Abstract
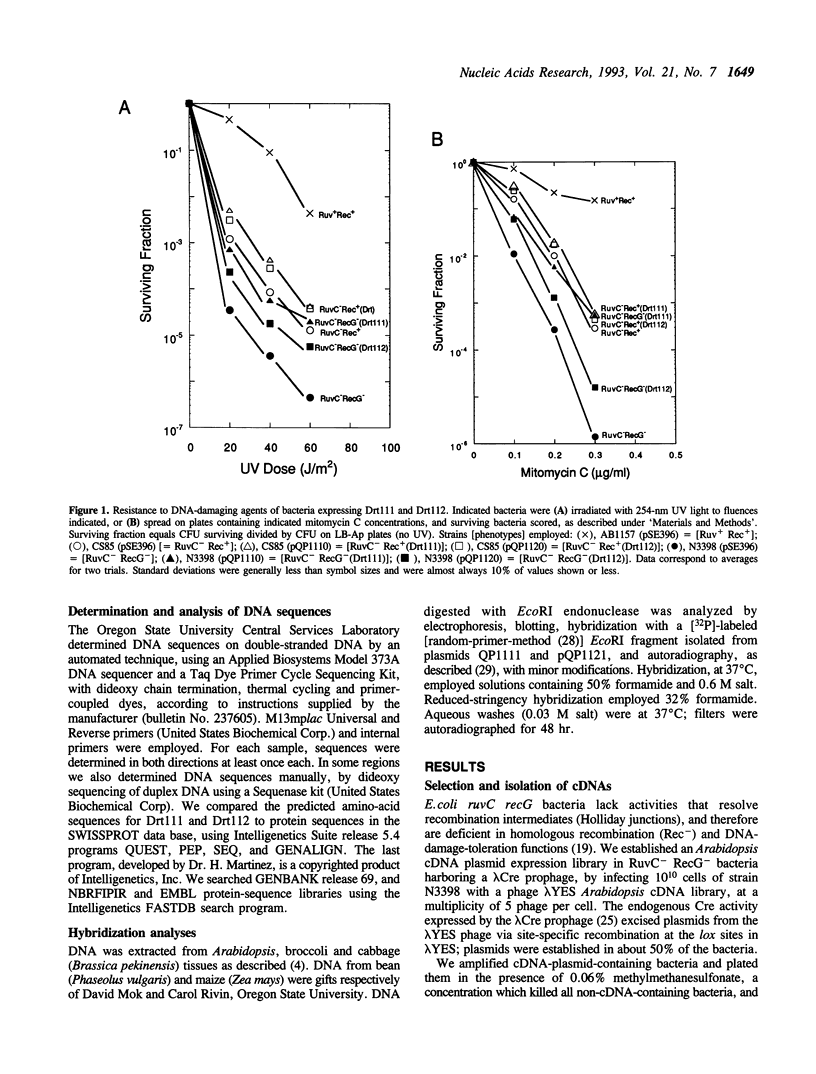
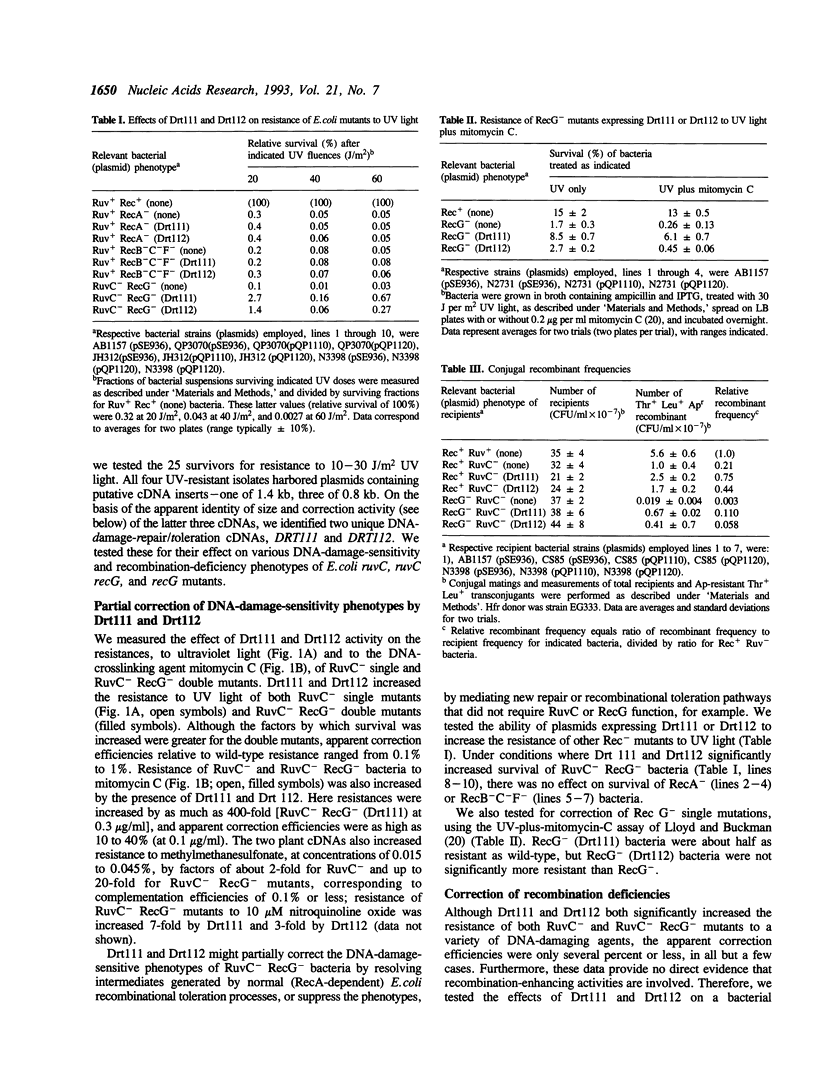
Escherichia coli ruvC recG mutants lack RuvC endonuclease, which resolves crossed-strand joint molecules (Holliday junctions) formed during homologous recombination into recombinant products, and an activity (RecG) thought to partially replace RuvC. They are therefore highly deficient in homologous recombination, and sensitive to UV light and chemical DNA-damaging agents, presumably because of inability to tolerate unrepaired DNA damage by recombinational mechanisms (Lloyd, R.G. (1991) J. Bacteriol. 173:5414-5418). We transformed these mutants with plasmids expressing cDNAs from the plant Arabidopsis thaliana. Selection for bacteria with increased resistance to methylmethanesulfonate yielded two cDNAs, designated DRT111 and DRT112 (DNA-damage-repair/toleration). Expression of these plant cDNAs, especially DRT111, restored conjugal recombination proficiencies in ruvC and ruvC recG mutants to nearly wild-type levels. Both plant cDNAs significantly increased resistance of both mutants to UV light and several chemical DNA-damaging agents, but did not fully correct the mutant phenotypes. Drt111 activity, but not Drt112, also increased, to nearly wild-type levels, resistance of recG single mutants to UV plus mitomycin C. The predicted Drt111 and Drt112 polypeptides, 383 and 167 amino acids respectively, show no similarity with one another or with prokaryotic Holliday resolvases. Both appear chloroplast targeted; Drt112 is highly homologous to Arabidopsis plastocyanin. DRT111 and DRT112 probes hybridize only to DNA from closely related plants.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth K. A., Powell D., Trupin M., Mosig G. Regulation of two nested proteins from gene 49 (recombination endonuclease VII) and of a lambda RexA-like protein of bacteriophage T4. Genetics. 1988 Oct;120(2):329–343. doi: 10.1093/genetics/120.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986 Aug;167(2):594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Osman M., Grandoni P., Jagendorf A. T. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8068–8072. doi: 10.1073/pnas.89.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Toward a metabolic interpretation of genetic recombination of E. coli and its phages. Annu Rev Microbiol. 1971;25:437–464. doi: 10.1146/annurev.mi.25.100171.002253. [DOI] [PubMed] [Google Scholar]
- Connolly B., Parsons C. A., Benson F. E., Dunderdale H. J., Sharples G. J., Lloyd R. G., West S. C. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B., West S. C. Genetic recombination in Escherichia coli: Holliday junctions made by RecA protein are resolved by fractionated cell-free extracts. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8476–8480. doi: 10.1073/pnas.87.21.8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunderdale H. J., Benson F. E., Parsons C. A., Sharples G. J., Lloyd R. G., West S. C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991 Dec 19;354(6354):506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Mulligan J. T., Ramer S. W., Spottswood M., Davis R. W. Lambda YES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990 Feb 26;261(2):455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Hays J. B., Martin S. J., Bhatia K. Repair of nonreplicating UV-irradiated DNA: cooperative dark repair by Escherichia coli uvr and phr functions. J Bacteriol. 1985 Feb;161(2):602–608. doi: 10.1128/jb.161.2.602-608.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensch F., Kosak H., Seeman N. C., Kemper B. Cruciform cutting endonucleases from Saccharomyces cerevisiae and phage T4 show conserved reactions with branched DNAs. EMBO J. 1989 Dec 20;8(13):4325–4334. doi: 10.1002/j.1460-2075.1989.tb08619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S., Kemper B., Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992 Feb;11(2):699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer C. S., Hays J. B., Windle B. E., Schaefer T. S., Lee E. H., Hays S. L., McClure M. R. Enhancement of Escherichia coli plasmid and chromosomal recombination by the Ref function of bacteriophage P1. Genetics. 1989 Nov;123(3):465–476. doi: 10.1093/genetics/123.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991 Feb;173(3):1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991 Sep;173(17):5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993 Jan;12(1):17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991 Nov;173(21):6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Pang Q., Hays J. B., Rajagopal I. A plant cDNA that partially complements Escherichia coli recA mutations predicts a polypeptide not strongly homologous to RecA proteins. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8073–8077. doi: 10.1073/pnas.89.17.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Q., Hays J. B. UV-B-Inducible and Temperature-Sensitive Photoreactivation of Cyclobutane Pyrimidine Dimers in Arabidopsis thaliana. Plant Physiol. 1991 Feb;95(2):536–543. doi: 10.1104/pp.95.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons C. A., Tsaneva I., Lloyd R. G., West S. C. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W. D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968 Jan 28;31(2):291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- Sharples G. J., Lloyd R. G. Resolution of Holliday junctions in Escherichia coli: identification of the ruvC gene product as a 19-kilodalton protein. J Bacteriol. 1991 Dec;173(23):7711–7715. doi: 10.1128/jb.173.23.7711-7715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Iwasaki H., Nakata A., Shinagawa H. SOS-inducible DNA repair proteins, RuvA and RuvB, of Escherichia coli: functional interactions between RuvA and RuvB for ATP hydrolysis and renaturation of the cruciform structure in supercoiled DNA. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8445–8449. doi: 10.1073/pnas.88.19.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. A., Hays J. B. Repair and recombination of nonreplicating UV-irradiated phage DNA in E. coli II. Stimulation of RecF-dependent recombination by excision repair of cyclobutane pyrimidine dimers and of other photoproducts. Mol Gen Genet. 1985;201(3):393–401. doi: 10.1007/BF00331329. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- Vorst O., Oosterhoff-Teertstra R., Vankan P., Smeekens S., Weisbeek P. Plastocyanin of Arabidopsis thaliana; isolation and characterization of the gene and chloroplast import of the precursor protein. Gene. 1988 May 15;65(1):59–69. doi: 10.1016/0378-1119(88)90417-9. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Meinkoth J. L., Kimmel A. R. Northern and Southern blots. Methods Enzymol. 1987;152:572–581. doi: 10.1016/0076-6879(87)52064-x. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Nie Z. Q., Yang J. The 18-kD protein that binds to the chloroplast DNA replicative origin is an iron-sulfur protein related to a subunit of NADH dehydrogenase. Plant Cell. 1989 May;1(5):551–557. doi: 10.1105/tpc.1.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagursky R. J., Hays J. B. Expression of the phage lambda recombination genes exo and bet under lacPO control on a multi-copy plasmid. Gene. 1983 Sep;23(3):277–292. doi: 10.1016/0378-1119(83)90018-5. [DOI] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]



