the translocation pore. Molecular dynamics has been employed to study the plasticity of the pore formed by a SecYEG monomer (14–16). By pushing virtual soft balls through a single SecY pore, a maximal functional size of the pore of 16 Å has been suggested without the need for lateral gate opening (16). On the other hand, experimental studies with microsomes harboring the eukaryotic Sec61 complex indicate a pore diameter of 40–60 Å in the active state (17). The SecYEG complex seems rather promiscuous as it can translocate preproteins that are chemically cross-linked to nonpolypeptide constituents (18, 19). Here, we have employed a preprotein conjugated to large rigid spherical molecules with defined molecular dimensions to probe the diameter of the translocation pore in its active state. The data indicate that the active pore diameter by far exceeds the estimate made by the molecular dynamics simulations of the monomeric pore suggesting a more complex pore geometry.
Results
Synthesis of Tetraarylmethanes.
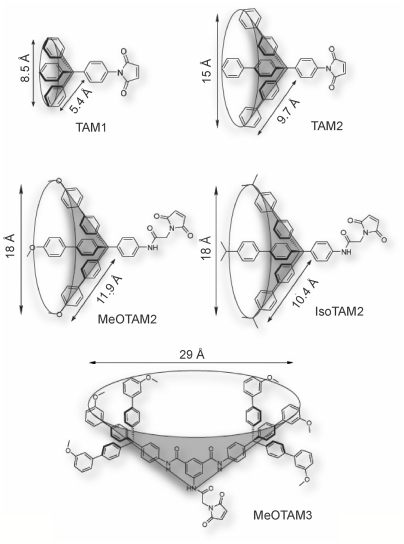
To access the size exclusion limit of the protein-conducting pore, organic compounds were synthesized with a precisely defined and systematically increasing size (Fig. 1 and SI Appendix). The nature of these compounds resembles the methane structure in which the carbon atom carries a sp3 hybridization. In this way, the phenyl and biphenyl groups, which are used as substituent, are oriented toward the x, y, and z axes giving the molecule the desired bulkiness and a spherical shape. Steric hindrance associated with the aromatic rings prevents coplanarity of the system and gives the desired rigidity. In addition, each molecule synthesized carries a maleimide group allowing the formation of a covalent protein-organic compound conjugate via a single cysteine present in the precursor of OmpA. The size of the compounds refers to the distance between the apical hydrogen atoms of the phenyl, biphenyl, and substituted biphenyl groups. The sizes are: approximately 8.5 Å for 1-(4-trityl-phenyl)pyrrole-2,5-dione (TAM1), approximately 15 Å for 1-[4-(tris-biphenyl-yl-methyl)phenyl]-pyrrole-2,5-dione (TAM2), approximately 18 Å for 2-(2,5-Dioxo-2,5-dihydro-pyrrol-1-yl)-N-{4-[tris-(4′-isopropyl-biphenyl-4-yl)methyl]-phenyl}-acetamide (IsoTAM2) and 2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-N-(4-(tris(4′-methoxybiphenyl-4-yl) methyl)phenyl) acetamide (MeOTAM2), and approximately 29 Å for 5-(2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)acetamido)-N1,N3-bis(4-(tris(3′-methoxybiphenyl-4-yl)methyl)phenyl)isophthalamide (MeOTAM3). Due to the presence of the aromatic component in the molecules the basic structure of the molecules has a high hydrophobicity. This hydrophobicity was decreased by modification of the diphenyl groups with methyl-oxy (MeO) groups as shown in MeOTAM2. The rigid conical shaped molecules conjugated to the preprotein proOmpA have a molecular weight of 415.15 (TAM1), 643.25 (TAM2), 849.70 (IsoTAM2), 790.90 (MeOTAM2), and 1608.1 (MeOTAM3), respectively.
Fig. 1.
Overview of the structures of the different tetraarylmethanes used to label proOmpAS245C. TAM1, 1-(4-trityl-phenyl)pyrrole-2,5-dione; TAM2, 1-[4-(tris-biphenyl-yl-methyl)phenyl]-pyrrole-2,5-dione; IsoTAM2, 2-(2,5-Dioxo-2,5-dihydro-pyrrol-1-yl)-N-{4-[tris-(4′-isopropyl-biphenyl-4-yl)methyl]-phenyl}-acetamide; MeOTAM2, 2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-N-(4-(tris(4′-methoxybiphenyl-4-yl)methyl)phenyl)acetamide; and MeOTAM3, 5-(2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)acetamido)-N1,N3-bis(4-(tris(3′-methoxybiphenyl-4-yl)methyl)phenyl)isophthalamide.
Conjugation of proOmpA with Spherical Tetraarylmethanes.
The tetraarylmethanes maleimide derivates were conjugated to a unique cysteine of the precursor protein proOmpA (S245C). A position in the main chain was chosen rather than at the C-terminal end of proOmpA to assure that the organic molecule passes the pore in combination with the polypeptide chain, which substantially adds to size to be translocated. Because the organic compounds are not readily soluble in water, the compounds were dissolved in an appropriate organic solvent and subsequently added to proOmpA that was denatured in urea. After labeling, the derivatized proOmpA was precipitated with trichloroacetic acid, washed with acetone and dissolved in urea buffer. To determine the extent of labeling, conjugated proteins were reduced with tris-(2-carboxyethyl) phosphine (TCEP) and labeled with fluorescein-5-maleimide (Fmal). The fluorescence intensity of the conjugated and subsequently Fmal labeled proOmpA was compared with that of proOmpA labeled with Fmal only (Fig. S1). The IsoTAM2, MeOTAM2, and MeOTAM3 derivatives were hardly labeled with Fmal indicting that conjugation of proOmpA (S245C) with these compounds was almost 100%. With TAM1 and TAM2 conjugated proOmpA an approximately 10% labeling with Fmal was observed (Fig. S1). However, control experiments with the cysteineless proOmpA indicate a 5% of nonspecific labeling with Fmal. Taking this into account we concluded that for all tetraarylmethane maleimide derivatives the degree of labeling of proOmpA (S245C) is ≥95%. Except for the largest MeOTAM3 (Fig. 2B), labeling of proOmpA (S245C) with the other tetraarylmethane maleimides did not result in a significant change in the mobility of proOmpA on SDS-PAGE. This is likely due to the small molecular size of the conjugates (400–800 Da) and because the derivatization does not affect the overall charge of proOmpA.
Fig. 2.
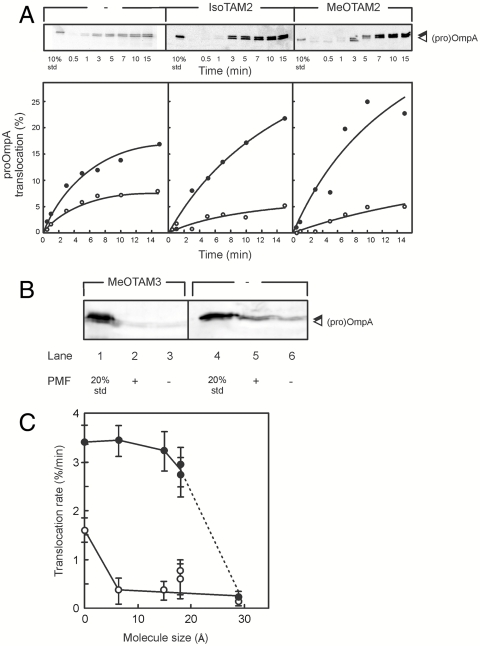
Translocation of proOmpA tetraarylmethane depends on the PMF. The different proOmpA conjugates were diluted into translocation buffer containing SecA (20 μg/mL), SecB (32 μg/mL), ATP (1 mM), and 10 μg IMVs. At different time intervals the translocation reaction was terminated by proteinase K treatment on ice. Samples were precipitated with trichloric acid (TCA) and protease protected material was analyzed by SDS-PAGE and immunoblotting using a polyclonal antibody against OmpA that recognizes the C-terminal end of proOmpA. Translocation reactions were performed in the presence and absence of a PMF. To dissipate the PMF nigericin and valinomycin (1 μM final concentration) was added to the reaction mix. (A) Translocation of wild-type proOmpA and the different proOmpA conjugates, in the presence (Left) and absence (Right) of a PMF. (B) Translocation of MeOTAM3-proOmpA in the presence and absence of a PMF. (C) Plot of the translocation rate versus the molecular size of the tetraarylmethane-conjugated to proOmpAS245C. Closed dots, +PMF; and open dots, −PMF.
Translocation of Tetraarylmethane proOmpA Conjugates by the SecYEG Complex.
The proOmpA conjugates labeled with the different tetraarylmethanes were assayed for translocation using inner membrane vesicles (IMVs) of E. coli strain UH203 containing overexpressed levels of SecYEG. Translocation assays were performed in the presence and absence of the ionophores nigericin and valinomycin to assess the role of the proton motive force (PMF). Under the conditions used unlabeled proOmpA translocated efficiently into the IMVs (Fig. 2A, WT) and translocation was two- to 2.5-fold stimulated by the PMF (Fig. 2A, −PMF vs. +PMF, open and filled dots, respectively). In the presence of a PMF the proOmpA tetraarylmethane conjugates translocated into UH203 IMVs as efficiently as unlabeled proOmpA as shown for proOmpA-IsoTAM2 (Fig. 2A, +PMF filled dots), except for the MeOTAM3-labeled proOmpA that was not translocated (Fig. 2B). In contrast to the unlabeled proOmpA, the translocation of the proOmpA tetraarylmethane conjugates was more dependent on the presence of a PMF (Fig. 2A, −PMF vs. +PMF). The translocation of MeOTAM3-labeled proOmpA was not restored by the PMF (Fig. 2B). Fig. 2C summarizes this data showing the translocation rate of the various proOmpA tetraarylmethane conjugates in the presence of the PMF plotted against the molecular size of the conjugate.
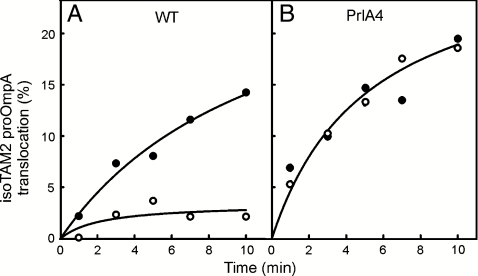
Previous studies have shown that the PMF-dependent translocation of proOmpA is suppressed in the PrlA4 mutant strain (20). The PrlA4 SecY protein contains two mutations, F286Y and I408N, where the latter is responsible for the suppressor effect (21). To determine if the strong PMF-dependent translocation of the proOmpA tetraarylmethane conjugates is suppressed by the PrlA4 strain we analyzed the translocation of proOmpA conjugated with IsoTAM2. Whereas translocation of IsoTAM2-proOmpA into wild-type IMVs is strongly dependent on the PMF (Fig. 3A) this PMF dependence is completely relieved with PrlA4 IMVs (Fig. 3B). As shown before for unconjugated proOmpA (22), translocation of IsoTAM2-proOmpA into PrlA4 IMVs is more efficient as compared to translocation into wild-type IMVs.
Fig. 3.
The SecY PrlA4 mutation relieves the strong PMF-dependent translocation of IsoTAM2-proOmpA. Translocation reactions were performed in the presence (black dots) and absence (white dots) of a PMF (A) with wild-type IMVs and (B) IMVs derived from the PrlA4 mutant.
Translocation Arrest by Tetraarylmethane-Conjugated proOmpA.
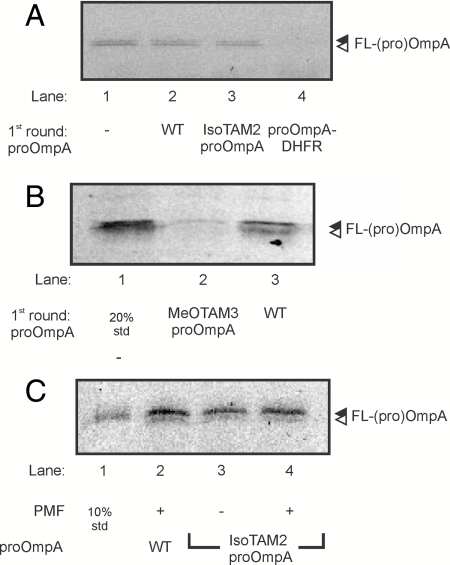
To determine if the tetraarylmethane conjugate arrests translocation because of blocking the translocation pore, we performed a translocation reaction using saturating concentrations of proOmpA, IsoTAM2-proOmpA, MeOTAM3, or proOmpA-dihydrofolate (DHFR). Addition of methotrexate and NADPH to the latter fusion construct leads to tight folding of the DHFR domain and results in an arrest in translocation of proOmpA-DHFR (23). After translocation of the different proteins, IMVs were recovered by centrifugation through a sucrose cushion and used in a second translocation reaction using Fmal-proOmpA as substrate. When the first translocation reaction was performed in the absence of a preprotein, Fmal-proOmpA was readily translocated into the IMVs in the second translocation reaction (Fig. 4A, lane 1). In contrast, IMVs used to translocate proOmpA-DHFR in the first translocation reaction were unable to translocate Fmal-proOmpA (lane 4). IMVs used in a translocation reaction with proOmpA or IsoTAM2-proOmpA showed similar levels of Fmal-proOmpA translocation as IMVs incubated without a preprotein (lanes 2 and 3). In contrast, when first MeOTAM3-proOmpA was translocated into the IMVs, translocation of Fmal-proOmpA in a second round of translocation was completely blocked (Fig. 4B, lane 2). These results indicate that even in the presence of a PMF, the largest molecule tested; i.e., MeOTAM3 causes a block of the translocation pore.
Fig. 4.
Translocation of MeOTAM3-proOmpA blocks the SecYEG pore. (A) IMVs containing overexpressed levels of SecYEG were used for a translocation reaction in the absence of proOmpA (lane 1), in the presence of wild-type proOmpA (lane 2), in the presence of IsoTAM2-proOmpA (lane 3), and in the presence of proOmpA-DHFR kept in its folded state by the addition of 1 mM NADPH and 50 μM methotrexate (lane 4). After 30 min at 37 °C the vesicles were recovered through a sucrose cushion and used for a second round of translocation with Fmal-labeled proOmpA. (B) Translocation reaction in the absence of proOmpA (lane 1), in the presence of wild-type proOmpA (lane 2), and in the presence of MeOTAM3-proOmpA (lane 3). After 30 min at 37 °C the vesicles were recovered through a sucrose cushion and used for a second round of translocation with Fmal-labeled proOmpA. All reactions were performed in the presence of a PMF. Samples were precipitated with TCA and protease protected material was analyzed by SDS-PAGE and in gel fluorescence. (C) Translocation of isoTAM2-proOmpA was performed with a limiting amount of IMVs in the presence and absence of a PMF. After 10 min at 37 °C, the reaction was stopped on ice and the PMF was dissipated in the reactions were translocation was performed in the presence of a PMF. Subsequently, the IMVs were isolated by centrifugation through a 0.8 M sucrose cushion and used in a second translocation reaction using Fmal-labeled proOmpA as substrate. After 10 min at 37 °C the reactions were stopped by the addition of proteinase K and incubated for 30 min on ice. Samples were precipitated with TCA and protease protected material was analyzed by SDS-PAGE (12% acrylamide) and in gel fluorescence. 10%: 10 percent of the Fmal-labeled proOmpA used in the translocation reaction.
As translocation of the proOmpA tetraarylmethane derivatives is strongly dependent on the PMF we investigated this requirement in further detail. To this end, IsoTAM2-proOmpA was translocated into IMVs both in the presence and in the absence of a PMF. A collapse of the PMF was induced by the addition of the ionophores valinomycin and nigericin. After the translocation reaction, reactions performed in the presence of a PMF were supplemented with valinomycin/nigericin and IMVs were recovered as described above. The reisolated IMVs were used in a second translocation reaction using Fmal-proOmpA as substrate. Both IMVs used to translocate IsoTAM2-proOmpA in the absence and presence of a PMF showed Fmal-proOmpA translocation (Fig. 4C, lane 3 and 4) with an efficiency comparable to IMVs that had not been incubated with proOmpA (lane 2). This result demonstrates that even in the absence of a PMF when translocation is slow (Fig. 2A), the smaller conjugates do not block the SecY pore (Fig. 4C).
Translocation by a SecYEG Pore that Is Constrained by a Cross-Linked Lateral Gate.
Tetraarylmethanes are relatively hydrophobic molecules. Therefore, the possibility exists that a large part of these molecules cross the membrane by sliding along the interface of the pore and the lipid bilayer, possibly at the lateral gate. To address this possibility, two strategies were adopted: First, we synthesized a tetraarylmethane in which the hydrophobic isopropyl groups were replaced by more hydrophilic methoxy groups. In this way, the outer shell of the molecule is more hydrophilic in nature thereby minimizing unwanted interactions with the lipid phase. Like the other conjugates, MeOTAM2-proOmpA was efficiently translocated into the IMVs in the presence of a PMF (Fig. 2A; MeOTAM2 and Fig. 2C). This indicates that the observed translocation characteristics of the proOmpA tetraarylmethane conjugates is due to the size of the tetraarylmethane and unrelated to their hydrophobicity.
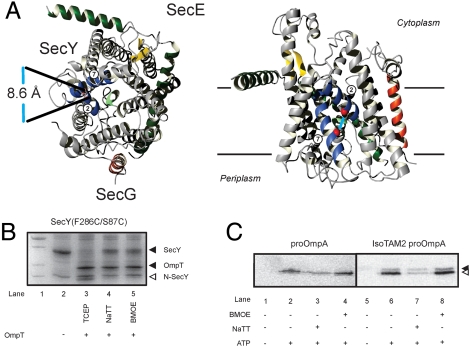
Second, a double cysteine SecY mutant (F286C and S87C) was used in which the lateral gate of the translocon at the interface of TM2 and TM7 can be closed by thiol-reactive cross linkers with different spacer lengths (Fig. 5A) (8) thereby forcing the passage of the tetraarylmethane through the membrane via the central hydrophilic pore. As described previously (8), IMVs containing SecY(S87C/F286C)EG were treated with the oxidizer tetrathionate (NaTT) to link TM2 and TM7 of the lateral gate by means of a disulfide bond, and by incubation with the cross-linker bis-maleimidoethane (BMOE) that introduces a spacer of approximately 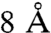 between the thiol groups. To determine the extent of crosslinking, the IMVs were treated with OmpT, an outer membrane protease that specifically cleaves SecY at the double arginine motif in the C4 loop. OmpT digestion of SecY that is not treated with NaTT or BMOE resulted in the formation of a typical 22 kDa N-terminal SecY fragment that can be visualized by SDS-PAGE and staining with Coomassie brilliant blue R250 (Fig. 5B, lane 3). In contrast, when SecY(S87C/F286C)EG IMVs were treated with NaTT or BMOE, SecY was cleaved by OmpT but the N- and C-terminal fragments of SecY remain cojoined and migrate on SDS-PAGE as a full length SecY protein with a more fuzzy appearance as compared to nondigested SecY (Fig. 5B, lanes 4 and 5). As virtually no 22-kDa fragment was detected we conclude that the crosslinking of the two cysteines in SecY was very efficient. Next, the different cross-linked IMVs were tested for translocation of the largest conjugated preprotein that still translocates, IsoTAM2-proOmpA. Translocation of IsoTAM2-proOmpA was as efficient as that of Fmal-proOmpA when reduced SecY(S87C/F286C)EG IMVs were used (Fig. 5C, lanes 2 and 6). In contrast, with IMVs treated with NaTT, translocation of both Fmal-proOmpA and IsoTAM2-proOmpA was drastically reduced (compare lane 3 vs. 2 and lane 7 vs. 6) to the levels observed with IMVs containing the native levels of wild-type SecYEG (8). The BMOE treated IMVs, however, showed translocation efficiencies for both substrates that are comparable to that of nontreated IMVs (compare lane 4 vs. 2 and lane 8 vs. 6). This indicates that translocation of the IsoTAM2 is not hindered by a cross-linker that fixes the lateral gate formed by TM2 and TM7, but that still allows opening of the central channel. Moreover, this result suggests that the tetraarylmethane is translocated via a single pore and that it does not cross the membrane at the interface of the lateral gate/pore region and lipid bilayer.
between the thiol groups. To determine the extent of crosslinking, the IMVs were treated with OmpT, an outer membrane protease that specifically cleaves SecY at the double arginine motif in the C4 loop. OmpT digestion of SecY that is not treated with NaTT or BMOE resulted in the formation of a typical 22 kDa N-terminal SecY fragment that can be visualized by SDS-PAGE and staining with Coomassie brilliant blue R250 (Fig. 5B, lane 3). In contrast, when SecY(S87C/F286C)EG IMVs were treated with NaTT or BMOE, SecY was cleaved by OmpT but the N- and C-terminal fragments of SecY remain cojoined and migrate on SDS-PAGE as a full length SecY protein with a more fuzzy appearance as compared to nondigested SecY (Fig. 5B, lanes 4 and 5). As virtually no 22-kDa fragment was detected we conclude that the crosslinking of the two cysteines in SecY was very efficient. Next, the different cross-linked IMVs were tested for translocation of the largest conjugated preprotein that still translocates, IsoTAM2-proOmpA. Translocation of IsoTAM2-proOmpA was as efficient as that of Fmal-proOmpA when reduced SecY(S87C/F286C)EG IMVs were used (Fig. 5C, lanes 2 and 6). In contrast, with IMVs treated with NaTT, translocation of both Fmal-proOmpA and IsoTAM2-proOmpA was drastically reduced (compare lane 3 vs. 2 and lane 7 vs. 6) to the levels observed with IMVs containing the native levels of wild-type SecYEG (8). The BMOE treated IMVs, however, showed translocation efficiencies for both substrates that are comparable to that of nontreated IMVs (compare lane 4 vs. 2 and lane 8 vs. 6). This indicates that translocation of the IsoTAM2 is not hindered by a cross-linker that fixes the lateral gate formed by TM2 and TM7, but that still allows opening of the central channel. Moreover, this result suggests that the tetraarylmethane is translocated via a single pore and that it does not cross the membrane at the interface of the lateral gate/pore region and lipid bilayer.
Fig. 5.
IsoTAM2-proOmpA is translocated by SecY with a lateral gate that is constrained by a 8.6 Å cross-linker. (A) Top view (Left) and side view (Right) of the crystal structure of the M. jannaschii SecYEβ. SecE and SecG are indicated in green and orange, respectively. TM 2 and 7 that form the lateral gate are indicated in blue. The red balls indicate the crosslinking sites. (B) OmpT assay performed on IMVs containing the cysteineless and SecY(F286C/S87C)EG complex incubated with different chemical cross linkers. In the presence of sodium tetrathionate (lane 4), or bis-maleimidoethane (lane 5), the OmpT-treated SecY migrates as the uncleaved protein (lane 2). In the presence of TCEP, SecY is cleaved (lane 3). The molecular mass standard is indicated in lane 1. (C) Translocation of Fmal-proOmpA (Left) and IsoTAM2-proOmpA (Right) into SecY(F286C/S87C)EG IMVs under reducing conditions (lanes 2 and 6), or upon treatment with sodium tetrathionate (lanes 3 and 7), or bis-maleimidoethane (lanes 4 and 8). As a control, no ATP was added to the translocation reaction (lanes 1 and 5).
Discussion
In this study we investigated the diameter of the active SecYEG pore. For this purpose, different tetraarylmethanes were synthesized and covalently linked, via a maleimide group, to a unique cysteine residue at position 245 of the preprotein proOmpA. The synthesized tetraarylmethanes have spherical dimensions ranging from 8.5 up to 29 Å (Fig. 1). Due to their rigid structure they can be used as molecular rulers to access the size of the functional translocation pore. Remarkably, all synthesized tetraarylmethanes conjugated to proOmpA were readily translocated into E. coli IMVs except for the largest molecule MeOTAM3 that has a molecular dimension of approximately 29 Å. When the size of the unfolded polypeptide is taken into account, assuming an extended conformation of 4–6 Å, the overall diameter of the translocation pore must be at least approximately 22–24 Å. Surprisingly, this exceeds the expected size for a monomeric pore without a lateral gate opening as determined by molecular dynamics simulations. To test if these molecules indeed pass through a single pore and/or whether lateral gate opening is required, we employed a SecY mutant in which the lateral gate opening was controlled through the use of a site-specific crosslink between TM2 and TM7 that together form the lateral exit site (8). Herein, two unique cysteines were introduced in the lateral gate. These were chemically cross-linked by oxidation or by the use of the chemical cross-linker BMOE that separates the thiols by approximately 8 Å. When the lateral gate was constrained by oxidation, translocation of both proOmpA and the tetraarylmethane conjugates was blocked. However, when the lateral gate was cross-linked with BMOE, translocation occurred unrestricted. In addition, the large proOmpA-MeOTAM3 blocked the pore for subsequent rounds of translocation, whereas the smaller IsoTAM2 did not. Therefore, we conclude that the translocation pore can accommodate relatively large structures, which indicates a more complex pore geometry than previously suggested by molecular dynamics simulations (16).
To exclude the possibility that the hydrophobic nature of the tetraarylmethanes influences the translocation of the conjugates we decreased the hydrophobicity of the tetraarylmethane (IsoTAM2) by substituting each aromatic unit with a methyl-oxy group (MeOTAM2). The translocation kinetics of this proOmpA derivative was nearly indistinguishable from that of the other tetraarylmethanes indicating that hydrophobicity is not a major factor (Fig. 2B). As translocation of the proOmpA derivatives was also undisturbed with a SecYEG complex containing a fixed lateral gate, an interface translocation model of the tetrarylmethane molecules can be ruled out. Rather, the additional space provided by the opened lateral gate may contribute to the size of the active pore. The experimentally determined pore size of approximately 22–24 Å will be closed to the maximal pore diameter, as a further expansion of the tetraarylmethane sphere to 29 Å arrested translocation. This size is substantially smaller than the previously size of approximately 40–60 Å based on fluorescent quenching techniques (17). In this respect, the recent structure of SecYEG from T. maritima with SecA bound in an intermediate state of ATP-hydrolysis shows in comparison to the M. jannaschii SecYEβ structure, a partial opening of the lateral gate region around TM2 and TM7/8 (7) that points at a more complex pore geometry possibly including an opened lateral gate as an extension of the central pore.
Another characteristic feature of the translocation of proOmpA derivatized with tetraarylmethanes is the much stronger PMF-dependence than of wild-type proOmpA. Our data support the hypothesis that the PMF modulates the opening or even the width of the pore during translocation (20, 24). To further investigate the strong PMF-dependent translocation of proOmpA tetraarylmethane derivatives, translocation of proOmpA-IsoTAM2 was investigated with IMVs containing the PrlA4 mutant of SecY. The SecYEG pore of this mutant is thought to be in a relaxed state, probably because of a destabilization of the closed state (25). In IMVs containing the Prl4 mutant, translocation of proOmpA-IsoTAM2 indeed is independent of the PMF. Also, the translocation kinetics of proOmpA-IsoTAM2 into PrlA4 IMVs is increased as compared to wild-type IMVs as shown previously for wild-type preproteins.
Summarizing, our data suggest a high plasticity of the SecYEG translocation pore that can accommodate large nonpolypeptide moieties. Importantly, the data suggest that the lateral gate opening contributes to the functional pore size and that the PMF modulates the width of the translocation pore.
Materials and Methods
Materials.
SecA (26) and SecB (27) were purified as described. IMVs with overexpressed levels of SecYEG were obtained from E. coli strain UH203 transformed with pET610 (28). IMVs containing overexpressed levels of SecY(F286C/S87C) were obtained from E. coli strain SF100 transformed with pFE-SecY16 plasmid (8). OmpT was expressed from plasmid pND9 in strain SF100 and expressed under its own temperature sensitive promoter (29). The proOmpA cysteine mutant S245C was constructed with the QuickChange site-directed mutagenesis kit (Stratagene) using pET2345 containing the cysteineless proOmpA as a template (30). Primers used introduced in addition a silent MluI cutting site for cloning purposes: S245C forward primer, ccgaccgcat cggttgtgac gcgtacaacc agggtctg; S245C reverse primer, cagaccctgg ttgtacgcgt cacaaccgat gcggtcgg. The introduced mutations were confirmed by sequencing. ProOmpA(S245C) was purified as described previously (30) and further referred to as proOmpA.
Crosslinking of Lateral Gate.
IMVs containing SecY(S87C/F286C)EG were isolated as previously described (8). IMVs (1 mg of protein/mL) were incubated for 30 min at 37 °C with Na2S2O8 or BMOE at a final concentration of 1 mM and 300 μM respectively. To test the efficiency of the crosslinking IMVs were treated with 1 mg/mL OmpT in 50 mM Tris/HCl pH 7, 0.1% Triton X100 for 30 min at 37 °C. Samples were analyzed by SDS-PAGE gel (12% acrylamide), and Coomassie brilliant blue staining.
Tetraarylmethane Synthesis, proOmpA Labeling, and Translocation Assays.
See SI Appendix.
Supplementary Material
Acknowledgments.
This work was supported by the Royal Academy of Arts and Sciences of the Netherlands (KNAW), NanoNed (a national nanotechnology program coordinated by the Dutch Ministry of Economic Affairs), and the Zernike Institute for Advanced Materials.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101705108/-/DCSupplemental.
References
- 1.Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 3.Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 4.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 5.Clemons WM, Jr, Menetret JF, Akey CW, Rapoport TA. Structural insight into the protein translocation channel. Curr Opin Struct Biol. 2004;14:390–396. doi: 10.1016/j.sbi.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Li W, et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJM. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheuring J, et al. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J Mol Biol. 2005;354:258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 10.Hanein D, et al. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 11.Mitra K, et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 13.Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumbart J, Schulten K. Molecular dynamics studies of the archaeal translocon. Biophys J. 2006;90:2356–2367. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gumbart J, Schulten K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 16.Tian P, Andricioaei I. Size, motion, and function of the SecY translocon revealed by molecular dynamics simulations with virtual probes. Biophys J. 2006;90:2718–2730. doi: 10.1529/biophysj.105.073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 18.Tani K, Mizushima S. A chemically cross-linked nonlinear proOmpA molecule can be translocated into everted membrane vesicles of Escherichia coli in the presence of the proton motive force. FEBS Lett. 1991;285:127–131. doi: 10.1016/0014-5793(91)80741-k. [DOI] [PubMed] [Google Scholar]
- 19.Kato M, Mizushima S. Translocation of conjugated presecretory proteins possessing an internal non-peptide domain into everted membrane vesicles in Escherichia coli. J Biol Chem. 1993;268:3586–3593. [PubMed] [Google Scholar]
- 20.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci USA. 1996;93:5953–5957. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osborne RS, Silhavy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Wolk JP, et al. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 1998;17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arkowitz RA, Joly JC, Wickner W. Translocation can drive the unfolding of a preprotein domain. EMBO J. 1993;12:243–253. doi: 10.1002/j.1460-2075.1993.tb05650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tani K, Tokuda H, Mizushima S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J Biol Chem. 1990;265:17341–17347. [PubMed] [Google Scholar]
- 25.Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 1999;18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabelli RJ, Chen L, Tai PC, Oliver DB. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 27.Weiss JB, Ray PH, Bassford PJ., Jr Purified secB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc Natl Acad Sci USA. 1988;85:8978–8982. doi: 10.1073/pnas.85.23.8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufmann A, Manting EH, Veenendaal AK, Driessen AJM, van der Does C. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry. 1999;38:9115–9125. doi: 10.1021/bi990539d. [DOI] [PubMed] [Google Scholar]
- 29.Kramer RA, Dekker N, Egmond MR. Identification of active site serine and histidine residues in Escherichia coli outer membrane protease OmpT. FEBS Lett. 2000;468:220–224. doi: 10.1016/s0014-5793(00)01231-x. [DOI] [PubMed] [Google Scholar]
- 30.Crooke E, Brundage L, Rice M, Wickner W. ProOmpA spontaneously folds in a membrane assembly competent state which trigger factor stabilizes. EMBO J. 1988;7:1831–1835. doi: 10.1002/j.1460-2075.1988.tb03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.