Abstract
Notch signaling is a central regulator of differentiation in a variety of organisms and tissue types1. Its activity is controlled by the multi-subunit γ–secretase complex (γSE) complex2. Although Notch signaling can play both oncogenic and tumor suppressor roles in solid tumors, in the hematopoietic system, it is exclusively oncogenic, notably in T cell acute lymphoblastic leukemia (T-ALL), a disease characterized by Notch1 activating mutations3. Here we identify novel somatic inactivating Notch pathway mutations in a fraction of chronic myelomonocytic leukemia (CMML) patients. Inactivation of Notch signaling in mouse hematopoietic stem cells (HSC) resulted in an aberrant accumulation of granulocyte/monocyte progenitors (GMP), extramedullary hematopoieisis and the induction of CMML-like disease. Transcriptome analysis revealed that Notch signaling regulates an extensive myelomonocytic-specific gene signature, through the direct suppression of gene transcription by the Notch target Hes1. Our studies identify a novel role for Notch signaling during early hematopoietic stem cell differentiation and suggest that the Notch pathway can play both tumor-promoting and suppressive roles within the same tissue.
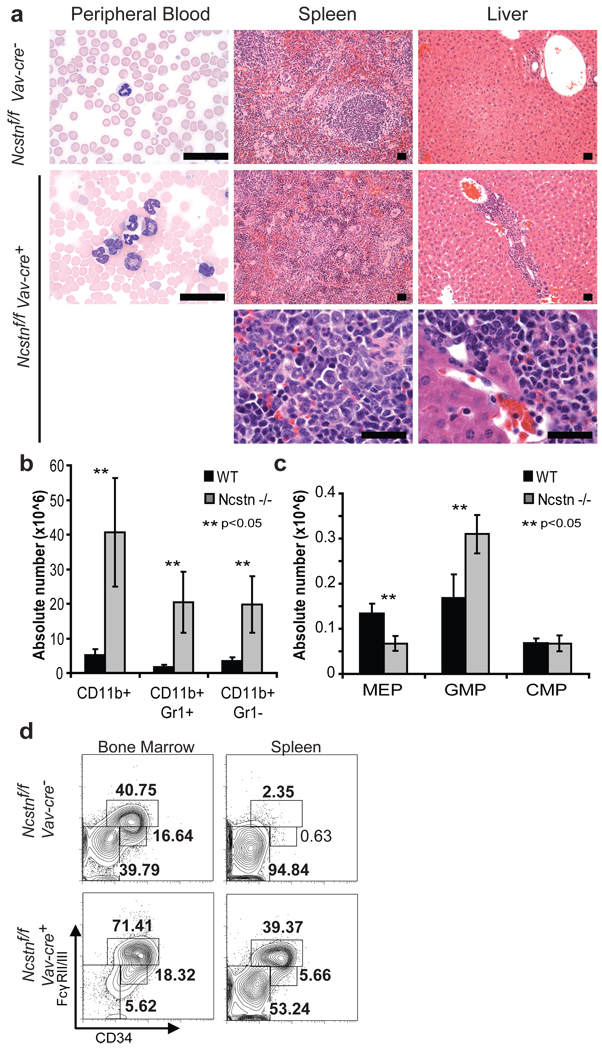
Notch is essential for the emergence of definitive hematopoiesis4, controls HSC differentiation to the T-cell lineage5,6 and is a major oncogene, as the majority of T-ALL patients harbor activating Notch1 mutations7. To study hematopoieis in the absence of any Notch-derived signal (as mammals express four different Notch receptors) we have targeted Nicastrin (Ncstn), a member of the γSE complex and one of the few non-redundant members of the pathway (Suppl. Fig. 1). We crossed the Ncstnf/f mice to both an inducible (Mx1-cre)8 and a hematopoietic-specific (Vav-cre)9 recombinase strain. Both modes of deletion (referred herein as Ncstn−/−) produced identical phenotypes. Unexpectedly, none of the Ncstn−/− mice survived longer than 20 weeks. Further analysis revealed a striking peripheral blood (PB) leukocytosis and monocytosis with enlargement of the spleen (Fig. 1a–b, Suppl. Fig. 2–3). Histological analysis of the spleen showed a marked expansion of the red pulp with diffuse infiltration by myeloid and monocytic cells. The infiltrating myeloid cells were partially myeloperoxidase positive and CD11b+ and/or Gr1+ (Fig. 1b, Suppl. Fig. 2–3). The increase in monocyte numbers was also observed in the bone marrow and liver (Fig. 1a and data not shown). Taken together, these findings were diagnostic for a myeloproliferative/myelodysplastic process and reminiscent of human chronic myelomonocytic leukemia (CMML). CMML is a myeloid malignancy, classified as a MPD/MDS overlap syndrome, which is characterized by monocytosis, myeloproliferation, and variable bone marrow dysplasia and by a high rate of progression to acute myeloid leukemia (AML)10.
Figure 1. Ncstn deficiency leads to CMML-like disease and a significant enlargement of the GMP progenitor population.
a, Histological analyzis showing accumulation of monocytes and granulocytes in peripheral blood (Wright Giemsa staining), spleen and liver (H&E staining). A magnification of each infiltrant is shown in the lower panel. b, Absolute numbers of each monocytic/granulocytic subset from the spleen of control and Ncstnf/f Vav-cre+ littermate animals (12 weeks of age, mean ±S.D. n=10). c, Detailed FACS analysis of bone marrow and spleen myeloid progenitors (MP: Lin−/c-Kit+/Sca-1−) populations of Ncstnf/f Vav-cre+ and Ncstnf/fVav-cre− littermates showing a significant enlargement of the GMP (FcγRII/III+, CD34=) subset. d, Absolute numbers of each progenitor subpopulation in the bone marrow (mean ±S.D. n=10).
Since monocytes and granulocytes originate from the GMP subset we examined the stem and progenitor cell populations in the bone marrow (BM). Ncstn deletion lead to an enlargement of the LineagenegSca1+c-Kit+ (LSK) and specifically the LSK CD150+CD48+ subset, a population shown to have a myeloid commitment bias11. This differentiation bias was coupled to a significant reduction of the lymphoid-biased multipotential progenitor population (L-MPP)12 (Suppl. Fig. 4). Moreover, there was a striking increase in the absolute numbers of both BM and spleen GMP cells (Fig. 1d) coupled to a decrease of the megakaryocyte-erythrocyte progenitor (MEP) population. This apparent predisposition towards GMP-derived lineages was also evident in vitro, as Ncstn−/− progenitors generated a larger number of CFU-GM and CFU-M colonies (Suppl. Fig. 5). Further studies also revealed a striking ability of the Ncstn−/− progenitors to serially replate (Suppl. Fig. 6), suggesting an increase in their self-renewal potential. Consistent with this idea, whole transcriptome profiling of Ncstn−/− GMP progenitors revealed enrichment of a “leukemic self-renewal” signature13. Finally, bone marrow transplantation assays demonstrated that the effects of Ncstn deletion were cell-autonomous (Suppl. Fig. 7)14.
Although the γSE complex has other described substrates2, we focused on Notch signaling due to our finding that Ncstn deletion led to known Notch−/− phenotypes, including a block in T-cell differentiation (Suppl. Fig. 8)15. To prove a connection to Notch signaling we have generated animals that conditionally lack the expression of three out of four Notch receptors (Mx1-cre+N1f/fN2f/fN3−/−)16. Strikingly, triple Notch1/2/3 deletion copied the Ncstn−/− phenotypes (Suppl. Fig. 9). Notch1–3−/− mice developed both CMML-like symptoms, and significant enlargement of the GMP population. Notch3 expression was dispensable, as simultaneous deletion of only Notch1 and Notch2 led to an identical CMML-like pathology. However, introduction of a single wild-type Notch1 or Notch2 allele was able to suppress the disease phenotype. Consistent with the importance of Notch1–2 receptor signaling in these stages, qPCR studies revealed expression of Notch1 and Notch2, but not Notch3 in wild-type stem and progenitors cells (Suppl. Fig. 10).
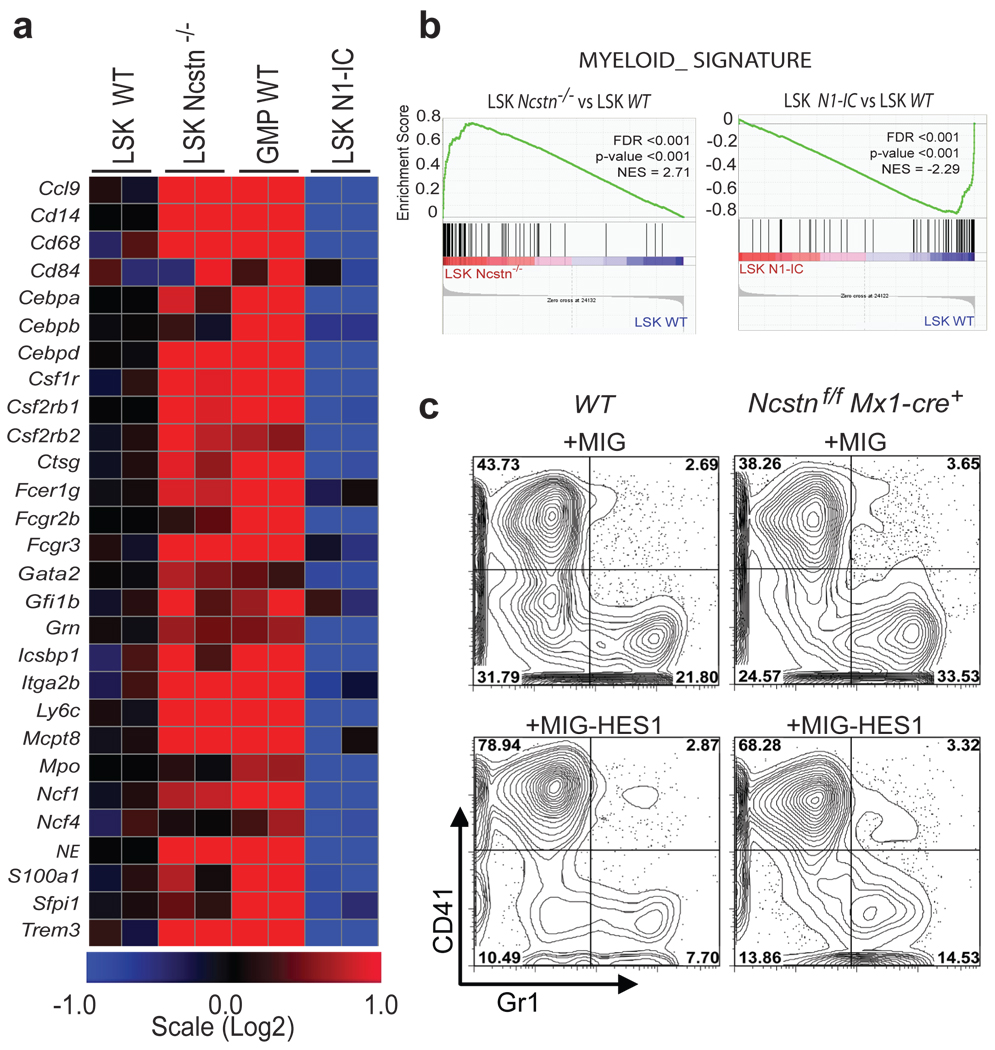
We next sought to delineate the mechanism by which Notch directs the regulation of early hematopoiesis. Deletion of the Ncstn did not lead to any alterations in the GMP cell cycle status or cell death rate (not shown). We hypothesized that γSE complex /Notch signals actively suppress a GMP-specific gene expression program. We sorted LSK and GMP cells and studied their transcriptome. This analysis revealed a statistically significant de-repression of an extended myeloid-specific program12 in Ncstn−/− LSK cells (Figure 2a and Suppl. Fig. 11). Gene-set enrichment analysis (GSEA) demonstrated a significant enrichment of myeloid-specific gene-sets within the Ncstn−/− LSK gene-signature (Fig. 2b and Suppl. Table 1). Further dissection of the LSK subset showed that the GMP gene expression program was initiated as early as the CD150+ HSC stage of differentiation, and persisted at the CD150− subset, which included multipotential progenitors (MPP) (Suppl. Fig. 11). Furthermore, we were able to show that gene expression in LSKs purified from an inducible Notch1 gain-of-function genetic model (Ef1α1-lsl-Notch1ICMx1-cre+)17 was inversely correlated with the loss-of-function signature seen in Ncstn−/− LSK progenitors (Figure 4a). Notch1IC expression led to suppression of myeloid-specific genes suggesting that activation of Notch signaling can alter the transcription and differentiation of uncommitted HSC and MPP cells. GSEA analysis further supported these findings as it showed a negative correlation between the Notch1IC LSKs and previously reported myeloid gene expression signatures (Fig. 2a–b, Suppl. Table 2 and Suppl. Fig. 11).
Figure 2. Notch signaling suppresses an extensive myeloid gene expression program through the induction of the transcriptional repressor Hes1.
a, Heat map showing regulation of genes representative of the myeloid-signature from the indicated cell populations and mice. b, Expression data were analyzed for lists of genes positively involved in myelopoiesis using GSEA analysis. Enrichment plots show up-regulation of myeloid-specific genes in Ncstnf/fMx1-cre+ and downregulation in Notch1IC Mx1-cre+ LSK cells (when compared to WT counterparts). c, Purified cKit+ progenitors from WT and Ncstnf/fMx1-cre+ mice were transduced with retroviruses encoding Hes-1 or empty vector, subsequently plated on methylcellulose for 7 days and analyzed for expression of myeloid or megakaryocyte differentiation markers (Gr1, CD41). A representative of four experiments is shown.
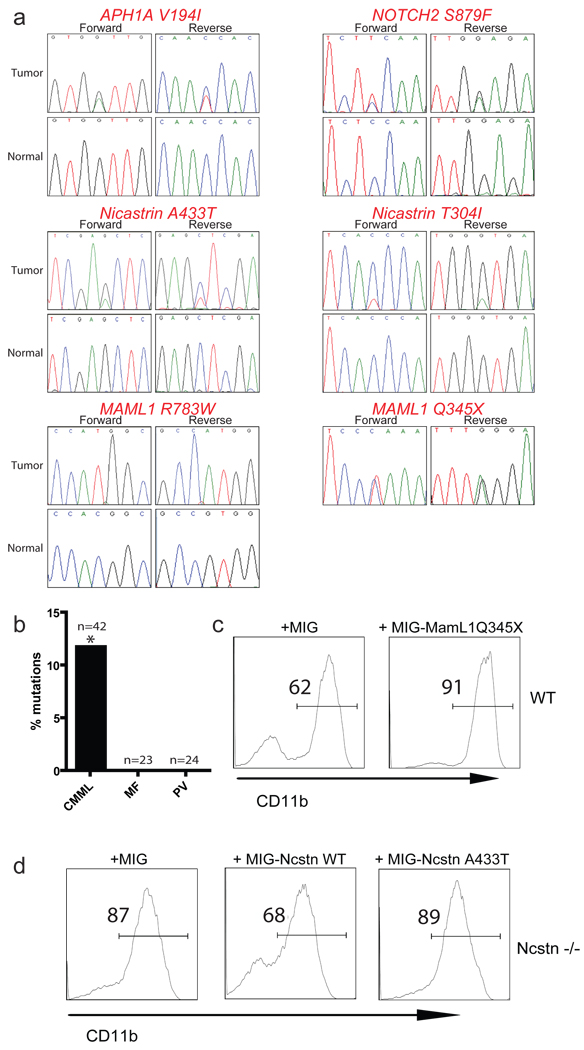
Figure 4. Novel, loss-of-function Notch pathway mutations in human CMML.
a, Sequence traces of identified Notch pathway mutations in tumor of CMML patients but not in normal tissues show somatic origin. b, Comparison of percentage of Notch pathway mutations in CMML, MF and PV patient speciments. The “*” denotes that only verified somatic CMML mutations are included. c, OP9-DL1 co-culture of wild-type LSK cells infected with specified constructs. Analysis of CD11b+ population was studied 14 days after the initiation of the culture. d, A similar experiments as in c utilizing LSK Ncstn−/− progenitors infected with the specified constructs. In all cases, a representative of more than three experiments is shown.
As Notch is thought to primarily function as a transcriptional activator, we hypothesized that its suppressive effects on GMP-specific gene expression could be explained by the induction of a transcriptional repressor. A search for such molecule revealed Hes1, a known Notch target and a transcriptional repressor (Suppl. Fig. 11a). Interestingly, Hes1 was previously suggested to play a role in myeloid leukemia, as a downstream effector of the Junb tumor suppressor in an animal model of CML18. To directly test the potential involvement of Hes1, we used in vitro differentiation assays and showed that Hes1 ectopic expression was sufficient to direct differentiation away from the GM lineage (Fig. 2c). Furthermore, Hes1 expression suppressed the expression of key GM commitment genes such as Cebpa and Pu.1 (Suppl. Fig. 12a). In agreement with this finding, we identified putative Hes (N-box) binding sites on the promoters of both genes. Reporter and chromatin immuno-precipitation (ChIP) assays proved direct binding of Hes1 on these promoters and suppression of transcription (Suppl. Fig. 12b–e).
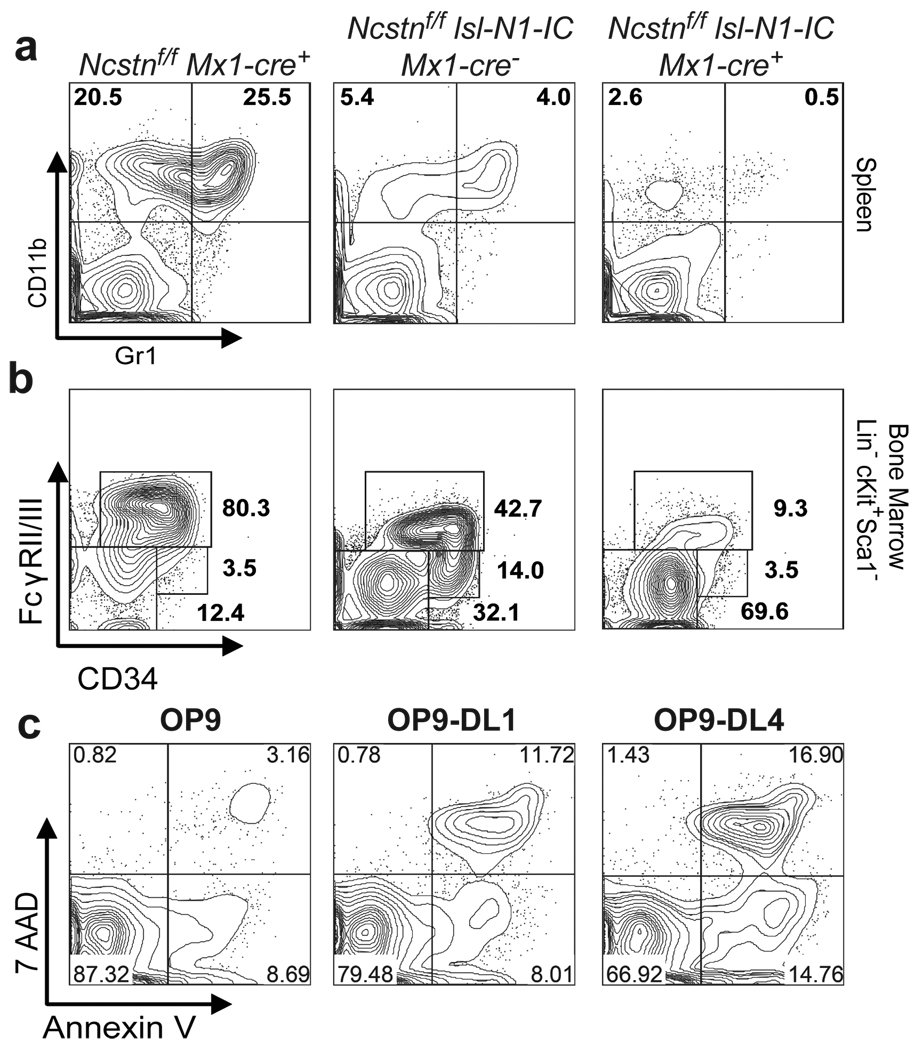
These findings suggested that Notch (or Hes1) hyper-activation could suppress the CMML-like disease developing in the Ncstn−/− animals. To test this hypothesis we used the previously described Ef1α1-lsl-Notch1IC+ mice17. We generated Notch1ICNcstnf/f-Mx1-Cre+ animals and analyzed both GMP accumulation and disease progression. Notch1IC expression was sufficient to significantly suppress both GMP expansion and disease development (Fig. 3a–b and Suppl. Fig. 13). Interestingly, Notch1IC expression drove progenitor commitment towards the lymphoid (T cell) and MEP lineages19. However, Notch1IC expression did not affect cell cycle kinetics within the GMP subset (Suppl. Fig. 13). To uncouple differentiation to the GMP subset from effects on GMP homeostasis we have purified wild-type GMP and plated them on stroma in the presence or absence of Notch ligands (Dll1 and Dll4). A striking increase of apoptosis rate was noted in the presence of Dll1–4 (Fig. 3c). These experiments suggested that Notch ectopic expression can both affect commitment to the GMP subset and the survival of already committed GMP progenitors.
Figure 3. Ectopic expression of Notch1-IC is able to prevent CMML-like disease in Ncstn−/− mice.
a, PolyI-polyC-induced Notch1-IC expression in Ncstnf/flsl-N1-IC Mx1-Cre+ animals suppresses myeloid cells in the Spleen. b, Notch1-IC expression suppresses GMP progenitor population in the bone marrow. c, Induction of cell death in wild-type GMP cells cultured in the presence (OP9-DL1-) or absence (OP9) of Notch ligands. Cell death was measure by the combination of 7AAD and Annexin-V staining 48h after coculture initiation. For a–c a representative of more than three experiments is shown.
Our studies thus far demonstrated that Notch controls murine myelopoiesis and its deletion leads to GMP expansion and monocytic disease. To prove that Notch is also important in human hematopoiesis and leukemogenesis we have initially cultured purified human CD34+CD38−Lin− bone marrow and cord blood stem and progenitor cells on stroma expressing different Jagged and Delta-like Notch ligands20. We found that expression of Notch ligands efficiently suppressed differentiation of human multipotential progenitors towards both the granulocyte (CD15+) and monocyte (CD14+) lineages (Suppl. Fig. 14 and 15). This suppression was prevented when the activity of the γSE complex was suppressed using either small molecule inhibitors or by the expression of a dominant negative MAML1 mutant.
To gain further insights into the role of Notch in human CMML, we have undertaken an extensive sequencing of a large number of γSE/Notch pathway genes. Exon resequencing of CMML patient specimens (Suppl. Table 3) identified a substantial fraction (6 novel mutations in 5 out of 42 patients) harboring somatic heterozygous mutations in multiple Notch pathway genes including NCSTN, APH1, MAML1 and NOTCH2 (Fig. 4 and Suppl. Table 4). In addition, several other putative mutations (single nucleotide variants, SNVs) were detected, which are not annotated as known germline SNPs but for which we could not prove a somatic origin (Suppl. Table 5). The validated somatic mutations were only observed in CMML, as resequencing of 47 samples from patients with myeloproliferative disorders (polcythemia vera or myelofibrosis) did not reveal somatic mutations in the Notch pathway (Fig. 4b). Importantly, CMML specimens with Notch mutations also had somatic alterations in well-characterized myeloid oncogenic lesions21, including JAK2, KRAS, TET2 and ASXL1, suggesting mutational co-operation between Notch signaling and other oncogenic pathways in CMML (Suppl. Table 4). We then asked if those mutations were causally-related with the disease using transcriptional reporter and in vitro differentiation assays. We demonstrated that selected mutations had the ability to negatively affect Notch activity either as dominant negative (MAML1Q345X) or as null (NCSTNA433T) alleles (Fig. 4c–d, Suppl. Fig. 16). This is the first description of somatic Notch pathway loss-of-function mutations in human cancer.
The presented studies identify novel inactivating Notch pathway mutations and suggest that γSI complex/ Notch signaling controls early HSC/MPP commitment decisions in the bone marrow. A significant portion of this regulation is controlled by the Hes family of transcriptional repressors. Most importantly, our studies suggest that silencing of Notch activity leads to the development of myeloid leukemia, suggesting a novel tumor suppressor function for the Notch pathway in hematopoiesis. Although mutation-mediated pathway silencing can be found in CMML, it is conceivable that there are additional control mechanisms, including epigenetic silencing of Notch pathway target genes. Whatever the mode of regulation, our observations suggest that reversible activation of the Notch pathway may represent an attractive future therapy, targeting specifically the progression and relapse of granulocytic and monocytic neoplasms.
METHODS SUMMARY
Animals
All mice were kept in specific pathogen-free animal facilities at the New York University School of Medicine. Mx1-Cre+ animals were injected with 20µg polyI:polyC/gram of body weight for a total of 3 injections. The injections were initiated 14 days after birth and done every two days. Animals were analyzed 4–6 weeks after the last injection unless indicated otherwise. All animal experiments were done in accordance to the guidelines of the NYU School of Medicine IACUC.
Antibodies and FACS analysis
Antibody staining and FACS analysis was performed as previously described22. All antibodies were purchased from BD-Pharmingen or e-Bioscience. We used the following antibodies: c-kit (2B8), Sca-1 (D7), Mac-1 (M1/70), Gr-1 (RB6-8C5), NK1.1 (PK136), TER-119, CD3 (145-2C11), CD19 (1D3), IL7Rα(A7R34), CD34 (RAM34), FcγRII/III (2.4G2), Flk-2/Flt-3 (A2F10.1), CD4 (RM4–5), CD4 (H129.19), CD8 (53–6.7), CD45.1 (A20), CD45.2 (104), CD150 (9D1), CD48 (HM481). Bone marrow lineage antibody cocktail includes: Mac-1, Gr-1, NK1.1, TER-119, CD3, CD19.
OP9-DL1/DL4 in vitro coculture
OP9-DL1 cells were maintained in MEM additioned of 10% Foetal Bovine Serum. 10 × 103 purified infected (GFP+) LSK or cKit+ BM progenitors were seeded into a 6-well plate with confluent OP9 cells in the presence of 10ng/ml SCF, 10ng/ml IL3, 5 ng/ml Flt3-L and 5 ng/ml Il-7. Flow cytometric analysis was performed on a LSRII (BDIS). Hematopoietic cells were gated using CD45 cell surface expression.
Microarray analysis
LSK or GMP cells from individual mice were used. In order to generate sufficient sample quantities for oligonucleotide gene chip hybridization experiments, we used the Ovation® RNA Amplification System V2 (Nugen) for cRNA amplification and labeling. The amplified cRNA was labeled and hybridized to the Mouse 430.2 microarrays (Affymetrix). The data was normalized using the previously published Robust Multi-array Average (RMA) algorithm using the GeneSpring GX software (Agilent, Palo Alto, CA).
Methods
Animals
Genotyping of N1f/f N2f/f N3−/− mice 23–26 was performed as previously reported. Ncstnf/fMx1-Cre+ and N1f/fN2f/fN3−/− Mx1-Cre+ animals were injected with 20µg polyI:polyC/gram of body weight for a total of 6 injections. The injections were initiated 14 days after birth and done every two days. Animals were analyzed 4–6 weeks after the last injection unless indicated otherwise. All animal experiments were done in accordance to the guidelines of the NYU School of Medicine IACUC.
Generation of the Ncstnf/f mice
To generate a conditional Ncstn allele we have used standard ES cell targeting approaches. Exons 5–7 of the Ncstn locus were targeted for Cre-dependent recombination. Removal of these exons generates an in-frame stop codon that prematurely terminates translation. As homology region we used a 7 Kb BsrGI-EcoRI genomic fragment that spans introns 2–11. A PGK-NEO cassette and a loxP site were cloned into an EcoRV site located in intron 4, while the second loxP site was subcloned into a unique PmlI site of intron 7. Embryonic stem (ES) cell colonies (129/sj) were screened by Southern blot of genomic DNA digested with BamHI (Figure 1B). Correctly targeted ES cells were injected in C57BL6 blastocysts and chimeras were generated. After verification of germline transmission the PGK-NEO cassette was removed using a germline Flp mouse 27. Ncstnf/fFlp+ mice were crossed to the Mx-cre strain. Genotyping primers and probes are available upon request.
Antibodies and FACS analysis
Antibody staining and FACS analysis was performed as previously described 22. All antibodies were purchased from BD-Pharmingen or e-Bioscience. We used the following antibodies: c-kit (2B8), Sca-1 (D7), Mac-1 (M1/70), Gr-1 (RB6-8C5), NK1.1 (PK136), TER-119, CD3 (145-2C11), CD19 (1D3), IL7Rα(A7R34), CD34 (RAM34), FcγRII/III (2.4G2), Flk-2/Flt-3 (A2F10.1), CD4 (RM4–5), CD4 (H129.19), CD8 (53-6.7), CD45.1 (A20), CD45.2 (104), CD150 (9D1), CD48 (HM481). Bone marrow lineage antibody cocktail includes: Mac-1, Gr-1, NK1.1, TER-119, CD3, CD19. For western blotting goat polyclonal anti-Nicastrin antibody ( N19, sc-14369, Santa Cruz) and mouse monoclonal anti-Actin antibody (Clone C4, MAB1501R, Millipore) were used. Annexin V/7AAD staining was done using AnnexinV PE detection kit (BD Pharmingen, 559763) following manufacturer’s protocol.
RT-PCR
Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) and cDNA was synthesized using the SuperScript First-Strand Kit (Invitrogen). Quantitative PCR was performed using SYBR green iMaster and a LightCycler 480 (Roche) using the following primer sequences (Tm=60°C used for all primers): Cebpa Forward TTACAACAGGCCAGGTTTCC, Cebpa Reverse CTCTGGGATGGATCGATTGT, Pu.1 Forward ATGGAAGGGTTTTCCCTCACCGCC, Pu.1 Reverse GTCCACGCTCTGCAGCTCTGTGAA, Gata2 Forward AACGCCTGTGGCCTCTACTA, Gata2 Reverse TCTCTTGCATGCACTTGGAG, Cebpd Forward ATCGCTGCAGCTTCCTATGT, Cebpd Reverse AGTCATGCTTTCCCGTGTTC, Hes1 Forward TCCAAGCTAGAGAAGGCAGAC, Hes1 Reverse TGATCTGGGTCATGCAGTTG, Gata1 Forward ACTGTGGAGCAACGGCTACT, Gata1 Reverse TCCGCCAGAGTGTTGTAGTG, Ncstn Forward CTGGCGCGTGCACTGTATGAG, Ncstn Reverse GGAGACGGCGATGTAGTGTTGAAG.
Bone marrow transplantation assays
5×105 bone marrow cells (Ly5.2+) were transplanted by retro-orbital i.v. injections into lethally irradiated (960 cGy) BL6SJL (Ly5.1+) recipient mice. 4 weeks after transplant mice were injected with 20µg polyI:polyC per gram of body weight for a total of 6 injections. Peripheral blood of recipient mice was collected at 4, 7, 9 and 12 weeks after transplant. Recipient mice were sacrificed 16 weeks after transplant for analysis.
Microarray analysis
LSK or GMP cells from individual mice were used. Microarray analysis was performed as previously described 28. Briefly, freshly isolated cells were sorted by surface marker expression, and total RNA was extracted using the RNeasy kit (QIAGEN, CA). In order to generate sufficient sample quantities for oligonucleotide gene chip hybridization experiments, we used the Ovation® RNA Amplification System V2 (Nugen) for cRNA amplification and labeling. The amplified cRNA was labeled and hybridized to the Mouse 430.2 microarrays (Affymetrix). The Affymetrix gene expression profiling data was normalized using the previously published Robust Multi-array Average (RMA) algorithm using the GeneSpring GX software (Agilent, Palo Alto, CA). The gene expression intensity presentations were generated with Matrix2png software (http://chibi.ubc.ca/matrix2png/bin/matrix2png.cgi) or Multi Experiment Viewer software (http://www.tm4.org/mev/).
GeneSet Enrichment Analysis
Geneset Enrichment Analysis was performed using GSEA software 29,30, http://www.broadinstitute.org/gsea/, using Gene set as permutation type, 1000 permutations and log2 ratio of classes as metric for ranking genes.
The “Myeloid Signature” geneset was generated using a systematic approach based on the comparison of gene expression arrays from WT LSK and WT GMP. Genes that were significantly upregulated in GMP compared to LSK (over 1.5 fold induction p-value<0.05) were used to define myeloid signature genes. The list was trimmed of unknown function genes and genes related to metabolism.
Other myeloid specific Genesets used in the analysis were taken from genesets already present in the MSig Database of the Broad Institute.
Retroviral infection of LinegaenegcKit+ bone marrow cells or LSK cells
Bone marrow cells were enriched for cKit-positive cells using either the EasySep kit (StemCell Technology) or the Dynabead kit (DYNAL, Invitrogen). LSK cells were flow sorted using lineage marquers Mac-1, Gr-1, NK1.1, TER-119, CD3, CD19, Sca1 and cKit. Cells were subsequently cultured in OPTI-MEM supplemented with 10% fetal bovine serum, 100 µ/ml penicillin, 100 µg/ml streptomycin, 50 ng/ml of SCF and Flt3l, 10 ng/ml of IL6 and IL7. For retroviral production, Plat-E cells were transfected with appropriate retroviral expression constructs by calcium phosphate method. Virus supernatant was collected 48 hr post transfection and used directly for spin infection of cKit positive-enriched bone marrow cells or sorted LSK cells at 2500 rpm for 90 minutes. Forty-eight hours after infection, lineage-negative GFP-positive cells were sorted for RT-PCR analysis, methylcellulose pating assays or OP9 coculture assay.
In vitro differentiation assays
Total bone marrow (15,000), sorted LSK (500) and GMP (500) were plated in triplicates into cytokine-supplemented methylcellulose medium (MethoCult 3434, Stem Cell Technologies). Colony type was scored after 10 days of culture. Replating was performed after 8 days of culture. For experiments involving Hes1 over-expression 10,000 GFP+ infected cells were plated in duplicate on 35mm cytokine-supplemented methylcellulose medium (MethoCult 3434, Stem Cell Technologies). Cells were recovered 8 days later, stained and analyzed by FACS as described.
Human progenitor/OP9 cocultures
BM samples were obtained and used according to the guidelines of the Medical Ethical Commission of Ghent University Hospital (Belgium). BM mononuclear cells were isolated through Lymphoprep density-gradient centrifugation and CD45+ glycophorin-A positive (Gly-A+) cells were enriched via micro-magnetic beads (Miltenyi Biotec). Subsequently, CD45+Gly-A+ cells were labelled with CD34-FITC, CD38-PE, CD19-APC and CD14-APC to sort CD34+CD38−Lin− cells on a FACSAria to a purity of 0>98%. OP9-control and OP9 cells expressing human ligands Dll1, Dll4, Jag1 and Jag2 were generated by retroviral transduction of OP9 cells (kindly provided by J.C. Zúñiga-Pflücker), followed by sorting for eGFP+ cells, as described previously31. 2.0 – 2.6 × 103 purified BM progenitors were seeded into a 24-well plate with confluent OP9 cells in the presence of 20 ng/ml SCF, 20 ng/ml Flt-3L, 20 ng/ml TPO, 10 ng/ml GM-CSF and 10 ng/ml G-CSF. Cocultures were performed and prepared for analysis as described previously32. Flow cytometric analysis was performed on a LSRII (BDIS). Human cells were gated using a CD45 monoclonal antibody and dead cells were excluded using propidium iodide.
OP9-DL1/DL4 in vitro coculture
OP9 cells were maintained in MEM additioned of 20% Foetal Bovine Serum. 10 × 103 purified infected (GFP+) LSK or cKit+ BM progenitors were seeded into a 6-well plate with confluent OP9 cells in the presence of 10ng/ml SCF, 10ng/ml IL3, 10ng/ml IL6, 5 ng/ml Flt3-L and 5 ng/ml Il-7. Flow cytometric analysis was performed on a LSRII (BDIS). Hematopoietic cells were gated using CD45 cell surface expression and GFP expression.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed with 80µg of genomic DNA from 32D-MIG-infected or 32D-HES1-infected cells following standard procedures. Briefly, chromatin was cross-linked with 1% formaldehyde, and sheared by sonication. Immunoprecipitation was performed with IgG or anti-HES1, followed by incubation with protein A magnetic beads/salmon sperm DNA (Invitrogen Dynal AS,Oslo, Norway). DNA isolated from antibody bound fraction was eluted after wash, extracted with phenol/chloroform, and precipitated with ethanol. Real-time PCR quantification of immunoprecipitated DNA was carried out with the SYBR Green PCR Master Mix (Roche) and primers designed to amplify regions covering each putative HES1 binding site in the Pu.1 and Cebpa promoters: Pu.1-1 For ACGTTCAAGGGTTGGAGAAA, Pu.1-1 Rev GCCAATTAGGGCCAACAGTA, Pu.1–2 For GACCAAAGTCCTCCACCTGA, Pu.1–2 Rev CTGGGAGGGAGAAAGGCTA, Cebpa-1 For CCAAAGCAGTCTCCAACCTC, Cebpa-1 Rev CCCACTTCCAGCCAACACTA, Cebpa-2 For CGCCTAACCACGGACCAC, Cebpa-2 Rev AGTAGGATGGTGCCTGCTG.
Histological Analyses
Mice were killed and autopsied, and then dissected tissue samples or tumors were fixed for 24 h in 10% buffered formalin, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 5 µm and stained with hematoxylin and eosin.
Luciferase reporter assays
pGL3 promoter reporter plasmid (Promega) was used to clone Pu.1, Cebpa or Hes1 promoter sequences. pGL3 promoter empty vector or the vector containing Pu.1, Cebpa or Hes1 promoter sequences, HA-HES1 expression plasmid or pcDNA3.1 control plasmid and Renillia expression vector were co-transfected into HEK293T cells using regular Calcium Phosphate transfection protocol. Luciferase activities were examined 24 hours after transfection using the Dual Luciferase reporter assay system (Promega) following manufacturer’s instructions, and normalized to Renillia activity.
Statistical analysis
The means of each data set were analyzed using the Student’s t test, with a two-tailed distribution and assuming equal sample variance.
Supplementary Material
Acknowledgements
We would like to thank Drs. G. Fishel, F. Radtke and R. Kopan for donating mouse strains; P. Lopez and the NYU Flow Facility for expert cell sorting; A. Heguy and the Geoffrey Beene Translational Core laboratory for assistance with DNA resequencing. The NYU Cancer Institute Genomics Facility for help with micro-array processing. Supported by the National Institutes of Health (RO1CA133379, RO1CA105129, R21CA141399, RO1CA149655 to I.A.; R01CA1328234 to R.L.L. and F.M.; U54CA143798 to F.M.), the Leukemia & Lymphoma Society (to I.A.), the American Cancer Society (to I.A.), the Irma T. Hirschl Trust, the Dana Foundation, The Mallinckrodt Foundation, the Alex’s Lemonade Stand Foundation (to I.A.), and the Fund for Scientific Research Flanders (FWO) and its Odysseus Research Program (to T.T.). A.E. was supported by the National Cancer Institute (1P01CA97403, Project 2) and a gift from the Berrie Foundation. A.K. was supported by a Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. I.V.d.W. is supported by Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT). S.C. is supported by the Hope Street Kids Foundations and P.O. by the NYU MSTP Program. T.T. is supported by the FWO. O.A.W. is supported by the Clinical Scholars Program at Memorial Sloan Kettering Cancer Center and by the American Society of Hematology. R.L.L. is an Early Career Award recipient of the Howard Hughes Medical Institute and is the Geoffrey Beene Junior Chair at Memorial Sloan Kettering Cancer Center. I.A is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Author Contributions
I.A. and C.L. conceived the study and designed all experiments. A.K. and A.E. helped with experimental planning and generated the Ncstnf/f mice. C.L. performed the majority of the mouse experiments and in vitro studies aided by P.O., S.B, S.C. T.T. and E.A. R.L.L, O.A-W. and M.B. performed and analyzed human leukemia sample exon sequencing. H.H. and F.F. helped with disease modeling and computational analysis of disease progression. I.vDW. and T.T. performed the human stem cell differentiation assays. S.L. and S.I. analyzed mouse disease pathology. J.Z. processed and analyzed gene expression data.
Author Information
The microarray data discussed in this study can be found in the Gene Expression Omnibus (GEO) of NCBI at http://www.ncbi.nih.gov/geo/ through accession numbers GSE27794, GSE27799, and GSE27811.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
REFERENCES
- 1.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. doi:S0955-0674(07)00026-9 [pii] 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B. Nicastrin: gatekeeper of the gamma-secretase complex. Cell. 2005;122:318–320. doi: 10.1016/j.cell.2005.07.021. doi:S0092-8674(05)00751-8 [pii] 10.1016/j.cell.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 3.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nature reviews. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 4.Robert-Moreno A, et al. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 2008;27:1886–1895. doi: 10.1038/emboj.2008.113. doi:emboj2008113 [pii] 10.1038/emboj.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuniga-Pflucker JC. T-cell development made simple. Nature reviews. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 6.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 7.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006:1–13. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 9.Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- 10.Emanuel PD. Juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia. Leukemia. 2008;22:1335–1342. doi: 10.1038/leu.2008.162. doi:leu2008162 [pii] 10.1038/leu.2008.162. [DOI] [PubMed] [Google Scholar]
- 11.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. doi:S1934-5909(10)00046-9 [pii] 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. doi:S1074-7613(09)00138-1 [pii] 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. doi:nature04980 [pii] 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 14.Dumortier A, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of notch signaling in the murine skin. PLoS ONE. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. doi:10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 16.Demehri S, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. doi:07-PLBI-RA-3899 [pii] 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonamici S, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. doi:nature08020 [pii] 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santaguida M, et al. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15:341–352. doi: 10.1016/j.ccr.2009.02.016. doi:S1535-6108(09)00072-5 [pii] 10.1016/j.ccr.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercher T, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3:314–326. doi: 10.1016/j.stem.2008.07.010. doi:S1934-5909(08)00344-5 [pii] 10.1016/j.stem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. doi:leu201069 [pii] 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aifantis I, Feinberg J, Fehling HJ, Di Santo JP, von Boehmer H. Early T cell receptor beta gene expression is regulated by the pre-T cell receptor-CD3 complex. J Exp Med. 1999;190:141–144. doi: 10.1084/jem.190.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 23.Yang X, et al. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. doi:10.1016/j.ydbio.2004.01.014 S0012160604000636 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Saito T, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell KJ, et al. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet. 2001;28:241–249. doi: 10.1038/90074. [DOI] [PubMed] [Google Scholar]
- 26.Leighton PA, et al. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410:174–179. doi: 10.1038/35065539. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. doi:10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 28.Thompson BJ, et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. doi:0506580102 [pii] 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. doi:10.1038/ng1180 ng1180 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Van de Walle I, et al. An early decrease in Notch activation is required for human TCR-alphabeta lineage differentiation at the expense of TCR-gammadelta T cells. Blood. 2009;113:2988–2998. doi: 10.1182/blood-2008-06-164871. doi:blood-2008-06-164871 [pii] 10.1182/blood-2008-06-164871. [DOI] [PubMed] [Google Scholar]
- 32.Taghon T, et al. Notch signaling is required for proliferation but not for differentiation at a well-defined beta-selection checkpoint during human T-cell development. Blood. 2009;113:3254–3263. doi: 10.1182/blood-2008-07-168906. doi:blood-2008-07-168906 [pii] 10.1182/blood-2008-07-168906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.