Abstract
We compare here the structural and functional properties of heparan sulfate (HS) chains from both male or female adult mouse liver through a combination of molecular sieving, enzymatic cleavage, and strong anion exchange-HPLC. The results demonstrated that male and female HS chains are significantly different by a number of parameters; size determination showed that HS chain lengths were ∼100 and ∼22 kDa, comprising 30–40 and 6–8 disaccharide repeats, respectively. Enzymatic depolymerization and disaccharide composition analyses also demonstrated significant differences in domain organization and fine structure. N-Unsubstituted glucosamine (ΔHexA-GlcNH3+, ΔHexA-GlcNH3+(6S), ΔHexA(2S)-GlcNH3+, and N-acetylglucosamine (ΔHexA-GlcNAc) are the predominant disaccharides in male mouse liver HS. However, N-sulfated glucosamine (ΔHexA-GlcNSO3) is the predominant disaccharide found in female liver. These structurally different male and female liver HS forms exert differential effects on human mesenchymal cell proliferation and subsequent osteogenic differentiation. The present study demonstrates the potential usefulness of gender-specific liver HS for the manipulation of human mesenchymal cell properties, including expansion, multipotentiality, and subsequent matrix mineralization. Our results suggest that HS chains show both tissue- and gender-specific differences in biochemical composition that directly reflect their biological activity.
Keywords: Glycosaminoglycan, Heparan Sulfate, Heparin, Heparin-binding Protein, Stem Cell
Introduction
Heparan sulfate proteoglycan structure classically consists of a core protein to which one or more linear HS glycosaminoglycan (GAG)2 chains are attached at specific serine-glycine residues. Heparan sulfates have complex sulfated domain substructures that are initially synthesized as non-sulfated polysaccharides of d-glucuronic acid-N-acetyl-d-glucosamine (GlcA-GlcNAc) repeats (1–4). These linear, sulfated glycosaminoglycans have molecular masses that typically range from 10 to 100 kDa (5). Concurrent with the polymerization of an HS chain is a series of enzymatic modifications that generate the diverse sulfated domain clusters at intervals along the growing chain. The non-template-driven diversity of HS structure is thought to be capable of giving rise to a wide range of biological functions through selective binding.
Several studies have demonstrated that the binding of growth factors to HS that gives rise to mitogenic or adhesive activity occurs only when specific structural features are present within the HS chain (6). Such features include sulfation at specific positions within a given sequence of disaccharides; 6-O-sulfated N-sulfate glucosamine and 2-O-sulfated iduronic acid residues are thought to be particularly important, and minimum binding sequences are generally at least 5 or 6, and often 8–14 disaccharides in length (7–9). The precise structures within HS sulfated domains that are involved in these interactions have remained elusive. Variations in composition and the organization of HS from different cells and tissues have confirmed the relationship between HS structure and bioactivity. Each tissue type bears a unique complement of HS structures that also vary at different stages of tissue development (4, 10). Differing complements of HS appear to change the way that heparin/HS-dependent growth factors, such as the FGFs, exert their mitogenic and differentiative effects within developing tissues (11).
We have previously shown that HS helps control the osteogenic differentiation of preosteoblast MC3T3-E1 cells (12) and that exogenous application of such HS to cultures of rat bone marrow stem cells can stimulate their proliferation, which then leads to an increased expression of osteogenic markers and enhanced bone nodule formation (13). In recent years human mesenchymal stem cells (hMSCs) have been shown to be an important source of cells for tissue engineering applications. They are relatively easy to isolate and can be greatly expanded with various tissue culture manipulations. These cells can be differentiated into a variety of tissues within the mesenchymal and other lineages (14), including osteoblasts (15, 16), adipocytes (17), myocytes, astrocytes, and neurons (18, 19).
Here we sought to exploit hMSCs in order to monitor the biological effects of adult liver-derived HS. Our reasoning was that liver is a highly regenerative tissue (20), with high levels of FGFs (21), and may thus contain enriched preparations of mitogenic HS (22). General liver HS structure has been explored previously (23), and organ and gender-specific HS chains have been purified from rat and murine tissues and shown to be compositionally distinct at the disaccharide level (24, 25). However, comparative structural and functional analyses of such gender-specific HS have not been undertaken to date. In the present study, male liver heparan sulfate (MML HS) and female liver heparan sulfate (FML HS) were shown to possess major gross and fine structural differences. These differentially sulfated HS chain species differed in their biological abilities to support hMSC proliferation and osteogenic differentiation.
EXPERIMENTAL PROCEDURES
Materials
Heparin lyase I, II, and III from Flavobacterium heparium, heparin oligosaccharides (dp4–dp26), 12 heparin disaccharide standards, and de-O-sulfated and de-N-sulfated heparin oligosaccharides were from Iduron (Manchester, UK). Chondroitin ABC lyase, neuraminidase, Pronase, actinase E, chloroform, and methanol were from Sigma-Aldrich. HiPrep 16/10 DEAE columns, HiPrep 26/10 desalting columns, Superdex 75 10/300 GL column, Superdex 200 10/300 GL column, and high molecular weight and low molecular weight gel filtration calibration kits were from GE Healthcare. Bio-Gel P2 was from Bio-Rad. The hMSCs were from Lonza. All other reagents used were of analytical grade.
Radiolabeling and Preparation of Liver Tissues
CBA male and female mice of 12–24 weeks were used in this study. Two male and female mice were each given an intraperitoneal injection of 0.1 ml of phosphate-buffered saline containing 1 mCi of d-[6-3H]glucosamine hydrochloride. After 24 h, the mice were killed by cervical dislocation, and whole livers were quickly excised, together with unlabeled livers from 20 mice. Harvested organs were rinsed with PBS, immediately frozen, and kept at −20 °C until processing.
Tissue Processing and Isolation and Purification of Heparan Sulfate
The livers were cut into small pieces (<1 mm3) and homogenized. Fat was removed by washing the homogenate with chloroform/methanol mixtures (2:1, 1:1, and 1:2 (v/v)) and was then freeze-dried under vacuum and stored at −20 °C until further use. This was then suspended in 0.05 m Tris acetate buffer (pH 8.0) and digested for 48 h with actinase E (10 mg/g) at 50 °C. Homogenate was then placed in a boiling water bath for 30 min to deactivate the protease and centrifuged (2500 × g) for 30 min at room temperature. The recovered supernatant was applied to a HiPrep DEAE column; the total glycosaminoglycans were first eluted with high salt and then desalted using a HiPrep desalting column and freeze-dried. The samples were measured for uronic acid content with the carbazole method (26). The total glycosaminoglycans were treated with neuraminidase and chondroitin lyase ABC at 37 °C for 24 h to remove sialic acid and chondroitin and dermatan sulfates. The reaction was terminated by heating in a boiling water bath for 15 min, whereupon the digested sample was diluted 1:10 with water. The diluted samples were applied to HiPrep DEAE columns, and HS was eluted with high salt. The eluted samples were desalted, and the resulting HS samples were freeze-dried. These were again quantified for uronic acid content. HS samples were then recovered by gel filtration chromatography on a Superdex 75 column (10/300) eluted with 10 mm HEPES buffer containing 150 mm NaCl. Fractions of 2 ml were collected at a flow rate of 30 ml/h and monitored for absorbance at 232 nm. Fractions were also monitored for their 3H content by liquid scintillation. The male and female HS fractions containing the 3H radiolabel were further analyzed on a Superdex 200 column to confirm their sizes. Protein high and low molecular weight protein standards and even-numbered oligosaccharides derived from highly sulfated heparin were used to calibrate the sizing column.
Enzymatic Depolymerization of HS
Purified, dried samples (100 μg) of HS were dissolved in 100 mm sodium acetate, 0.2 m calcium acetate, pH 7.0, and incubated with 10 milliunits/ml heparinase I, II, or III in the same buffer at 37 °C for 16 h, with a second aliquot of enzyme added for a further 6 h. The digested HS samples were analyzed on a BioGel P-10 column (1 × 120 cm) equilibrated with 0.25 m NH4HCO3. Even-numbered oligosaccharides derived from heparin were used to calibrate the column.
Disaccharide Analysis Using Strong Anion Exchange Chromatography (SAX-HPLC)
Each sample was sequentially digested for disaccharide recovery in preparation for SAX-HPLC analysis. Samples were digested at 37 °C as follows: heparinase I for 3 h, heparinase III for 2 h, heparinase II for 16 h, and finally a combined aliquot of each lyase for a further 6 h. Samples were run on a Bio-Gel P-2 column (1 × 120 cm) equilibrated with 0.25 m NH4HCO3 to recover the disaccharides and then freeze-dried prior to analysis. The lyase-derived disaccharides were resolved by SAX-HPLC on a ProPac PA1 analytical column (4 × 250 mm) linked to a Dionex ICS-3000 HPLC system. After equilibration in the mobile phase (double-distilled water adjusted to pH 3.5 with HCl) at 1 ml/min, samples were injected, and disaccharides eluted with a linear gradient of sodium chloride from 0 to 1 m over 60 min in the same mobile phase. The elution was monitored for UV absorbance at 232 nm. Disaccharides were identified by comparison with the elution positions of 12 known disaccharide standards (Iduron).
HS Binding Assay
The interaction between HS and growth factor was determined using GAG binding plates (Iduron) according to the manufacturer's instructions. Briefly, HS was coated on the plate prior to adding growth factors. Bound growth factor was detected using biotinylated antibodies (R&D Systems).
Effect of Male and Female Liver HS on hMSC Proliferation
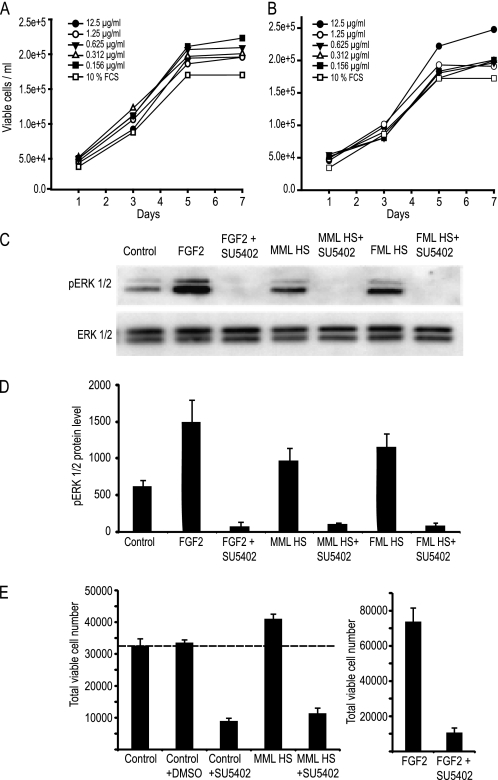
hMSCs were plated in maintenance medium consisting of DMEM (1000 mg/liter glucose), 10% fetal calf serum (FCS), 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin sulfate at 37 °C in a humidified atmosphere with 5% CO2. To assess their proliferation, assays were performed to determine cell number utilizing the GUAVA PCA-96 system (Millipore) as per the manufacturer's instructions. Briefly, cells were seeded at 3000 cells/cm2 in 48-well plates and allowed to adhere overnight. The following day, the cells were refed with the same medium with or without different concentrations of HS (12.5, 1.25, 0.625, 0.312, and 0.156 μg/ml), and the medium was changed every 3 days. Proliferation of cells was assessed on days 1, 3, 5, and 7 to determine total viable cells. Cells were washed in PBS, pelleted with 0.125% trypsin, the trypsin was neutralized, the pellet was resuspended in 400 μl of PBS with the addition of 4 μl of Flex reagent, the pellet was incubated for 10 min, and cell number and viability were determined utilizing the GUAVA Viacount software.
Cell Lysis and Immunoblotting
MSCs were seeded in designated media at 5000/cm2 in 6-well plates and cultured until subconfluent. Cells were washed and deprived of FCS for 24 h and then treated with HS or FGF-2 alone or together with the FGFR1-blocking drug SU5402 (Calbiochem) at 10 μm in DMSO for 60 min. Cells were lysed in 300 μl of ice-cold lysis buffer (150 mm NaCl, 10 mm Tris, pH 7.4, 2 mm EDTA, 1% Triton X-100, 0.1% SDS, 0.5% Igepal) supplemented with 0.2 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 30 μl of protease inhibitor mixture (Sigma). The lysate was incubated on ice for 10 min, passed through a 21-gauge needle, and then centrifuged at 10,000 × g for 10 min at 4 °C to remove cellular debris. Protein content was determined using a protein assay kit (Bio-Rad) following the manufacturer's instructions. Protein (20 μg) was mixed with an equal volume of 2× Laemmli buffer, boiled for 5 min, and then separated by SDS-PAGE on 8% gels. The protein was then transblotted onto nitrocellulose membranes (Amersham Biosciences) using a Trans-Blot® SD cell semidry transfer apparatus (Bio-Rad) for 15 min at 20 V. The membranes were blocked in 5% nonfat milk in Tris-buffered saline Tween 20 (TBST) (1.5 m NaCl, 1 m Tris, pH 7.4, 1% Tween) for 1 h and incubated overnight at 4 °C with rabbit anti-actin, rabbit anti-ERK1/2, monoclonal anti-diphosphorylated ERK1/2 (Sigma). The membranes were washed with TBST and incubated for 1 h with anti-rabbit IgG- or anti-mouse IgG-HRP-conjugated secondary antibodies (Southern Biotech, Birmingham, AL). Following washes, the membranes were covered with SuperSignal® West Pico chemiluminescent substrate (Pierce) for 5 min and then exposed on film.
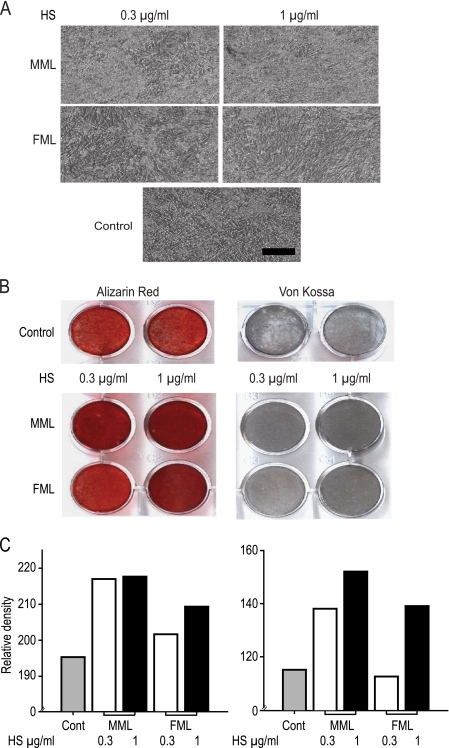
Effect of Liver HS on hMSC Osteogenic Differentiation
To confirm the effect of osteogenic potential of male and female mouse liver HS, mineralization assays were performed on triplicate cultures of hMSCs. Cells were seeded in triplicate at a density of 3000 cells/cm2 in 12-well plates in osteogenic medium (maintenance medium supplemented with 10 nm dexamethasone, 25 μg/ml l-ascorbic acid 2-phosphate, and 10 mm β-glycerophosphate) and left to adhere for 24 h. The following day the cells were refed with the same medium with or without two different concentrations of HS (1 or 0.3 μg/ml) and grown for 3 weeks with the medium being changed every 3 days. To stain for the accumulation of calcium within the matrix, cell monolayers were washed three times with PBS and incubated with 4% paraformaldehyde for 10 min at room temperature. The monolayers were then washed three times with double-distilled water (ddH2O), incubated with 1% alizarin red stain for 10 min, washed again three times with ddH2O, and air-dried. Images were taken using an Olympus BX51 microscope, DP70 camera, and DPControler software version 1.1.1.65. For phosphate nodule staining, cell monolayers were then washed three times with PBS and incubated with 4% paraformaldehyde for 10 min at room temperature. Cell monolayers were then washed three times with ddH2O and incubated for 30 min in 1% silver nitrate under UV light. The monolayers were then washed three times with ddH2O, incubated with 5% sodium thiosulfate for 2 min, and washed again three times with ddH2O and air-dried. Images were taken using an Olympus BX51 microscope, DP70 camera, and DPControler software version 1.1.1.65.
Histomorphometry Image Analysis
Bioquant Image Analysis® software (Bioquant Image Analysis Corp., Nashville, TN) was used to quantify the average density of Alizarin red S- and von Kossa-stained wells. Briefly, cultures were performed in triplicate for 21 days, with MML HS and FML HS treatments at each concentration performed on the same plate. To ensure the same light intensity and exposure within the samples, images of the plates were taken using an Epson Perfection 1670 scanner. Digital images from image analysis software were loaded into the field of view, and a consistent region of interest was placed over each well. The average density of the wells was recorded for each sample and is reported in units of grayscale, wherein darker stain correlates with minimal light transmission, thus yielding a higher density value. The data are presented as this value subtracted from the value for white light, giving apparent stain signal intensity.
Reverse Transcription-Polymerase Chain Reaction
hMSCs were seeded in triplicate at a density of 3000 cells/cm2 in 12-well plates in osteogenic medium and left to adhere for 24 h. The following day, the cells were refed with the same medium with or without two different concentrations of HS (1 or 0.3 μg/ml) and grown for 3 weeks with medium changed every 3 days. Total RNA was extracted at days 7, 14, and 21 using a Nucleospin® RNA II kit according to the manufacturer's instructions (Macherey-Nagel, Easton, PA). First strand cDNA synthesis was carried out on total RNA using Superscript III reverse transcriptase according to the manufacturer's instructions (Invitrogen). Primers and probe sets for quantitative real-time PCR (described in Table 1) were designed using Primer Express (version 1.0; Applied Biosystems) and synthesized by Proligo (Proligo LLC, Boulder, CO). Quantitative PCR was then performed to assess the relative expression of the target genes in hMSCs. This was carried out on an ABI Prism 7000 sequence detection system (Applied Biosystems, Warrington, UK) using 600 nmol/liter forward and reverse primers, 250 nmol/liter probe, and Taqman PCR Master Mix (ABI Applied Biosystems). Reactions were run in triplicate using the thermal profile, with an initial 10-min activation step at 95 °C followed by 45 cycles of 95 °C for 20 s, 55 °C for 10 s, 60 °C for 30 s, and 72 °C for 40 s. Data were normalized to 18 S ribosomal RNA expression in each sample.
TABLE 1.
Primers for real-time RT-PCR
| Genes | Oligonucleotide/probe | Sequences (5′ to 3′)a |
|---|---|---|
| 18 S rRNA | 18S_F | TTCGAGGCCCTGTAATTGGA |
| 18S_R | GCAGCAACTTTAATATACGCTATTGG | |
| 18S_P | AGTCCACTTTAAATCCTT | |
| Alkaline phosphatase | AP_F | ATGCCCTGGAGCTTCAGAAG |
| AP_R | TGGTGGAGCTGACCCTTGAG | |
| AP_P | ACG T+GG +CT+A A+GA +AT+G T+CA +TC | |
| Bone sialoprotein 2 | BSPII_F | AGAGGAAGCAATCACCAAAATGA |
| BSPII_R | TTGAGAAAGCACAGGCCATTC | |
| BSPII_P | ct g+Ct +T+Ta a+Tt +Ttg +Ct+C agc | |
| Osteopontin | OP_F | ACATCACCTCACACATGGAAAGC |
| OP_R | GCTGACTCGTTTCATAACTGTCCT | |
| OP_P | ctt c+Tg +At+T ggg +Ac+A gc+C gt | |
| RUNX2 | RX_F | AGGCATGTCCCTCGGTATGTC |
| RX_R | GAAGGGTCCACTCTGGCTTTG | |
| RX_P | aca c+Ct a+Cc tg+C ca+C ca+C cc |
Plus signs designate LNA nucleotides incorporated during probe design.
RESULTS
Purification and Sizing of Intact Gender-specific Mouse Liver HS Chains
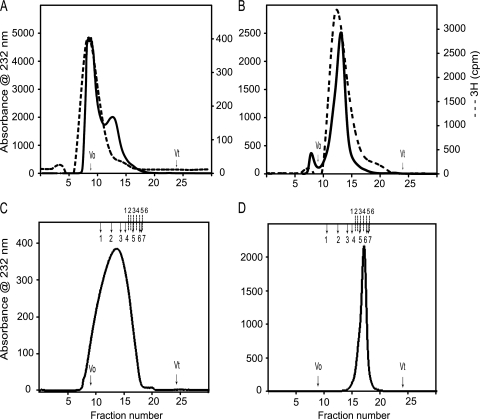
Male and female mouse HS-GAGs chains isolated with DEAE anion exchange chromatography were subsequently separated on a Superdex 75 column, which demonstrated that male chains eluted with a molecular weight peak that was similar both when excited at 232 nm and after incorporation of 3H radiolabel (Fig. 1A). In contrast, female liver HS chains eluted at a distinctly smaller molecular weight peak than the male HS chains (Fig. 1B). Importantly, however, as in the male, chain molecular weight was similar when assessed either by UV at either 206 nm (data not shown) or 232 nm or after 3H incorporation. We next reran the separated, radiolabeled MML and FML HS chains on Superdex 75 columns to confirm their homogeneity. The separated, HS chains were then treated with sodium borohydrate to release the linker oligosaccharide from the peptide backbone and subsequently fractionated on Superdex 200 columns to determine their size. This demonstrated that the MML HS eluted with an apparent Kav of 0.31 and the FML HS with a Kav of 0.54 (Fig. 1, C and D). The relative molecular weights of male and female liver HS chains thus appeared to be ∼100,000 and ∼22,000, respectively. In addition, the Superdex 200 column was calibrated with even-numbered heparin oligosaccharides ranging from Dp2 to Dp26. The FML HS chains have a Kav corresponding to Dp12, whereas the MML HS chains appear greater than Dp26. From these data, the intact FML HS chains appear to be composed of ∼6–8 disaccharide repeats, and MML HS chain appears to be composed of ∼30–40 disaccharides repeats. There is thus significant variation in the average chain lengths of MML HS and FML HS.
FIGURE 1.
High performance size exclusion chromatography of liver heparan sulfates. A Dionex HPLC system was used to equilibrate a prepacked Superdex 75 HR column (10 × 300 mm; GE Healthcare) at 0.5 ml/min in 10 mm HEPES buffer, 150 mm NaCl (pH 7.2). The anion exchange chromatography-purified liver HS samples were fractionated to check the intact full chains. Then the high molecular weight 3H-labeled male liver HS chains (A) and low molecular weight 3H-labeled female liver HS chains (B) were separated and run again on the same column to confirm the homogeneity. The male liver HS (C) and female liver HS (D) were run on the Superdex 200 HR column (10 × 300 mm) to check the size of the HS chain. The Superdex 200 column was calibrated using gel filtration high and low molecular weight protein calibration marker proteins and even-numbered heparin oligosaccharides derived from heparin. The column void (V0) and total (Vt) volumes were determined using blue dextran 2000 and sodium dichromate, respectively. The elution volumes (Ve) of protein standards were converted into a calibration chart of Kav against molecular mass (Kav = (Ve − Vo)/(Vt − Vo). A line of best fit was fitted to the calibration data using Microsoft Excel, and the equation of this line was used to estimate mass according to observed Kav. The total number of disaccharide repeats was calculated based on the elution position of the known heparin oligosaccharides (dp2–dp26). The completed arrows indicate the elution positions of high and low molecular weight protein standards (1, ferritin; 2, aldalose; 3, conalbumin; 4, ovalbumin; 5, carbonic anhydrase; 6, ribonuclease A; 7, aprotinin), and the dotted lines indicate the elution positions of heparin oligosaccharide standards (1, dp26; 2, dp20; 3, dp16; 4, dp12; 5, dp8; 6, dp6).
Enzymatic Depolymerization of HS Chains
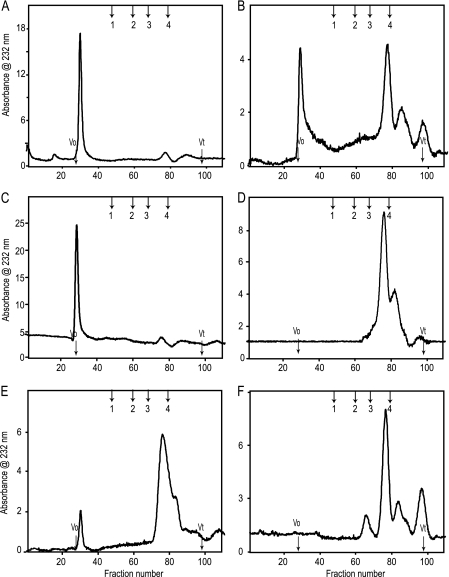
Heparinase-treated male and female liver HSs were applied to a Bio-Gel P10 column (1 × 120 cm) to determine the distribution and ratios of the resulting oligosaccharides. Representative patterns after heparinase I digestion are shown in Fig. 2, A and B. Heparinase I cleaves HS chains with sulfate-rich regions at an N-sulfated-glucosamine/sulfated-iduronic acid (GlcNS(±6S)-IdoUA(2S)) (27–31). The size of the fragments generated, compared with the undigested chain, reveals the frequency of these areas of high sulfation. Digestion by heparinase I was more extensive for FML HS than for MML HS. Male depolymerization profiles showed ∼10% linkage susceptibility, with a corresponding majority of resistant regions of larger size excluded from the Bio-Gel P10 column (Fig. 2A). However, female HS depolymerization revealed 60% linkage susceptibility to heparinase I, yielding di- and tetrasaccharides. There was a correspondingly smaller percentage of resistant regions of larger oligosaccharides eluting in the void volume (Fig. 2B). The splitting of the disaccharide peak is due to the almost complete resolution of the disaccharides into di- and trisulfated species. This reflects small clusters of the heparinase I-susceptible sites within the HS chains.
FIGURE 2.
Bio-Gel P10 size exclusion chromatography profiles of male and female liver HS after cleavage with heparinase I, heparinase II, or heparinase III. A, male liver HS after digestion with heparinase I; B, female liver HS after digestion with heparinase I; C, male liver HS after digestion with heparinase II; D, female liver HS after digestion with heparinase II; E, male liver HS after digestion with heparinase III; F, female liver HS after digestion with heparinase III. The columns void (V0) and total (Vt) volumes were determined using hemoglobin and sodium dichromate, respectively. The column was calibrated using even-numbered heparin oligosaccharides derived from heparin. The enzyme-digested peaks were identified by the elution positions of the known heparin oligosaccharides. The arrows indicate the elution positions of heparin oligosaccharide standards (1, dp10; 2, dp6; 3, dp4; 4, dp2).
Heparinase II has broad substrate specificity (28, 32, 33), with two distinct activities, one of which is heparinase I-like and the other of which is heparinase III-like (34). Heparinase II also has relative substrate specificity for unsaturated disaccharides that are O-sulfated at the C2 of uronic acid or C6 of GlcNH3+ (35). Heparinase II-digested MML HS profiles were similar to those of heparinase I (Fig. 2C). In contrast, FML HS completely digested into di- and tetrasaccharides with almost no material remaining in the void volume (Fig. 2D). Heparinase III cleaves HS chains within N-acetylated or N-sulfated regions (GlcNAc/GlcNS-HexA), with a preference for GlcA over IdoA. It tolerates 6-O-sulfation of the amino sugar but is inhibited by 2-O-sulfation of IdoA (33, 36). Heparinase III also cleaves non-sulfated, N-unsubstituted disaccharide (GlcA-GlcNH3+) (37). Heparinase III digested ∼80% of the MML HS chains, with the resulting digested fragments mainly di- and tetrasaccharides and a smaller portion eluting in the void volume (Fig. 2E). The FML HS chains completely digested into di-, tetra-, and hexasaccharides, with almost no material remaining in the void volume (Fig. 2F). These data demonstrated significant gross structural differences between the MML HS and FML HS.
SAX-HPLC Disaccharide Analysis
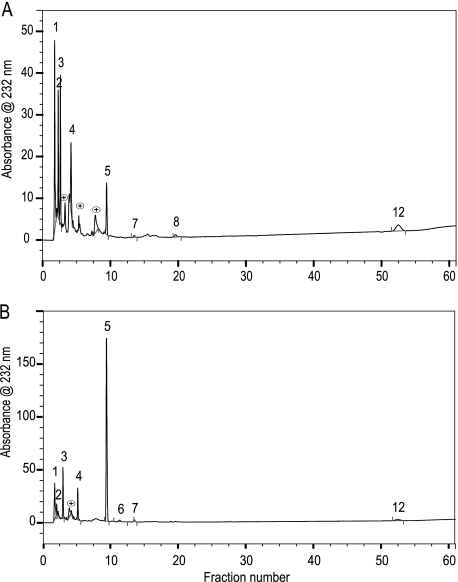
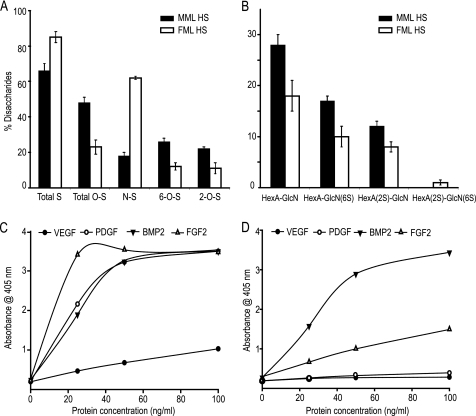
To investigate the fine structural differences in the disaccharides of the male and female liver HS chains, exhaustive digestion with a mixture of heparinases I, II, and III and separation on a Bio-Gel P2 column (1 × 120 cm) were followed by strong anion exchange HPLC; the resulting disaccharide peaks were identified by reference to well characterized disaccharide standards (Fig. 3, A and B). The area under each peak was used to obtain the disaccharide content of each sample. The N-unsubstituted GlcNH3+ (ΔHexUA-GlcNH3+, ΔHexUA-GlcNH3+(6S), and ΔHexUA(2S)-GlcNH3+) and N-acetylated (ΔHexUA-GlcNAc) forms are predominant in the MML HS, whereas the N-sulfated (ΔHexUA-GlcNSO3) form is predominant in the FML HS (Table 2). The total sulfation of the MML HS was 66%, with 18% N-sulfation, 26% 6-O-sulfation, and 22% 2-O-sulfation; FML HS total sulfation was 85%, with 62% N-sulfation, 12% 6-O-sulfation, and 11% 2-O-sulfation (Fig. 4A). MML and FML HS were composed of 57 and 30% N-unsubstituted disaccharides, respectively. This is higher than the 1.2–7.5% previously reported from various porcine, bovine, and rat tissues (24, 38, 39). Non-sulfated, monosulfated, and disulfated N-unsubstituted disaccharides (ΔHexUA-GlcNH3+, ΔHexUA-GlcNH3+(6S), ΔHexUA(2S)-GlcNH3+, and ΔHexUA(2S)-GlcNH3+(6S)) had different proportions in male and female liver HS (Fig. 4B). These results confirmed the major structural differences between gender-specific liver HS chains at the fine as well as the gross level. GAG binding plates revealed that male liver HS binds to all growth factors tested (FGF2, BMP2, PDGF, and VEGF; Fig. 4C), whereas female live HS selectively bound only FGF2 and BMP2 (Fig. 4D).
FIGURE 3.
Strong anion exchange high performance liquid chromatography profiles of male and female liver HS after cleavage with heparin lyases. Disaccharides were prepared by exhaustive digestion with a combination of heparinases I, II, and III; disaccharides were purified by Bio-Gel P2 gel filtration chromatography and then resolved on a Pro-Pac PA1-SAX-HPLC column eluted with a gradient of NaCl as described under “Experimental Procedures.” The elution was monitored using a UV detector at 232 nm. A, male liver HS after digestion with heparinase I, II, and III; B, female liver HS after digestion with heparinase I, II, and III. The column was calibrated using 12 unsaturated heparin disaccaride standards from Iduron (1, ΔHexUA-GlcN (Δdi-nonS); 2, ΔHexUA-GlcNAc (aΔdi-nonS); 3, ΔHexUA-GlcN,6S (Δdi-mono6S); 4, ΔHexUA,2S-GlcN (Δdi-mono2S); 5, ΔHexUA-GlcNSO3 (Δdi-monoNS); 6, ΔHexUA-GlcNAc,6S (aΔdi-mono6S); 7, ΔHexUA,2S-GlcNAc (aΔdi-mono2S); 8, ΔHexUA,2S-GlcN,6S (Δdi-di(2,6)S); 9, ΔHexUA-GlcNSO3,6S (Δdi-di(6,N)S); 10, ΔHexUA,2S-GlcNSO3 (Δdi-di(2,N)S); 11, ΔHexUA,2S-GlcNAc,6S (aΔdi-di(2,6)S); 12, ΔHexUA,2S-GlcNSO3,6S (Δdi-tri(2,6,N)S)). The numbers on the peaks correspond to the elution positions of known disaccharide standards.
TABLE 2.
Lyase-derived disaccharide percentage compositions of soluble, cell surface, and matrix HS
The area under each peak was integrated to calculate the percentage of each disaccharide. ND, not detected.
| Number | Disaccharide standards | Suggested terminology | Composition |
|
|---|---|---|---|---|
| Male liver HS | Female liver HS | |||
| % | % | |||
| 1 | ΔHexUA-GlcN | Δdi-nonS | 28 ± 2 | 12 ± 3 |
| 2 | ΔHexUA-GlcNAc | aΔdi-nonS | 14 ± 1 | 2 ± 1 |
| 3 | ΔHexUA-GlcN,6S | Δdi-mono6S | 17 ± 1 | 10 ± 2 |
| 4 | ΔHexUA,2S-GlcN | Δdi-mono2S | 12 ± 1 | 8 ± 1 |
| 5 | ΔHexUA-GlcNSO3 | Δdi-monoNS | 10 ± 1 | 60 ± 2 |
| 6 | ΔHexUA-GlcNAc,6S | aΔdi-mono6S | 0.5 ± 1 | ND |
| 7 | ΔHexUA,2S-GlcNAc | aΔdi-mono2S | 1.5 ± 1 | 0.5 ± 0.5 |
| 8 | ΔHexUA,2S-GlcN,6S | Δdi-di(2,6)S | ND | 0.5 ± 0.5 |
| 9 | ΔHexUA-GlcNSO3,6S | Δdi-di(6,N)S | ND | ND |
| 10 | ΔHexUA,2S-GlcNSO3 | Δdi-di(2,N)S | ND | ND |
| 11 | ΔHexUA,2S-GlcNAc,6S | aΔdi-di(2,6)S | ND | ND |
| 12 | ΔHexUA,2S-GlcNSO3,6S | Δdi-tri(2,6,N)S | 8 ± 2 | 2 ± 1 |
| Unknown | 9 ± 2 | 5 ± 1 | ||
FIGURE 4.
The percentage composition of sulfated and N-unsubstituted disaccharides of male and female liver tissues analyzed by SAX-HPLC. A, the percentage of total, O-, N-, 6-O-, and 2-O-sulfated disaccharides among total disaccharides of male and female liver HS. B, the percentage of N-unsubstituted disaccharides among total disaccharides of male and female liver HS. C and D, growth factors binding to male and female mouse liver HS. Shown is growth factor binding ability of male (C) and female (D) mouse liver HS coated on Iduron heparin/GAG binding plates. Error bars, ± S.E.
Effect of Gender-specific Liver HS on hMSC Proliferation
To assess whether the structural differences identified between the male and female HS underpin differences in their bioactivities, varying concentrations of HS were used to supplement tissue culture media, and hMSC proliferation was monitored. Viable cell numbers were assessed over a 7-day culture period in either normal maintenance or differentiation (osteogenic) media. In growth medium, progressive MML HS dose dependently increased the cell number, up to 12.5 μg/ml. Notably, however, for FML HS, there was an inverse relationship, with decreasing doses giving a greater proliferative effect (Fig. 5, A and B). In all cases, liver HS, irrespective of gender, stimulated proliferation of hMSCs in the presence of 10% FCS. Taken separately, FML-HS was maximally proliferative at 0.156 μg/ml, whereas MML-HS was maximally proliferative at 12.5 μg/ml. Notably, despite this dosing effect, both HSs produced similar numbers of hMSCs over a 7-day period (2–2.5 × 105). The GAG binding plates suggested that the liver HS isolates had much greater affinities for FGF2 than PDGF-BB, VEGF165, or BMP2, factors known to be physiologically relevant for hMSCs (supplemental Fig. 1). In order to confirm whether the mechanism involved in triggering the proliferation by the liver HS was via FGF2, because HS is a co-receptor for the FGF-FGFR complex and because FGFR1 is a high affinity receptor for FGF-2 binding, we next sought to verify whether the inhibition of this receptor with the chemical inhibitor SU5402 could affect the intracellular signaling activity of liver HS. Activation of the ERK cascade is required for FGF-induced mitogenesis. Thus, we sought to verify if liver HS, like FGF-2, could induce ERK activation in low serum conditions. As expected if the HS was involved directly with FGFR1, both male and female HS were able to stimulate the phosphorylation of ERK1/2 (Fig. 5C; protein levels quantified in Fig. 5D); the effects for both liver HS species could be blocked with the SU5402. Knowing that MML HS maximally binds FGF2 and increases hMSC proliferation, we next examined whether this effect was mediated by FGFR signaling. Pulsing with SU5402 had an inhibitory effect on the basal growth of hMSCs while also preventing HS- and FGF2-induced proliferation (Fig. 5E). This result further confirmed that the mechanism involved in triggering the proliferation by the liver HS was via FGF2-FGFR signaling.
FIGURE 5.
Effect of male and female liver HS on the growth of hMSC. Cells were cultured for 7 days in normal maintenance medium in the presence of different concentrations (12.5, 1.25, 0.625, 0.312, or 0.156 μg/ml) of male liver HS (A) and female liver HS (B). Viable cell counts were determined at days 1, 3, 5, and 7. C, the importance of FGFR signaling for HS activity was investigated by supplementing serum-starved subconfluence cultures with male or female HS (2.5 μg/ml) or FGF2 (2.5 ng/ml) in the presence or absence of an FGFR1 inhibitor (SU5402 at 20 μm) for 60 min. Total protein was extracted for Western blotting and first probed for activated ERK1/2 expression (pERK1/2). Membranes were then striped and reprobed with anti-ERK1/2 antibody. D, the amount of pERK1/2 protein was determined by densitometry. E, cells were cultured for 5 days with male HS (2.5 μg/ml) or FGF2 (2.5 ng/ml) in the presence or absence of an FGFR1 inhibitor (SU5402 at 20 μm). Error bars, ± S.E.
In Vitro Osteogenic Differentiation and Mineralization of hMSC
We next examined whether these differential effects were maintained when the cells were induced to differentiate down the osteoblast lineage. Growth rates of cells were confirmed in osteogenic medium over 7 days (supplemental Fig. 2). Photomicrography revealed the osteogenic differentiation of hMSC with or without liver HS in osteogenic media after 21 days (Fig. 6A); it also clearly showed that both male and female liver HS increase hMSC mineralization in a dose-dependent manner. MML HS was considerably more potent because at lower concentrations (0.3 μg/ml) it induced as much mineralization as FML HS at higher concentrations (1 μg/ml). The effect of liver HS on osteogenic differentiation was also visualized by Alizarin red S and von Kossa staining. MML HS at both 0.3 and 1 μg/ml induced an extensively mineralized matrix after 21 days, whereas FML HS at 0.3 μg/ml concentration did not show increased mineralization over controls (Fig. 6B). At 1 μg/ml, female liver HS increased mineralization to a level comparable with that seen with 0.3 μg/ml MML HS. Measurement of calcium-bound Alizarin red S dye showed higher calcium accumulation in male HS-treated samples than FML HS (Fig. 6C). Measurement of mineralized bone nodules by von Kossa staining similarly showed higher bone nodules in MML HS-treated cells than FML HS (Fig. 6C).
FIGURE 6.
Osteogenic differentiation of hMSCs 21 days after induction with osteogenic medium. A, phase-contrast morphology of hMSCs grown in osteogenic medium with or without (0.3 or 1 μg/ml) liver HS after a 21-day culture period. All pictures are shown in ×10 magnification. B, mineralized nodules formed by controls and HS-treated hMSCs were stained with Alizarin red S and Von Kossa. Representative dishes of different concentrations of HS on hMSC at 21 days of culture were stored. C, quantitative analysis of Alizarin red S and Von Kossa staining density was measured with BioQuant software. Representative examples of triplicate wells are shown. Scale bar, 100 μm.
Expression of Osteogenic Markers
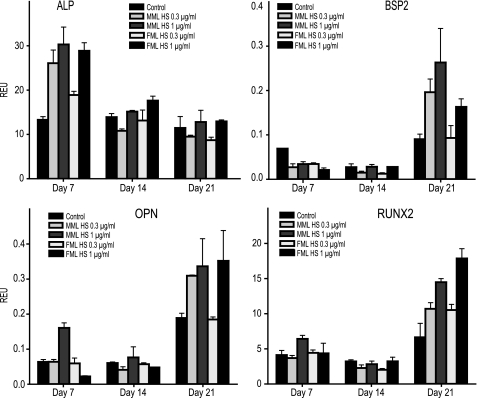
We next investigated the effect of liver HS on the expression of mRNA transcripts related to osteoblast differentiation with quantitative PCR. The expression level of alkaline phosphatase, a biomarker of early stage osteoblast differentiation, was markedly increased after 7 days culture in the presence of liver HS. The MML HS (0.3 or 1 μg/ml)-treated cells showed higher expression of alkaline phosphatase when compared with controls. Again, only the higher concentrations (1 μg/ml) of FML HS could trigger increased alkaline phosphatase expression. In MML HS-treated hMSCs, the expression levels of BSP2 (bone sialoprotein), osteopontin, and Runx2 were up-regulated at both concentrations tested over a 21-day culture period compared with controls but only by the higher concentration of FML HS (Fig. 7). Notably, irrespective of the type of HS, there was an increase in the osteogenic biomarkers following exogenous dosing with HS.
FIGURE 7.
The expression profile of osteogenic marker genes after 7, 14, and 21 days of HS treatment. hMSCs were seeded at 3000 cells/cm2 and grown in osteogenic medium with or without liver HS for 21 days with medium changes every 3 days. Total RNA was extracted at days 7, 14, and 21, and quantitative real-time polymerase chain reaction was performed using sequence-specific primers (Table 1) and probes as markers of osteoblast differentiation. The expression levels of osteogenic markers alkaline phosphatase (ALP), BSP2, osteopontin (OPN), and Runx2 were normalized to universal 18 S ribosomal RNA. The data represent mean relative expression units (REU) ± S.E. (error bars) (n = 3).
DISCUSSION
In this study, we analyze the structural and functional properties of the HS chains purified from male and female mouse liver tissue and show they are structurally and functionally distinct. Our SEC-HPLC data suggest that MML HS was ∼100 kDa, with ∼30–40 disaccharide repeats, comparable with the chain lengths observed from seven other rat tissues (38). The FML HS had an apparent molecular mass of 22 kDa, with ∼6–8 disaccharide repeats. This is consistent with a previous report that fully sulfated heparin 12-mers also have a molecular mass of ∼22 kDa (40). We also purified HS from both male and female mouse liver from rats of different ages, but the results remained consistent (data not shown). This confirmed that the HS chain size and structure was gender- rather than age-specific.
MML HS showed less sensitivity to heparinase I, suggesting that it consists primarily of relatively small clusters at its non-reducing end. This is similar to skin HS, where the IdoUA residue essential for heparinase I recognition is present only at low concentrations (41–43). Female liver HS had a greater sensitivity to heparinase I; thus, heparinase-susceptible sites are closer in FML HS than MML HS. Heparinase III cleaved 80% of the MML HS into di- and tetrasaccharides. This is similar to the 78% obtained for endothelial HS (44) but substantially different from the 63% obtained for skin fibroblast HS (41). Female liver HS in contrast was completely degraded into di- and tetrasaccharides. This is unusual when compared with other HS. The susceptible sites were highly contiguous, and the resistant fragments alternated to form tetrasaccharides. These results suggest that FML HS is composed of mostly N-acetylated, N-sulfated, and N-unsubstituted disaccharides. MML HS showed less sensitivity and FML HS showed more sensitivity to heparinase II. This must reflect the close proximity of heparinase II-susceptible sites in the FML HS.
The disaccharide composition analyses also confirmed that MML HS is less sulfated than the FML HS. This is consistent with the results of a heparinase I digestion profile of MML HS and FML HS. Comparing the male with the female liver HS, it can be seen that ΔHexUA(2S)-GlcNS(6S) and ΔHexUA-GlcNS are the most likely to be the sulfated disaccharides at the non-reducing ends of both HSs. This is consistent with a previous report that the non-reducing end of bovine kidney HS is heavily sulfated and especially N-sulfated (45). MML HS had ∼70% disaccharides that were less sulfated, but heparinase II digestion produced only ∼10 disaccharides, presumably because the N-acetylated saccharides (GlcA-GlcNAc) continuously present at the reducing end cleave into disaccharides, but the N-sulfated, N-unsubstituted, and 6-O-N-unsubstituted saccharides present within the N-acetyl/N-sulfated region in the middle of the HS chain do not. This enzyme cleaves these saccharides, but because of the longer chain length, they are excluded into the void volume of the P10 column. However, in the FML HS, ∼90% of saccharides are less sulfated, so the complete chain is cleavable by heparinase II into disaccharides. This again must reflect the close proximity of heparinase II-susceptible sites within the FML HS chains.
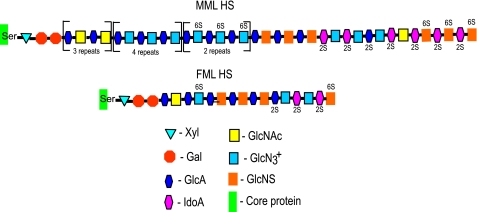
The overall structures of male and female mouse liver intact HS chains can be predicted with the combined data obtained from chain length; heparinase I, II, and III digestion; and percentage composition of the differentially sulfated disaccharides (Fig. 8). Intact MML HS and FML HS chains are composed of 30–40 and 6–8 disaccharide repeats, respectively. MML HS chains may consist of a substantial proportion of alternating acetylated glucosamine sequence (GlcA-GlcNAc) at the reducing end (NA domain), with N-unsubstituted glucosamines the major proportion of mixed sequences in the middle of the HS chain, containing ΔHexA-GlcNH3+, ΔHexA-GlcNH3+(6S), ΔHexA(2S)-GlcNH3+, ΔHexA(2S)-GlcNAc, and a minor portion of ΔHexA-GlcNSO3 (NA/NS domains). There is also a relatively minor proportion of ∼3–4 trisulfated disaccharides (ΔHexA(2S)-GlcNS(6S) in an S-domain at the non-reducing end. FML HS chains consist of a major proportion of N-sulfated glucosamines (ΔHexA-GlcNSO3) with a substantial proportion of N-unsubstituted glucosamines in the middle of the HS chain, which contains ΔHexA-GlcNH3+, ΔHexA(2S)-GlcNH3+ (NA/NS domain). A minor proportion of ΔHexA-GlcNAc is present at the reducing end (N-acetyl domain) with a minor proportion of ΔHexA(2S)-GlcNS(6S) present at the non-reducing end (N-sulfated domain).
FIGURE 8.
Proposed structural model of male and female mouse liver intact HS chains. The structure of male and female mouse liver HS chains has been predicted with combined data obtained from chain length (Fig. 1), heparin lyase digestion patterns (Fig. 2), SAX-HPLC disaccharide profile (Fig. 3), percentage of disaccharide composition (Table 1), and percentage of sulfated and N-unsubstituted disaccharides (Fig. 4). Note that there are three repeats of duplicated GlcA-GlcNAc, four repeats of triplicated GlcA-GlcNH3+, and two repeats of triplicated GlcA-GlcNH3+(6S).
The structural data immediately raise the question of what functional differences are manifested by the different chains. We selected human mesenchymal stem cells for functional studies because hMSCs are able to manifest several different fates, including osteogenic, depending on the extracellular milieu. We have previously shown that HS isolated from neuroepithelial cells can increase the proliferation and osteogenic differentiation of rat bone marrow stem cells (13). Male and female liver HS also act to increase the proliferation and differentiation of hMSCs, albeit the FML preparations were of substantially less potency. Interestingly, it cannot be just the degree of sulfation that is the determinant of activity because the shorter, more highly sulfated FML HS were overall less active. Blocking FGFR1 with SU5402 indicates that the major mechanism of action for the liver HS preparations involves FGF signaling through the ERK1/2 pathway.
In control experiments, the effect of different de-O-sulfated heparins on hMSC growth in normal and osteogenic differentiation medium was also assessed. Only de-N-sulfated heparin failed to improve hMSC proliferation; 2-de-O-sulfated or 6-de-O-sulfated heparin had less effect on proliferation, and neither de-N-sulfated, 2-de-O-sulfated, nor 6-de-O-sulfated heparin had any effect on hMSC proliferation in osteogenic medium (data not shown). Lower concentrations of MML HS are presumably able to induce greater levels of osteogenic differentiation in hMSCs because cell proliferation was greater over the 5 days, so resulting in faster rates of confluence, which in turn triggered more efficient osteoblast phenotypic development through better matrix mineralization; Yang et al. (46) have also shown that the degree of HS sulfation is an important determinant of rates of hMSC osteogenic differentiation and mineralization. Bovine aortic smooth muscle cell mineralization is inhibited by highly sulfated heparin, but N-desulfated heparin or the less sulfated dermatan sulfate has no inhibitory activity. Our data, however, show that MML HS, with its lesser sulfation levels (65% with only 11% N-sulfation) was better at triggering osteogenic differentiation and mineralization than the FML HS (85% with 61% N-sulfation). These findings further confirm that specific HS structure, and not charge density, is the significant determinant for osteogenic differentiation and mineralization. We show here that HS modulates the expansion and osteogenic differentiation of hMSCs, thus offering the potential to develop a novel strategy for the ex vivo expansion of adult stem cells for therapeutic use.
Supplementary Material
Acknowledgments
We thank Dr. A. Sawyer for help with Bioquant analysis and Zophia Lim for assistance with the animal work.
This work was supported by the Biomedical Research Council A*STAR (Agency for Science Technology and Research), Singapore, and the Institute of Medical Biology, Singapore.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GAG
- glycosaminoglycan
- HS
- heparan sulfate
- MML HS
- male mouse liver heparan sulfate
- FML HS
- female mouse liver heparan sulfate
- hMSC
- human mesenchymal stem cell
- dp
- degree of polymerization
- SAX
- strong anion exchange.
REFERENCES
- 1. Casu B., Lindahl U. (2001) Adv. Carbohydr. Chem. Biochem. 57, 159–206 [DOI] [PubMed] [Google Scholar]
- 2. Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 4. Lindahl U., Kusche-Gullberg M., Kjellén L. (1998) J. Biol. Chem. 273, 24979–24982 [DOI] [PubMed] [Google Scholar]
- 5. Gandhi N. S., Mancera R. L. (2008) Chem. Biol. Drug. Des. 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 6. Walker A., Turnbull J. E., Gallagher J. T. (1994) J. Biol. Chem. 269, 931–935 [PubMed] [Google Scholar]
- 7. Lyon M., Rushton G., Askari J. A., Humphries M. J., Gallagher J. T. (2000) J. Biol. Chem. 275, 4599–4606 [DOI] [PubMed] [Google Scholar]
- 8. Ostrovsky O., Berman B., Gallagher J., Mulloy B., Fernig D. G., Delehedde M., Ron D. (2002) J. Biol. Chem. 277, 2444–2453 [DOI] [PubMed] [Google Scholar]
- 9. Pye D. A., Gallagher J. T. (1999) J. Biol. Chem. 274, 13456–13461 [DOI] [PubMed] [Google Scholar]
- 10. Brickman Y. G., Ford M. D., Gallagher J. T., Nurcombe V., Bartlett P. F., Turnbull J. E. (1998) J. Biol. Chem. 273, 4350–4359 [DOI] [PubMed] [Google Scholar]
- 11. Nurcombe V., Ford M. D., Wildschut J. A., Bartlett P. F. (1993) Science 260, 103–106 [DOI] [PubMed] [Google Scholar]
- 12. Jackson R. A., Murali S., van Wijnen A. J., Stein G. S., Nurcombe V., Cool S. M. (2007) J. Cell. Physiol. 210, 38–50 [DOI] [PubMed] [Google Scholar]
- 13. Dombrowski C., Song S. J., Chuan P., Lim X., Susanto E., Sawyer A. A., Woodruff M. A., Hutmacher D. W., Nurcombe V., Cool S. M. (2009) Stem Cells Dev. 18, 661–670 [DOI] [PubMed] [Google Scholar]
- 14. Mauney J. R., Volloch V., Kaplan D. L. (2005) Tissue Eng. 11, 787–802 [DOI] [PubMed] [Google Scholar]
- 15. Bruder S. P., Jaiswal N., Haynesworth S. E. (1997) J. Cell. Biochem. 64, 278–294 [DOI] [PubMed] [Google Scholar]
- 16. Haynesworth S. E., Goshima J., Goldberg V. M., Caplan A. I. (1992) Bone 13, 81–88 [DOI] [PubMed] [Google Scholar]
- 17. Pittenger M. F., Martin B. J. (2004) Circ. Res. 95, 9–20 [DOI] [PubMed] [Google Scholar]
- 18. Hofstetter C. P., Schwarz E. J., Hess D., Widenfalk J., El Manira A., Prockop D. J., Olson L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanchez-Ramos J., Song S., Cardozo-Pelaez F., Hazzi C., Stedeford T., Willing A., Freeman T. B., Saporta S., Janssen W., Patel N., Cooper D. R., Sanberg P. R. (2000) Exp. Neurol. 164, 247–256 [DOI] [PubMed] [Google Scholar]
- 20. Gennero L., Roos M. A., Sperber K., Denysenko T., Bernabei P., Calisti G. F., Papotti M., Cappia S., Pagni R., Aimo G., Mengozzi G., Cavallo G., Reguzzi S., Pescarmona G. P., Ponzetto A. (2010) Cell Biochem. Funct. 28, 178–189 [DOI] [PubMed] [Google Scholar]
- 21. Touboul T., Hannan N. R., Corbineau S., Martinez A., Martinet C., Branchereau S., Mainot S., Strick-Marchand H., Pedersen R., Di Santo J., Weber A., Vallier L. (2010) Hepatology 51, 1754–1765 [DOI] [PubMed] [Google Scholar]
- 22. Jastrebova N., Vanwildemeersch M., Lindahl U., Spillmann D. (2010) J. Biol. Chem. 285, 26842–26851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lyon M., Deakin J. A., Gallagher J. T. (1994) J. Biol. Chem. 269, 11208–11215 [PubMed] [Google Scholar]
- 24. Warda M., Toida T., Zhang F., Sun P., Munoz E., Xie J., Linhardt R. J. (2006) Glycoconj. J. 23, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi X., Zaia J. (2009) J. Biol. Chem. 284, 11806–11814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berry D., Shriver Z., Venkataraman G., Sasisekharan R. (2004) Biochem. Biophys. Res. Commun. 314, 994–1000 [DOI] [PubMed] [Google Scholar]
- 27. Hovingh P., Linker A. (1970) J. Biol. Chem. 245, 6170–6175 [PubMed] [Google Scholar]
- 28. Linhardt R. J., Turnbull J. E., Wang H. M., Loganathan D., Gallagher J. T. (1990) Biochemistry 29, 2611–2617 [DOI] [PubMed] [Google Scholar]
- 29. Linker A., Sampson P. (1960) Biochim. Biophys. Acta 43, 366–368 [DOI] [PubMed] [Google Scholar]
- 30. Lohse D. L., Linhardt R. J. (1992) J. Biol. Chem. 267, 24347–24355 [PubMed] [Google Scholar]
- 31. Perlin A. S., Mackie D. M., Dietrich C. P. (1971) Carbohydr. Res. 18, 185–194 [DOI] [PubMed] [Google Scholar]
- 32. Moffat C. F., McLean M. W., Long W. F., Williamson F. B. (1991) Eur. J. Biochem. 202, 531–541 [DOI] [PubMed] [Google Scholar]
- 33. Nader H. B., Porcionatto M. A., Tersariol I. L., Pinhal M. A., Oliveira F. W., Moraes C. T., Dietrich C. P. (1990) J. Biol. Chem. 265, 16807–16813 [PubMed] [Google Scholar]
- 34. Rhomberg A. J., Shriver Z., Biemann K., Sasisekharan R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12232–12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moffat C. F., McLean M. W., Long W. F., Williamson F. B. (1991) Eur. J. Biochem. 197, 449–459 [DOI] [PubMed] [Google Scholar]
- 36. Desai U. R., Wang H. M., Linhardt R. J. (1993) Biochemistry 32, 8140–8145 [DOI] [PubMed] [Google Scholar]
- 37. Wei Z., Lyon M., Gallagher J. T. (2005) J. Biol. Chem. 280, 15742–15748 [DOI] [PubMed] [Google Scholar]
- 38. Zhang J., Wang Y., Shi X. (2009) Comput. Med. Imaging Graph. 33, 602–607 [DOI] [PubMed] [Google Scholar]
- 39. Toida T., Yoshida H., Toyoda H., Koshiishi I., Imanari T., Hileman R. E., Fromm J. R., Linhardt R. J. (1997) Biochem. J. 322, 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson C. J., Harmer N. J., Goodger S. J., Blundell T. L., Gallagher J. T. (2005) J. Biol. Chem. 280, 42274–42282 [DOI] [PubMed] [Google Scholar]
- 41. Turnbull J. E., Gallagher J. T. (1990) Biochem. J. 265, 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Turnbull J. E., Gallagher J. T. (1991) Biochem. J. 273, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turnbull J. E., Gallagher J. T. (1991) Biochem. J. 277, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lindblom A., Fransson L. A. (1990) Glycoconj. J. 7, 545–562 [DOI] [PubMed] [Google Scholar]
- 45. Wu Z. L., Lech M. (2005) J. Biol. Chem. 280, 33749–33755 [DOI] [PubMed] [Google Scholar]
- 46. Yang L., Butcher M., Simon R. R., Osip S. L., Shaughnessy S. G. (2005) Atherosclerosis 179, 79–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.