Summary
Exposure of DNA to UV radiation causes covalent linkages between adjacent pyrimidines. The most common lesion found in DNA from these UV-induced linkages is the cis-syn cyclobutane pyrimidine dimer (CPD). Human DNA polymerase κ (Polκ), a member of the Y-family of DNA polymerases, is unable to insert nucleotides opposite the 3’T of a cis-syn T-T dimer, but it can efficiently extend from a nucleotide inserted opposite the 3’T of the dimer by another DNA polymerase. We present here the structure of human Polκ in the act of inserting a nucleotide opposite the 5’T of the cis-syn T-T dimer. The structure reveals a constrained active site cleft that is unable to accommodate the 3’T of a cis-syn T-T dimer but is remarkably well adapted to accommodate the 5’T via Watson-Crick (WC) base pairing, in accord with a proposed role for Polκ in the extension reaction opposite from CPDs in vivo.
Introduction
Cellular DNA is continually subjected to damaging agents such as ultraviolet (UV) and ionizing radiation externally and oxidation and hydrolysis internally. The most common UV-induced DNA lesions are cis-syn cyclobutane pyrimidine dimers (CPDs) in which the two adjacent pyrimidines become linked by inter-base covalent bonds 1; 2. Although CPDs can be removed by nucleotide excision repair 3, some lesions escape repair and present a strong block to replication. The recently discovered Y-family of DNA polymerases permit the continuity of the replication fork by allowing replication through lesions that impede the replicative polymerases 4. Humans have four Y-family polymerases – Polη, Polι, Polκ, and Rev1 – each with a unique DNA damage bypass and fidelity profile. Polη can replicate through a cis-syn T-T dimer by inserting As opposite the two Ts of the dimer with the same efficiency and accuracy as opposite undamaged Ts 5; 6; 7, and genetic studies in human and mouse cells have provided strong evidence that Polη replicates through CPDs in a highly error-free manner 8. Because of the involvement of Polη in promoting error-free replication through CPDs, its inactivation in humans causes the variant form of xeroderma pigmentosum 9; 10, a genetic disorder characterized by a greatly enhanced predisposition to sun induced skin cancers. Polι, on the other hand, is unable to replicate through a cis-syn T-T dimer but it can proficiently incorporate nucleotides opposite N2-adducted guanines and opposite adducts such as 1, N6-ethanodeoxyadenosine which impair the ability of the purine to engage in Watson-Crick (W-C) base-pairing 11; 12; 13; 14; 15. Rev1 is highly specialized for incorporation of C opposite template G 16; 17, and promotes efficient dCTP incorporation opposite bulky N2-dG adducts via a protein-template directed mechanism of DNA synthesis 18; 19. In all, Y-family polymerases in eukaryotes display a large degree of functional divergence, rendering them highly specialized for specific roles in lesion bypass.
Polκ is the only human Y-family polymerase with homologues in prokaryotes and archaea, including DinB (PolIV) in Escherichia coli and Dbh and Dpo4 in Sufolobus solfataricus 20; 21; 22. However, the amino acid (aa) sequence of Polκ differs from PolIV and Dpo4 (and other Y-family polymerases) by an extension at the N-terminus of approximately 75 amino acids 23. This N-terminal extension is indispensable for Polκ activity and is conserved only amongst eukaryotic Polκ proteins. The crystal structure of Polκ in ternary complex with undamaged DNA and an incoming nucleotide reveals encirclement of the DNA by this unique N-terminal extension, referred to as the N-clasp 23. The N-clasp effectively locks the polymerase around the template-primer, perhaps as a means to keep it engaged on a sugar-phosphate backbone distorted by a DNA lesion.
Polκ is specialized for the extension step of lesion bypass. On undamaged DNA, for example, Polκ misincorporates nucleotides with a frequency of ~10−3 24, whereas it extends mispaired termini with a frequency of ~10−1 to 10−2 25. Also, Polκ is unable to insert nucleotides opposite the 3’T of a cis-syn T-T dimer, but it can efficiently extend from a nucleotide inserted opposite the 3’T of the dimer by another DNA polymerase 24; 25; 26; 27. In keeping with these biochemical studies indicating a role for Polκ in extension opposite from a cis-syn T-T dimer, genetic studies in human and mouse cells have provided evidence for a role of Polκ in the mutagenic bypass of CPDs during replication 8, and Polκ−/− mouse embryonic stem cells display enhanced UV sensitivity 28.
To understand the basis of Polκ’s ability to extend efficiently opposite from a cis-syn T-T dimer, we determined the structure of Polκ in the act of inserting a nucleotide opposite the 5’T of the T-T dimer. The structure reveals a constrained active site cleft that is unable to accommodate the 3’T of a T-T dimer but is well adapted to accommodate the 5’T via Watson-Crick (WC) base pairing, in accord with the proposed role for Polκ in extension opposite from CPDs in vivo 8.
Results
Structure determination
We crystallized the Polκ catalytic core (aa 19–526) in ternary complex with a 13-nt/18-nt primer/template presenting the 5’T of the cis-syn T-T dimer as the templating base and with dATP as the incoming nucleotide. The cocrystals diffract to ~3.33Å resolution with synchrotron radiation (Argonne National Laboratory) and there are two ternary complexes (A and B) in the crystallographic asymmetric unit (Table 1). The structure was determined by molecular replacement using the polymerase from the Polκ ternary complex with undamaged DNA as a search model 23. Electron density maps showed clear densities for the bound template/primer and incoming dATP. For ternary complex A, the final model consists of residues 25–224 and 281–520 of Polκ, nucleotides 4–18 of the template, nucleotides 1–13 of the primer, incoming dATP, and 2 Mg2+ ions. For ternary complex B, the final model consists of residues 19–224 and 282–518 of Polκ, nucleotides 4–17 of the template, nucleotides 2–13 of the primer, incoming dATP, and 2 Mg2+ ions. The two complexes in the asymmetric unit are similar in structure, though complex A is slightly more ordered than complex B (average B-factor of ~109Å2 versus ~130Å2). We describe below the structure of complex A and refer to complex B as needed.
Table 1.
Data collection and refinement statistics
| Data Collection | ||
|---|---|---|
| Wavelength (Å) | 0.97949 | |
| Resolution (Å) | 3.33 | |
| Number of measured reflections | 118,834 | |
| Number of unique reflections | 28,984 | |
| Data coverage (%)a | 99.9 (99.2) | |
| Rsym (%)b | 10.3 (75.2) | |
| I/σ | 15.9 (2.0) | |
| Refinement Statistics | ||
| Resolution range (Å) | 50.0–3.33 | |
| Reflections | 27,432 | |
| Rcryst (%)c | 24.2 | |
| Rfree (%)d | 28.7 | |
| Rms Deviations | ||
| Bonds (Å) | 0.011 | |
| Angles (°) | 1.681 | |
| Nonhydrogen atoms | Molecule A | Molecule B |
| Protein | 3,352 | 3,195 |
| DNA | 584 | 543 |
| dATP | 30 | 30 |
| Mg2+ | 2 | 2 |
| Water | 5 | 3 |
| B-factors (Å2) | ||
| Protein | 108.9 | 130.0 |
| DNA | 111.0 | 123.9 |
| dATP | 75.9 | 117.1 |
| Mg2+ | 73.6 | 84.2 |
| Water | 75.6 | 82.7 |
| Ramachandran plot quality | ||
| Most favored (%) | 85.9 | 82.2 |
| Additional allowed (%) | 13.2 | 17.0 |
| Generously allowed (%) | 4.0 | 0.2 |
| Disallowed (%) | 0.0 | 0.5 |
Values for outermost shells are given in parentheses.
Rsym=∑|I−<I>|/∑I, where I is the integrated intensity of a given reflection.
Rcryst=∑‖Fobserved| − |Fcalculated‖ / ∑|Fobserved|.
Rfree was calculated using 5% random data omitted from the refinement.
Overall arrangement
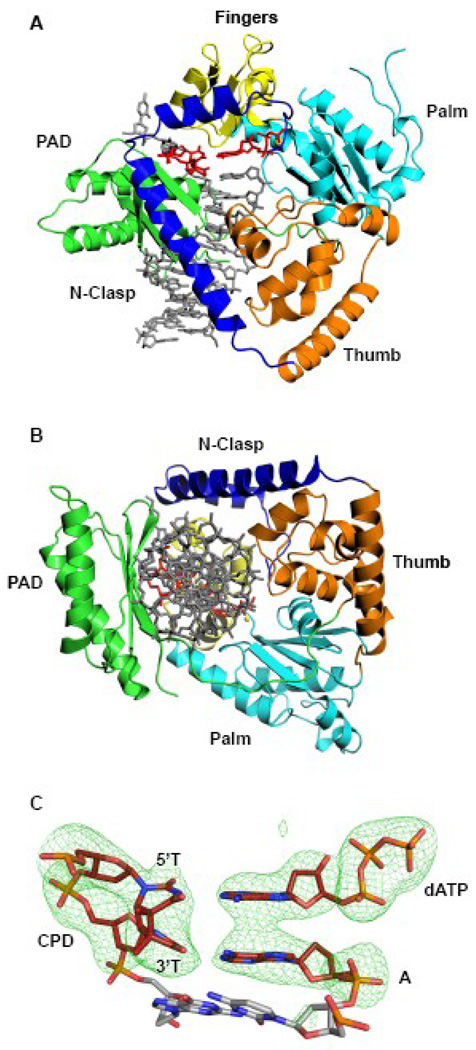
Polκ encircles the CPD adducted DNA in much the same manner as in the ternary complex with undamaged DNA 23 (Fig. 1). That is, the conventional right-handed grip on the template-primer by the palm, fingers, and thumb domains, and the PAD (polymerase associated domain), is augmented by an N-clasp subdomain (aa 19–74) that extends from the thumb domain and traverses across the template-primer to the PAD side of the DNA (Fig. 1). The palm and fingers domains interact primarily with the replicative end of the template-primer, wherein the palm (aa 101–109 and 171–338) carries the active site residues (Asp107, Asp198 and Glu199) that catalyze the nucleotidyl transfer reaction, and the fingers domain (aa 110–170) lies over the nascent base pair in the active site formed between the 5’T of the T-T dimer and incoming dATP (described below). The thumb and the PAD straddle the duplex portion of the template-primer, connected by a long linker that cradles one side of the DNA. The thumb (aa 79–100 and 339–401) skims the minor groove surface, while the PAD (aa 401–518) anchors in the major groove (Fig. 1). The majority of DNA interactions are mediated by the PAD, wherein the main chain amides on the “outer” β-strands of the PAD β-sheet make a series of hydrogen bonds with the template and primer strands. Additional DNA contacts are made by the thumb and the N-clasp, with the N-clasp effectively “locking” the thumb, fingers, palm domains and the PAD around the DNA (Fig. 1B).
Figure 1.
Polκ/T-T dimer/dATP ternary complex. (A) Ribbon diagram showing the overall structure of the ternary complex. The palm, fingers, thumb, PAD and N-clasp domains are shown in cyan, yellow, orange, green, and blue, respectively. DNA is shown in gray and the T-T dimer and incoming dATP are shown in red. A putative Mg2+ ion is shown as a yellow sphere (B) A view of the ternary complex looking down the DNA helix to show encirclement of the adducted DNA by the N-clasp. (C) Simulated annealing Fo-Fc map contoured at 5σ, showing the cis-syn TT dimer, incoming dATP, and the primer terminus.
Extension opposite from a cis-syn T-T dimer
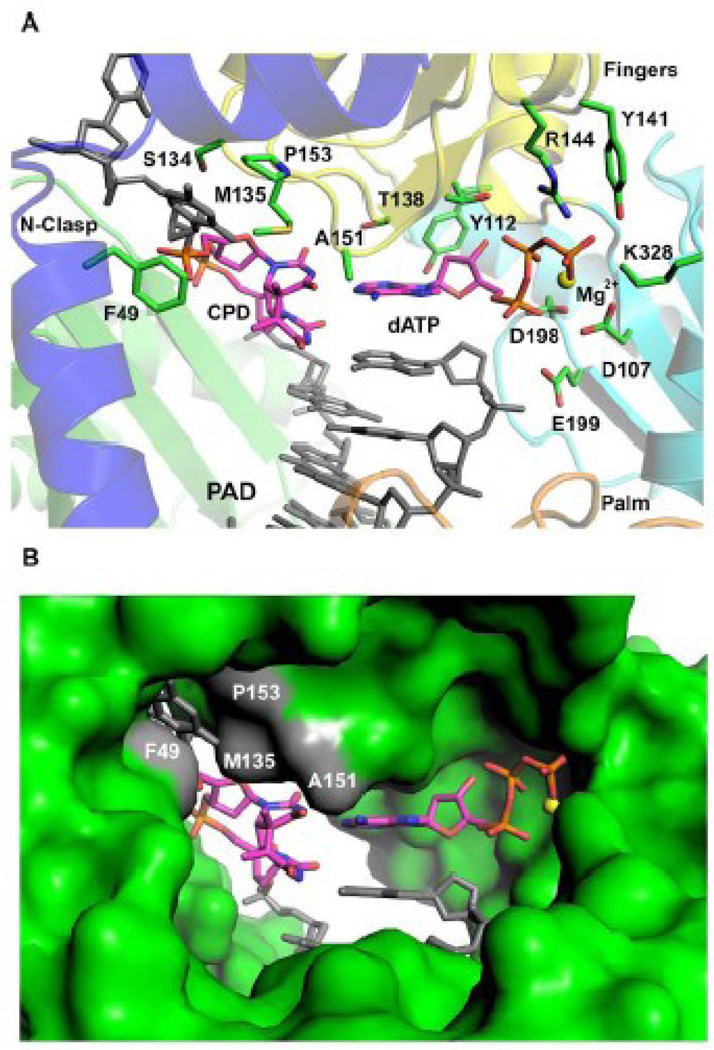
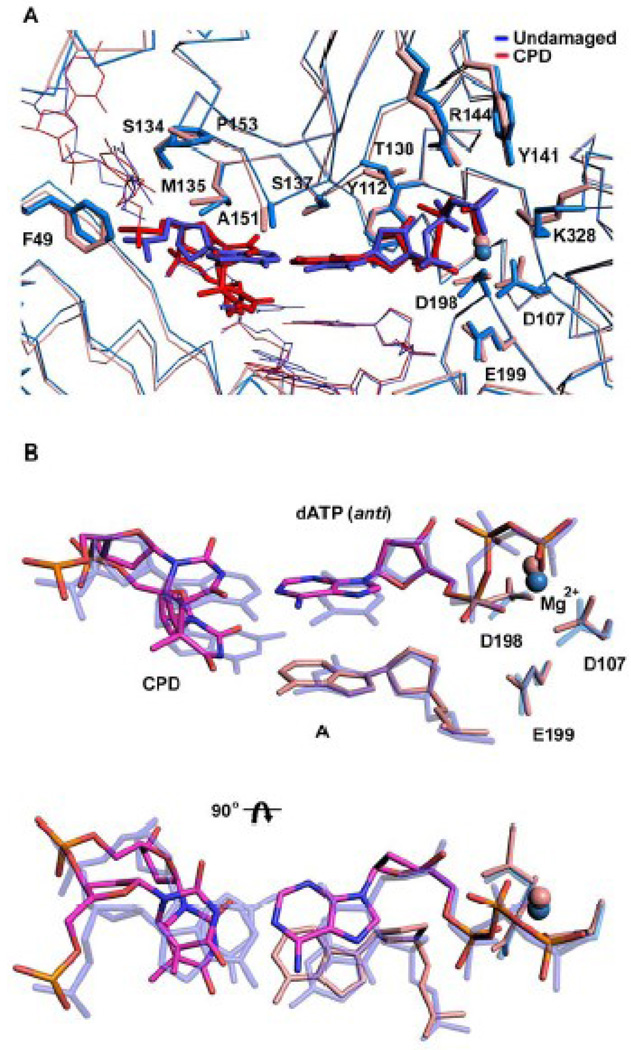
The structure reveals the 5’T of the T-T dimer making a Watson-Crick (WC) base pair with incoming dATP (Figs. 1C & 2). The C1’-C1’ distance across the 5’T(CPD):A nascent base pair is ~10.89Å, which is almost identical to the distance (~10.86Å) in the nascent A:T base pair in the Polκ undamaged ternary complex 23. However, the 5’T(CPD):A base pair is substantially more propeller twisted (−26.4°) as compared to the A:T base pair in the undamaged complex (which has a propeller twist of only −0.03°). The 3’T of the T-T dimer also forms a WC base pair with an A at the primer terminus (Figs. 1C & 2), but it is substantially more buckled when compared to the nascent 5’T(CPD):A base pair (a buckle angle of ~38.1° as compared to 1.5° for the 5’T(CPD):A base pair) (Fig 2A). Thus, despite being covalently-linked and rotationally constrained both T’s of the cis-syn T-T dimer are engaged in WC base pairing. There is no major alteration in the polymerase structure, which superimposes with an rms deviation of 0.55Å (for 427 Cαs) when compared to the polymerase in the undamaged complex (Fig. 3A). The template-primer binds in the same register as in the undamaged complex, and there is little or no movement of the N-clasp in accommodating 5’T of the CPD adduct at the templating position. Incoming dATP binds with its triphosphate moiety interlaced between the fingers and palm domains, making hydrogen bonds with Tyr141 and Arg144 from the fingers domain and Lys328 from the palm domain (Fig. 2 & 3B). The catalytic residues, Asp107, Asp198 and Glu199, are clustered between the triphosphate moiety and the primer terminus (Fig. 2A). A Mg2+ ion occupies a position corresponding to "metal B" in replicative polymerases 29; 30; 31, and is coordinated in the basal octahedral plane by the unesterified oxygens of dATP β- and γ-phosphates and the carboxylates of Asp107 and Asp198, and at the apical positions by the α-phosphate and the main chain carbonyl of Met108. There is no density for a Mg2+ ion at a position analogous to "metal A" in replicative polymerases or in Y-family polymerases. However, as in the ternary complex with undamaged DNA 23, there is density for a water molecule, located ~2Å from the site normally occupied by metal A in replicative polymerases.
Figure 2.
Watson-Crick base pairing between 5’T of the T-T dimer and incoming dATP. (A) Close-up view of the Polκ active site cleft. Highlighted and labeled are the catalytic residues (D107, D198, and E199), residues apposed close to incoming dATP (Y112, T138, R144, Y141, and K328), 5’T of the T-T dimer (M135 and A151), and the base upstream to the 5’T (F49, S134 and P153). A putative Mg2+ ion is shown as a yellow sphere. (B) Molecular surface representation of the Polκ active site cleft. Highlighted in gray and labeled are residues apposed close to 5’T of the T-T dimer and the base upstream to it. A putative Mg2+ ion is shown as a yellow sphere.
Figure 3.
Comparison between T-T dimer and undamaged DNA ternary Polκ complexes. (A) Superimposition of the T-T dimer complex (red) and the complex with undamaged DNA (blue; PDB code 2OH2) (blue). Highlighted are the template bases (T-T dimer and A), incoming dNTPs (dATP and dTTP) and residues within the active site cleft. The putative Mg2+ ion is shown as a sphere in each structure. (B) Close-up views of the superposition of the T-T dimer (magenta) and undamaged DNA (blue), and the incoming nucleotides dATP (magenta) and dTTP (blue), respectively. The figure also shows the catalytic triad and the putative Mg2+ ion in each structure. The figure shows that the active site geometry is conserved in the damaged and undamaged structures.
A Mg2+ is also present within a loop between helices αM and αN in the thumb domain, coordinated by main chain carbonyls of residues Arg353, Val354 and Ile357 and the phosphate group of the penultimate base at the primer terminus. There is an analogous ion in the human Rev1 ternary complex 19, as well as (at a nearby position) in ternary complexes of Dpo4 20; 32. Moreover, there are analogous interactions in yeast Rev1 18, human Polι11; 13; 33, and Polκ ternary complexes 23, although in these cases, the ion was assigned as a water molecule. It has been suggested that these conserved bridging interactions may help to fix the primer terminus for the nucleophilic attack on incoming dNTP 19.
Compared to Polη, the Polκ active site cleft is much more constricted with its fingers domain impinging directly on the templating base. In particular, Met135, emanating from the fingers domain, bears down on the 5’T of the T-T dimer and prevents the next 5’ nucleotide from stacking above it (Fig. 2). Consequently, only the templating base is held in the active site, whereas the rest of the 5’ unpaired template strand is directed out of the active site cleft. Thus, when we model the 3’T of a T-T dimer at the templating position, the 5’T of the T-T dimer overlaps not only with the Met135 side chain but also with the main chain of the β-strand (residues 134–136) carrying Met135 on the fingers domain. In contrast, the Polκ active site is well-adapted to accommodate the 5’T of the T-T dimer at the templating position for base pairing with incoming dATP. Met135 makes extensive van der Waals interactions with the 5’T of the T-T dimer and appears to play a significant role in positioning the base for the insertion reaction (Fig. 2). The stabilizing interactions of Met135 with the 5’T of CPD, in addition to the sphere of interactions of other residues (Tyr141, Arg144 and Lys328) with the incoming nucleotide may account for the high rate of incorporation opposite to the 5’T of CPD (extension step) seen in Polκ25(Fig. 2).
Discussion
Exposure of DNA to UV radiation causes covalent linkages between adjacent pyrimidines 1; 2. Since the two Ts of a cis-syn T-T dimer are covalently linked and cannot be separated, the active site clefts of most DNA polymerases – with the exception of Polη 6; 7- are not equipped to handle a T-T dimer. The inability of Polκ to insert a nucleotide opposite the 3’T of a cis-syn T-T dimer derives from a constricted active site cleft that – unlike Polη - is unable to accommodate two unpaired nucleotides. In contrast, we show here that the 5’T of a T-T dimer is comfortably accommodated at the templating position in the Polκ active site, making a WC base pair with incoming dATP. Neither the fingers domain or the N-clasp interferes with the binding of the 5’T of the T-T dimer, and the nucleotide 5’ to the lesion is kinked and accommodated outside of the active site cleft, between the N-clasp and the fingers domain.
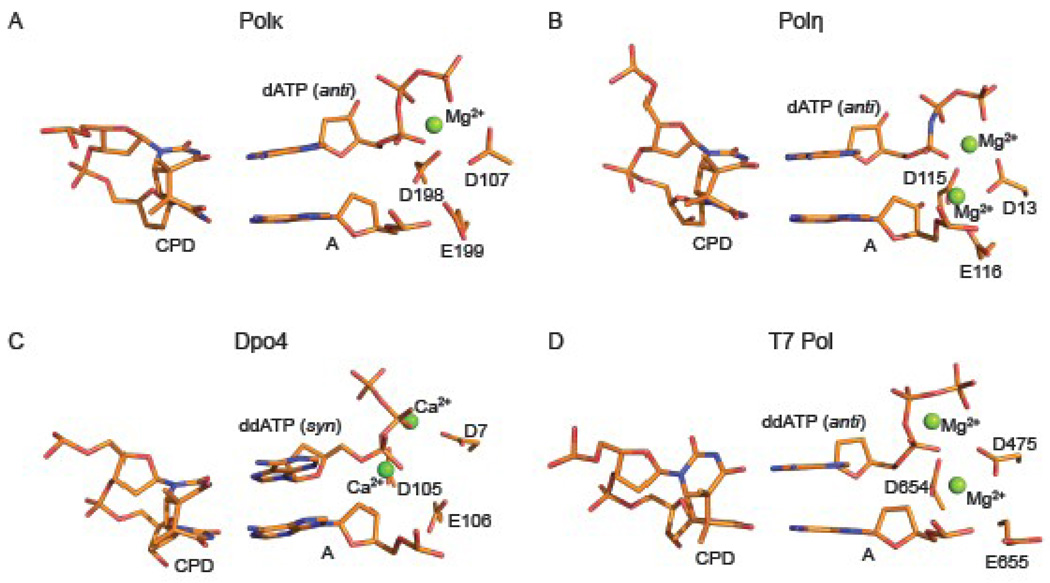
Van der Waals interactions with Met135 maintain the 5’T in a relatively coplanar arrangement with the base of incoming dATP. Most of the inclination of bases associated with a cis-syn T-T dimer is taken up by the 3’T, which is involved in a buckled WC base pair at the primer junction (Fig. 4A). Intriguingly, in a recent structure of human Polη inserting a nucleotide opposite the 5’T of a cis-syn T-T dimer 6, the nascent 5’T(CPD):A base pair is again relatively co-planar while the 3’T(CPD):A base pair is buckled (Fig. 4b). Thus, both human Polκ and Polη insert dATP opposite the 5’T of the T-T dimer by similar mechanisms, wherein both Ts of the T-T dimer are engaged in WC base pairing and most of the distortion associated with the cyclobutane linkage is absorbed by the 3’T. In contrast, the structure of archaeal Dpo4 inserting dATP opposite the 5’T of a T-T dimer shows Hoogsteen base pairing 34, wherein the incoming dATP is in a syn conformation and it is offset from the position of incoming nucleotide observed in other Dpo4 ternary complexes 20; 32; 35 (Fig. 4C). The structure of A-family phage T7 replicative polymerase (T7 Pol) inserting ddATP opposite the 5’T of the T-T dimer has also been determined36. However, unlike Polκ and Polη, most of the inclination of bases associated with a CPD lesion is taken up the 5’T(CPD):A base pair which is both buckled and propeller twisted 36(Fig. 4D).
Figure 4.
Insertion of dATP opposite the 5’T of a T-T dimer by different polymerases. Close up views of the Polκ (A), Polη (PDB ID 3MR4) (B), Dpo4 (PDB ID 1RYS) (C) and T7 Pol (PDB ID 1SL2) (D) active site regions with a cis-syn T-T dimer. Highlighted and labeled are the catalytic residues and metal ions (green spheres).
Taken together, Met135 in Polκ appears to play a dual role: first, it sterically impedes the binding of 3’T of a T-T dimer at the templating position and then via van der Waals contacts it appears to facilitate the binding of the 5’T of a T-T dimer. Met135 is unique to Polk; the equivalent residue in other Y-family polymerases is typically smaller. In yeast and human Polη, for example, the equivalent residues are Gly46 and Ser58, respectively, which because of their small size (and the fact that fingers domain in Polη is further away from the templating base than in Polκ) do not impinge on the templating base to the same extent as Met135 in Polκ6; 7. However, whereas in human Polη the side chain of Gln38 makes a direct hydrogen bond with O2 of the 5’T of CPD 6, there are no specific hydrogen bonds to the 5’T of CPD in the Polκ structure.
In summary, the structure we present here of Polκ reveals an active site cleft that is unable to accommodate the 3’T of a T-T dimer but is well adapted to accommodate the 5’T of a T-T dimer. The ability of Polκ to insert dATP opposite the 5’T of a T-T dimer is in accord with the proposed role for Polκ in extension opposite from CPDs in vivo, in an alternative pathway to Polη8.
Materials and Methods
Protein and DNA Preparation
Polκ19–526 was purified from yeast strain BJ5464 harboring plasmid pBJ943 as was described previously 23. The N-terminal fusion GST tag was removed by incubation with PreScission Protease (GE Healthcare) after an initial affinity chromatography step. For crystallization, the Polκ19–526 protein was further purified by ion exchange (SP sepharose) and size exclusion (SD200) chromatography. The 13-nt primer for crystallization was synthesized (Integrated DNA Technologies) with a dideoxyadenosine at its 3’ end (5’-GGGGGAAGGACCddA-3’) and annealed to an 18-nt template bearing a cis-syn thymine-thymine dimer (5’-TTCCTTGGTCCTTCCCCC-3’) (W.M. Keck Facility-Yale University). The primer and template strands were purified by ion exchange chromatography and reverse phase chromatography, respectively.
Cocrystallization
Polκ19–526 at a final concentration of 0.5 mM was incubated with the 13-nt/18-nt primer/template in a buffer containing 25 mM HEPES (pH 7.0), 200 mM NaCl, 1 mM TCEP (Tris [2-carboxyethyl] phosphine hydrochloride), 10mM MgCl2 and 10mM dATP. Cocrystals were obtained from solutions containing 14% PEG 5000 monomethyl ether (w/v) (MME), 200mM potassium acetate, 100 mM NaCl and 100 mM sodium cacodylate (pH 7.0). The cocrystals belong to the same space group (C2221) as the cocrystals reported previously with undamaged DNA 23, and have similar cell dimensions of a = 117.1 Å, b = 154.5 Å, and c = 217.9 Å. The cocrystals were cryoprotected for data collection by soaking them in mother liquor solutions containing increasing percentages of PEG 350 MME (5–30%), followed by flash-freezing in liquid nitrogen.
Structure determination and refinement
X-ray data were measured at the Advanced Photon Source (APS, beam line 17-ID). The data to 3.33Å resolution were indexed, integrated and scaled using HKL2000 37. Molecular Replacement (MR) with the program PHASER 38 using the previously solved Polκ structure in complex with DNA as a search model revealed a unique solution with two protein/DNA complexes per asymmetric unit. The program CNS 39 was subsequently used for initial rigid body refinement and electron density maps showed clear densities for the DNA and incoming dATP. Subsequent energy minimization and B-factor refinement in CNS, iterative model building with COOT 40, and a final restrained refinement with TLS in Refmac5 41 reduced the Rcryst and Rfree to 24.2% and 28.7% respectively. The final model includes residues 25–224 and 281–520 for protein molecule A; residues 19–224 and 282–518 for protein molecule B; nucleotides 4–18 for template (T) and 1–13 for primer (P) bound to protein A, and nucleotides 4–17 for template (U) and 2–13 for primer (Q) bound to protein B; two incoming dATPs; 4 Mg2+; and 8 water molecules were also positioned in the density. Approximately 7% and 16% of the amino acids were built as alanines in molecule A and B (primarily at the N-terminus), respectively, due to the lack of density to accurately build the corresponding side chains
Structural Analysis
The Polκ-TT model has good stereochemistry, as shown by PROCHECK 42, with >99.5% of residues in the allowed regions of the Ramachandran plot. Figures were prepared using PyMol 43.
Acknowledgements
We thank the staff at APS (beamline 17ID) for facilitating X-ray data collection. This work was supported by grant CA141209 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Coordinates
Coordinates and structure factors have been deposited in the PDB with an accession code 3PZP.
References
- 1.Taylor JS. Unraveling the molecular pathway from sunlight to skin cancer. Acc. Chem. Res. 1994;27:76–82. [Google Scholar]
- 2.You YH, Lee DH, Yoon JH, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J Biol Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 6.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;466:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 10.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 11.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 12.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA Polymerase iota Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair. Structure (Camb) 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 15.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2006;26:381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 17.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 18.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 19.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J Mol Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 22.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure (Camb) 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta [see comments] Proc Natl Acad Sci U S A. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci U S A. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yagi Y, Ogawara D, Iwai S, Hanaoka F, Akiyama M, Maki H. DNA polymerases eta and kappa are responsible for error-free translesion DNA synthesis activity over a cis-syn thymine dimer in Xenopus laevis oocyte extracts. DNA Repair (Amst) 2005;4:1252–1269. doi: 10.1016/j.dnarep.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Polkappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc Natl Acad Sci U S A. 2002;99:15547–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution [see comments] Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 32.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 35.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Dutta S, Doublie S, Bdour HM, Taylor JS, Ellenberger T. Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7. Nat Struct Mol Biol. 2004;11:784–790. doi: 10.1038/nsmb792. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve R, Jiang W, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;A47:110–119. [Google Scholar]
- 43.Delano WL. The PyMol Molecular Graphics System. Delano Scientific LLC. San Carlos, USA: 2002. [Google Scholar]