Abstract
The β-globin locus undergoes dynamic chromatin interaction changes in differentiating erythroid cells that are thought to be important for proper globin gene expression. However, the underlying mechanisms are unclear. The CCCTC-binding factor, CTCF, binds to the insulator elements at the 5′ and 3′ boundaries of the locus, but these sites were shown to be dispensable for globin gene activation. We found that, upon induction of differentiation, cohesin and the cohesin loading factor Nipped-B-like (Nipbl) bind to the locus control region (LCR) at the CTCF insulator and distal enhancer regions as well as at the specific target globin gene that undergoes activation upon differentiation. Nipbl-dependent cohesin binding is critical for long-range chromatin interactions, both between the CTCF insulator elements and between the LCR distal enhancer and the target gene. We show that the latter interaction is important for globin gene expression in vivo and in vitro. Furthermore, the results indicate that such cohesin-mediated chromatin interactions associated with gene regulation are sensitive to the partial reduction of Nipbl caused by heterozygous mutation. This provides the first direct evidence that Nipbl haploinsufficiency affects cohesin-mediated chromatin interactions and gene expression. Our results reveal that dynamic Nipbl/cohesin binding is critical for developmental chromatin organization and the gene activation function of the LCR in mammalian cells.
Keywords: Cell Differentiation, Chromatin Immunoprecipitation (ChiP), Chromatin Regulation, Development, Gene Regulation, CTCF, Nipbl, β-globin, Cohesin, Locus Control Region
Introduction
An emerging aspect of epigenetics is the three-dimensional organization of chromatin determined by long-distance chromatin interactions. Evidence suggests that this has critical influence on the nuclear positioning and distal regulatory element-promoter interactions important for developmentally coordinated and cell type-specific gene expression (1). However, our knowledge of the factors involved in these processes is limited.
The β-globin locus has been characterized extensively as a prime example of such chromatin organization. The 80-kb locus is flanked by 5′ and 3′ insulator elements that interact to form a large chromatin loop mediated by the CCCTC-binding factor (CTCF)3 (2). The interactions between the distal enhancer within the locus control region (LCR) and the developmental stage-specific β-globin genes strongly correlate with proper activation (3, 4). Despite the identification of transcriptional activators, such as LIM domain binding 1 (5), that critically influence this process, the molecular mechanism responsible for this bridging was not well understood (6).
Cohesin is a conserved protein complex essential for sister chromatid cohesion and proper chromosome segregation during cell division (7). It consists of SMC1, SMC3, Rad21 (or Scc1), and SA (or Scc3). Chromatin binding of cohesin requires the loading factor Nipbl (Scc2 or delangin). Recent evidence indicates that cohesin also plays a role in gene regulation. A major mechanism of the role of cohesin in gene regulation is thought to involve CTCF (8). CTCF is a zinc finger DNA-binding protein that acts as a transcriptional activator/repressor as well as an insulator (9). CTCF recruits cohesin to many of its binding sites, and over 70% of cohesin binding sites identified in unique regions in the mouse and human genome overlap with those of CTCF (8). Cohesin depletion impairs CTCF-mediated insulator function and chromatin loop formation (10–12). These studies suggested that cohesin plays an architectural role in chromatin domain organization in the context of CTCF binding sites, which is important for developmental gene regulation. Recent studies, however, also provided evidence for CTCF-independent cohesin recruitment to various genomic regions, suggesting different roles of cohesin in gene regulation (13–15). However, the extent of the involvement of cohesin in gene regulation is not fully understood. Here we report the identification of cohesin as an important mechanistic mediator of chromatin interactions at the β-globin locus. We found that cohesin and Nipbl bind not only to the expected CTCF insulator sites but also at other regions of the β-globin LCR as well as at active globin genes. Their binding occurs in a cell type-specific and differentiation-induced manner, which is critical for LCR-globin gene interactions and globin gene expression. Our results demonstrate that cohesin/Nipbl is an integral structural component that mediates gene regulation via chromatin interactions at the β-globin locus, which provides a paradigm for CTCF insulator-dependent and independent functions of cohesin in gene regulation.
EXPERIMENTAL PROCEDURES
Cells and Cell Lines
The mouse erythroleukemia (745A MEL) cell line was cultured at 37 °C and 5% CO2 in RPMI (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin (50 units/ml). To induce adult β-globin expression, dimethyl sulfoxide (DMSO) was added to 2% and incubated for 4 days. The human leukemia cell line K562 was cultured in RPMI with 10% fetal bovine serum and penicillin/streptomycin (50 units/ml). Mouse fetal livers were collected at E11.5 through E15.5 (16).
Antibodies
Antigen affinity-purified rabbit polyclonal antibodies specific for Rad21, Nipbl, and the preimmune IgG control for chromatin immunoprecipitation (ChIP) were published previously (15, 17). Antibodies against CTCF (Millipore, 07–729), P45 NF-E2 (Santa Cruz Biotechnology, sc291), and RNA polymerase II (Abcam, ab5408) were also used.
ChIP
ChIP was performed according to the Millipore (Upstate) ChIP protocol with slight modifications. Approximately 5 × 106 cells or 30 mg of tissue were used per IP. Samples were cross-linked with 1% formaldehyde for 10 min at room temperature. Glycine was added to a final concentration of 0.125 m to stop cross-linking. Cells were collected and washed twice with PBS. Cells were lysed with SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl (pH 8.1)) and protease inhibitors at a concentration of 2 × 107 cells/ml. Extracts were sonicated using a Bioruptor (Diagenode) to obtain 500- to 1000-bp fragments. The extracts were diluted with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl) with protease inhibitors and centrifuged for 10 min at 10,000 × g at 4 °C. Extracts were precleared for 1 h with protein A-Sepharose (GE Healthcare), BSA, and salmon sperm DNA. Ten percent of the extract for each sample was taken as input DNA. Approximately 1 μg of antibody was added to the extracts for each IP and incubated overnight on a nutator at 4 °C. The next day, the antibody-bound complexes were immunoprecipitated with protein A-Sepharose beads for 1 h and subsequently washed with low-salt (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), 150 mm NaCl), high-salt (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), 500 mm NaCl), lithium salt (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl (pH 8.1)), and twice with TE (10 mm Tris-HCl, 1 mm EDTA (pH 8.0)) buffers. DNA was eluted off the beads with 250 μl of elution buffer (1% SDS, 0.1 m NaHCO3) on a nutator at room temperature for 15 min. Elution was repeated for a total of 500 μl of sample. Twenty microliters of 5 m NaCl was added, and each sample was reverse cross-linked overnight at 65 °C. ChIP DNA was purified with Qiagen PCR purification kits. Quantitative PCR (Q-PCR) was performed using the iCycler iQ real-time PCR detection system (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad). Standards were generated for each primer using serial dilutions of genomic input DNA. The ChIP PCR signal was normalized by the subtraction of the preimmune IgG ChIP PCR signal, which was further divided by input genomic PCR. PCR reactions were repeated multiple times. Experiments were repeated for validation.
Chromosome Conformation Capture (3C) and ChIP-loop
The 3C protocol was performed as described previously (18). Approximately 1 × 107 cells were cross-linked with 1% formaldehyde at 37 °C for 10 min. Cross-linking was stopped by adding glycine to a final concentration of 0.125 m. Cells were centrifuged at 500 × g and resuspended in 3 ml of cell lysis buffer (10 mm Tris (pH 8.0), 10 mm NaCl, 0.2% Nonidet P-40, protease inhibitors) on ice for 10 min. Nuclei were washed with 500 μl of 1.2× restriction enzyme buffer and resuspended with another 500 μl of 1.2× restriction enzyme buffer. SDS was added to 0.3% and incubated at 37 °C for 1 h. All incubations were performed with shaking. Triton X-100 was added to 2% and incubated for 1 h. At this point, 800 units of restriction enzyme (HindIII for mouse liver and EcoRI for K562, New England Biolabs) was added and incubated overnight at 37 °C. The next day SDS was added to 1.6% and incubated at 65 °C for 25 min. The digested nuclei were added into 7 ml 1× ligation buffer with 1% Triton X-100, followed by 1 h incubation at 37 °C while gently shaking. T4 DNA ligase (2000 units) (New England Biolabs) was added and incubated for 4 h at 16 °C followed by 30 min at room temperature. Proteinase K (300 μg) was added, and the sample was reverse cross-linked at 65 °C overnight. Qiagen gel purification kits were used to purify DNA. Approximately 250 ng of template was used for each PCR reaction. PCR products were run on 2% agarose gels with SYBRSafe (Invitrogen), visualized on a Fujifilm LAS-4000 imaging system, and quantified using Multigauge (Fujifilm).
To calculate interaction frequencies, 3C products were normalized to interactions at the excision repair cross-complementing rodent repair deficiency, complementation group 3 (ercc3) locus (2, 19). A control template was made to control for primer efficiencies across the β-globin locus as described (20). PCR fragments spanning the restriction sites examined were gel-purified, and equimolar amounts were mixed (roughly 15 μg total), digested with 600 units of restriction enzyme overnight, and subsequently ligated at a high DNA concentration (>300 ng/μl). The template was purified with Qiagen gel purification kit and mixed with an equal amount of digested and ligated genomic DNA. The resulting control template (250 ng) was used for each PCR for normalization against PCR primer efficiencies.
The ChIP-loop (ChIP combined with 3C) procedure was performed as described previously (21) with modifications. After cell lysis, chromatin was digested by BglII restriction enzyme in the same manner as the 3C samples. Chromatin fragments were then precleared and immunoprecipitated following the ChIP protocol. Approximately 1 × 107 cells were used per immunoprecipitation. After washes, immunoprecipitated materials on beads were resuspended in 150 μl of ligation buffer and ligated at 16 °C overnight with 2000 units of T4 DNA ligase (New England Biolabs). DNA was purified after reverse cross-linking with Qiagen PCR purification kits.
All 3C and ChIP-loop products were cloned and sequenced to confirm their identities.
siRNA Transfection
K562 cells were transfected using HiPerFect (Qiagen) following the manufacturer's protocol with 10 nm siRNA. A mixture of 60 μl of HiPerFect, 3 μl of 20 μm siRNA, and 1 ml of RPMI was incubated for 10 min and added to 2 × 106 cells in 1 ml of RPMI. After 6 h, 4 ml of fresh RPMI with 10% FBS was added. Transfection was repeated the next day. Cells were harvested 48 h after the first transfection. For siRNA information, see supplemental procedures. SiRNAs against hSMC1 (22) and CTCF (CTCF #1) (23) were described previously. AllStars negative control siRNA was obtained from Qiagen.
Q-RT-PCR Analysis
Total RNA was extracted using the Qiagen RNeasy Plus kit. First-strand cDNA synthesis was performed with SuperScript II (Invitrogen). Q-PCR was performed using the iCycler iQ real-time PCR detection system (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad). Standard curves were generated for each primer pair using a linear dilution of a PCR 2.1 plasmid (Invitrogen) containing each cloned PCR product. Values were generated based on threshold cycles (Ct) with respect to the standard curve of each primer. Values were normalized to respective control genes.
PCR Primers
PCR primers are listed in the supplementary information.
RESULTS
Differentiation-induced Cohesin and Nipbl Binding at the β-globin Locus Is Associated with LCR Enhancer-Globin Gene Interactions
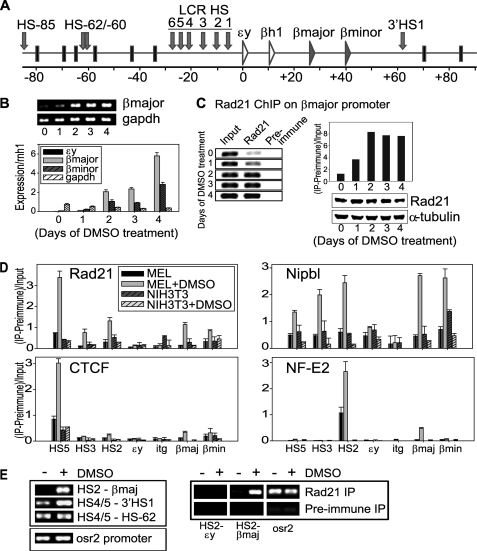
To investigate the role of cohesin at the β-globin locus, we used mouse erythroleukemia (MEL) cells, which are arrested at a proerythroblast stage. Upon DMSO treatment, MEL cells terminally differentiate into mature erythroid cells that produce high levels of adult β-globin (Fig. 1, A and B) (24). Interestingly, using antibody specific for Rad21, we found that cohesin binding at the adult β-major globin gene promoter is also strongly induced following DMSO treatment, paralleling β-globin gene activation (Fig. 1C). The presence of the cohesin holo-complex was confirmed by ChIP using antibodies specific for SMC1 and SA1 (supplemental Fig. S1).
FIGURE 1.
Adult β-globin gene induction in DMSO-treated MEL cells. A, schematic diagram of the mouse β-globin locus. The 80-kb β-globin locus is flanked by olfactory receptors and contains an upstream LCR and a globin gene cluster 12–50 kb downstream of the LCR. Several HS are located in the LCR. Most notably, HS2 functions as an enhancer, and the outer HS5 functions as an insulator. B, adult β-globin gene induction following DMSO treatment. End point RT-PCR of β-major gene induction and q-RT-PCR analysis of β-major and β-minor genes in comparison to GAPDH were examined over 4 days of DMSO treatment. Q-RT-PCR values were normalized to ribonuclease/angiogenin inhibitor 1 (rnh1). C, ChIP analysis of cohesin binding to the β-major promoter following DMSO induction from 0 to 4 days. End point PCR and semi-quantitation (Quantity One, Bio-Rad) of anti-Rad21 ChIP DNA are shown. Preimmune IgG ChIP was used as the negative control. Western blot analysis showed that the level of total Rad21 remains constant. α-tubulin was used as a loading control. D, ChIP analysis of cohesin (Rad21), Nipbl, CTCF, and NF-E2 binding to the β-globin locus. The locations of the primers are indicated at the bottom. HS5, CTCF insulator; HS2, enhancer; itg, intergenic region downstream of ϵy (see Fig. 1A and supplemental Fig. S2). NIH-3T3 fibroblast cells were used as a negative control. Arbitrary values were given for the y axis ((IP signal-Preimmune)/Input). E, 3C and ChIP-loop analysis of the HS2 and β-major promoter interaction at 4 days after DMSO treatment. The interactions of a region containing HS4 and HS5 (HS4/5) with either 3′HS1 or HS-62 are compared. An uncut region without 3C digestion sites on the odd-skipped-related 2 (osr2) promoter was used as loading control. For ChIP-loop analysis using anti-Rad21 antibody, the HS2-β-major interaction was specifically detected after DMSO induction. The HS2-ϵy interaction was examined for comparison. Preimmune IgG was used as the negative control for IP. The osr2 promoter is used again as loading control, as cohesin constantly binds to the osr2 promoter in both untreated and DMSO-induced MEL.
To test whether cohesin and Nipbl binding mirrors that of other known factors across the β-globin locus, we compared cohesin and Nipbl binding with CTCF and NF-E2 binding (Fig. 1D). HS5 contains the CTCF insulator element (25) and NF-E2, which is important for globin gene activation, binds to the HS2 enhancer element (26). Among the seven regions tested, only HS5 contains a consensus CTCF binding site (Fig. 1D and supplemental Fig. S2), and the only significant induction of CTCF binding was observed at this site. NF-E2 binding was induced at HS2 and, to a lesser extent, at the β-major promoter. In contrast, a robust increase of cohesin binding was observed not only at the HS5 insulator region but also at the HS3 and HS2 regions in the LCR (Fig. 1D). Cohesin binding was also increased at the β-major and β-minor promoters (Fig. 1D). Thus, the observed dynamic changes in binding are unique to cohesin and are distinct from CTCF and NF-E2. The results demonstrate that cohesin binding to non-CTCF sites is altered during cellular differentiation in a manner that correlates with gene expression.
We found no enrichment of cohesin binding at the silent embryonic ϵy gene promoter and at the intergenic region between βh1 and β-major (itg). DMSO failed to induce any significant cohesin binding at the locus in NIH-3T3 cells, which do not express β-globin, indicating that this is not a DMSO-induced nonspecific phenomenon (Fig. 1D). Thus, cohesin binding induction is specifically associated with gene activation. It should be noted that Nipbl binding is more clearly induced at HS3, HS2, and at the β-major and β-minor promoters than at HS5 (and not at ϵy or itg) following DMSO treatment (Fig. 1D). Thus, although both cohesin and Nipbl binding is induced at overlapping regions in the β-globin locus, the intensity of their binding signals do not always correlate in a linear fashion.
The active β-globin genes interact with distal enhancer regions in the 5′ LCR during gene activation (3, 4). We examined the HS2-β-major interaction using 3C. The HS2 interaction with the β-major region was observed only in the presence of DMSO, whereas the interactions between the CTCF insulator sites (HS4/5 and 3′HS1, HS4/5, and HS-62) were detected without DMSO induction, indicating that the HS2-β-major interaction is strictly associated with gene activation (Fig. 1, A and E, left). The ChIP-loop experiments using anti-Rad21 antibody led to enrichment of the HS2-β-major interaction product (Fig. 1E, right). The results indicate that cohesin binds to these regions when they interact. Taken together, cohesin and Nipbl are recruited not only to the HS5 insulator, but also to the enhancer and the active β-globin gene regions. This correlates with the induction of the interaction of these domains and β-globin gene activation.
Nipbl Is Required for Cohesin Binding and Chromatin Interactions at the β-globin Locus in Vivo
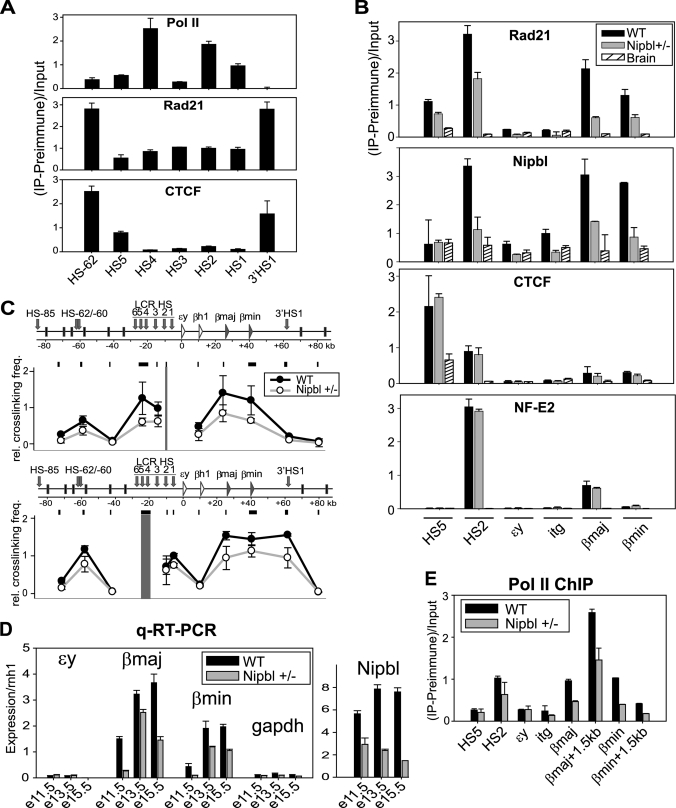
We next examined whether cohesin plays a similar role in vivo using mouse fetal liver tissues in which active erythropoiesis occurs. At E15.5, the adult β-globin genes are the most abundant globin transcripts in the liver (Fig. 2D) (27). Although cohesin (i.e. Rad21) and CTCF exhibited similarly strong binding at the upstream HS-62 and at the downstream 3′HS1, their binding patterns in the LCR were distinct (Fig. 2A). CTCF binding was restricted to HS5 in the LCR, whereas cohesin binding was observed throughout the LCR. The binding patterns of Rad21 and Nipbl at the β-globin locus are more similar to each other in liver cells than in MEL cells, with prominent binding at HS2 and at the β-major and β-minor promoters rather than at HS5 (Fig. 2B, WT). As predicted, the strongest binding site for CTCF was at HS5 but not at the globin promoters. There was also minor binding at HS2 despite the absence of consensus CTCF motifs (supplemental Fig. S2). Similar to MEL cells, NF-E2 binding was restricted to HS2 and the β-major promoter. Thus, cohesin and CTCF exhibit distinct binding patterns at the β-globin locus both in vivo and in vitro.
FIGURE 2.
Analysis of the effect of Nipbl/cohesin binding to the β-globin locus in vivo. A, ChIP analysis of RNA polymerase II, cohesin (Rad21), and CTCF at the DNase l hypersensitive sites in the β-globin region of E15.5 liver tissues. The PCR-amplified regions are indicated at the bottom (see also Fig. 1A). Q-PCR analysis of ChIP samples were normalized as in Fig. 1C. B, ChIP analysis of E15.5 liver using antibodies specific for Rad21, Nipbl, CTCF, and NF-E2. Q-PCR analysis of ChIP samples was normalized as in Fig. 1C. The age-matched Nipbl mutant liver and the wild-type brain were used for comparison. Similar results were obtained at E13.5. C, 3C analysis of the β-globin locus in E13.5 liver. Two different baits (HS2 and HS4/5) were used for the analysis. The results were compared with the age-matched liver tissue from the Nipbl mutant mice as indicated. D, Q-RT-PCR of the β-globin and Nipbl expression analysis of the wild-type and Nipbl mutant fetal liver at E11.5, E13.5, and E15.5. GAPDH was used for comparison. All genes were normalized to rnh1. E, ChIP analysis of RNA polymerase II (pol II) at the β-globin locus in the wild-type and Nipbl mutant E13.5 liver.
For comparison, we examined age-matched brain tissue of wild-type mice in which the β-globin genes are not expressed and do not adopt a chromatin loop structure (3). As we anticipated, no NF-E2 binding was observed at the β-globin locus (Fig. 2B, Brain). Although a small peak of CTCF was detected at the HS5 insulator, no significant cohesin or Nipbl peak was observed throughout the locus. Thus, the prominent cohesin and Nipbl binding at the β-globin locus is tissue-specific.
To address the role of Nipbl on cohesin binding at these sites, age-matched liver tissue from Nipbl heterozygous knockout mice was analyzed (Fig. 2B, Nipbl +/−). This mutant system is particularly useful because the partial reduction of Nipbl (Fig. 2D) is insufficient to cause mitotic defects but does generate developmental abnormalities accompanied by transcriptional misregulation (16). As expected, Nipbl binding was itself decreased, confirming the specificity of the Nipbl ChIP (Fig. 2B). Consistent with decreased Nipbl binding, cohesin binding was also diminished at all examined sites in the β-globin locus. CTCF and NF-E2 binding was not affected by the Nipbl mutation. Taken together, the results indicate that cohesin binding in the β-globin locus in vivo is dependent on Nipbl that binds to overlapping sites.
Consistent with the decreased binding of cohesin, LCR interactions with the rest of the locus were also negatively impacted (Fig. 2C). Using either HS2 or a region containing HS4 and HS5 (HS4/5) as bait, the interactions observed in the wild-type liver were all compromised in the Nipbl mutant liver, correlating with decreased β-major and β-minor globin gene expression (Fig. 2D). This was accompanied by a corresponding reduction in RNA polymerase II binding despite a comparable presence of the NF-E2 transcription activator (Fig. 2, B and E). Taken together, our results indicate that Nipbl is required for cell type-specific and developmental induction of cohesin binding, which is critical for both the chromatin interactions involving the LCR as well as β-globin gene expression in vivo.
Differential Effects of Cohesin at CTCF and Non-CTCF Sites in the β-globin Locus in Human Hematopoietic Leukemia Cells
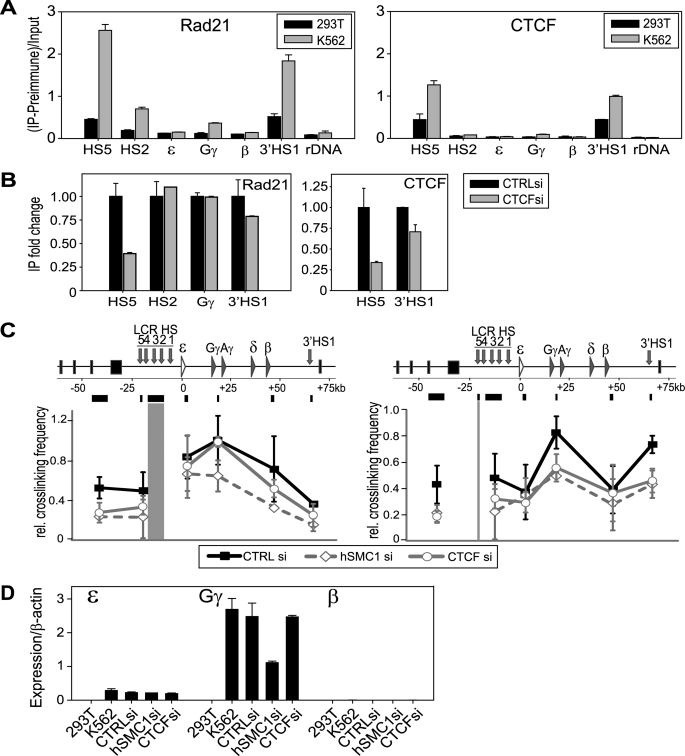
We examined whether the role of cohesin at the β-globin locus is conserved in human cells by using K562 human erythroleukemia cells and comparing the effects of CTCF and cohesin depletion. The human β-globin locus undergoes chromatin interactions that are similar to the mouse locus (3, 28), and the role of the LCR in β-globin gene expression is conserved (29). K562 cells produce embryonic (ϵ) and fetal (γ) globins, but not adult (β) globins (30). Cohesin was recently shown to bind to the CTCF insulator elements at HS5 in the LCR and at the 3′HS1 in K562 cells, mediating the 5′ and 3′ insulator interaction (31). However, the cohesin binding pattern within the locus was undefined. We observed prominent binding of cohesin at HS5 and 3′HS1 in K562 but only weakly in 293T cells (Fig. 3A), as was published (31). Importantly, we also found cohesin peaks at HS2 and the active Gγ promoter but not, in this case, at the inactive adult β-globin promoter. For comparison, we also examined cohesin binding in primary human CD34+ cells during erythropoietin-mediated differentiation (32). There is minimal adult β-globin gene expression on day 4 of differentiation, whereas it is highly expressed on day 15 (supplemental Fig. S3A). Correlating with this expression pattern, cohesin binding was increased at the adult β-globin gene region on day 15 compared with day 4 (supplemental Fig. S3B). Thus, the specific binding of cohesin to active globin genes is a conserved phenomenon in mouse and human cells. Similar to what was observed in MEL cells, cohesin binding to the HS2 and globin promoter appears to be less prominent compared with the insulator sites. This may be a common tendency for cells cultured in vitro.
FIGURE 3.
Differential effect of CTCF and cohesin in chromatin interactions in human erythroid cells. A, ChIP analysis of cohesin and CTCF at the β-globin locus in 293T and K562 cells. Locations in the β-globin locus are indicated at the bottom. B, the effect of CTCF depletion on cohesin binding at the β-globin locus. Fold-change of cohesin binding at each site was compared between control siRNA and CTCF siRNA depletion. CTCF ChIP at its binding sites (HS5 and 3′HS1) was also examined for siRNA specificity. Western blot analysis of CTCF depletion is shown in supplemental Fig. S4A. C, 3C analysis of the effect of hSMC1 or CTCF siRNA depletion at the β-globin locus compared with control siRNA. Left, HS2/3 as bait; right, HS5 as bait. D, Q-RT-PCR analysis of embryonic (ϵ), fetal (Gγ), and adult (β) β-globin gene expression in 293T, K562, and K562 treated with control, hSMC1, or CTCF siRNA (see also supplemental Fig. S4). Expression was normalized against β-actin.
To assess the functional interplay between CTCF and cohesin, we depleted CTCF or hSMC1 (cohesin) using siRNA (Fig. 3B and supplemental Fig. S4A). We confirmed that transient depletion of either one did not result in any significant mitotic defects (supplemental Fig. S4B). CTCF binding to HS5 and 3′HS1 was reduced accordingly after CTCF depletion (confirming the specificity of the siRNA and ChIP) (Fig. 3B, right). Cohesin binding was also decreased at HS5 and 3′HS1 but not at the HS2 and Gγ regions (Fig. 3B, left). The results suggest that cohesin is recruited to the HS2 and Gγ sites in a CTCF-independent manner.
Consistent with this, CTCF depletion failed to affect the 3C interaction between HS2/3 and Gγ, whereas hSMC1 depletion significantly impaired this interaction (Fig. 3C, left). This is in stark contrast to the interaction between HS5 and the rest of the locus, which was similarly suppressed by either CTCF or hSMC1 depletion (Fig. 3C, right). This agrees with the fact that cohesin requires CTCF for its binding to the insulator regions (Fig. 3B) and indicates that CTCF requires cohesin for chromatin interactions at this site. Consistent with these differential effects on the HS2/3-Gγ interaction, cohesin depletion resulted in a significant decrease of Gγ expression. We failed to observe any distinct effect of CTCF depletion on Gγ expression under these conditions (Fig. 3D). Taken together, the results indicate that cohesin plays a conserved role in mediating the chromatin interactions involving the LCR at the β-globin locus in mouse and human cells. Although cohesin is important for the insulator interactions to partition the β-globin locus in a CTCF-dependent manner, its insulator-independent role in mediating enhancer-promoter interactions is critical for proper expression of the β-globin genes.
DISCUSSION
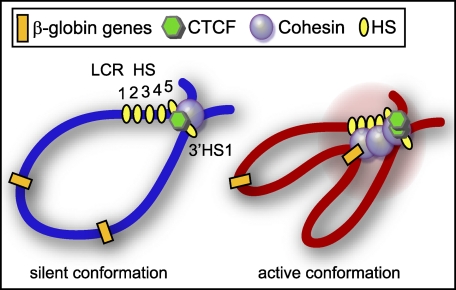
In this study, we examined the role of cohesin and the cohesin loading factor Nipbl in β-globin gene expression in mouse and human cells. We demonstrate the induction of Nipbl and cohesin binding during cellular differentiation at the β-globin locus. Cohesin/Nipbl binding is induced not only at CTCF insulator sites but also at other regions of the LCR and at active globin genes in an apparently CTCF-independent manner. A deficiency of cohesin or Nipbl disrupted the LCR enhancer-promoter interactions and inhibited gene expression. This strongly supports the critical role of cohesin/Nipbl in chromatin interaction-mediated gene regulation at the β-globin locus (Fig. 4). Our results provide important insight into the role of mammalian cohesin in LCR function.
FIGURE 4.
A schematic diagram of cohesin function at the β-globin locus. In the silent β-globin locus of erythroid progenitors, cohesin and CTCF bind at the HS5 and 3′HS1 insulator sites, setting the boundaries for later activation. When the locus becomes active, more transcription factors are recruited, and they can strengthen the chromatin interactions for optimal gene expression. Cohesin has a dual function, as it mediates both gene-enhancer interactions and boundary insulator interactions.
Cell Type-specific and Differentiation-induced Cohesin Binding Involves Specific Induction of Nipbl Binding
In Saccharomyces cerevisiae, the relationship between cohesin and the cohesin loading factor Scc2 (Nipbl homolog) binding sites is controversial. One study demonstrated the complete colocalization of cohesin with Scc2 (33), whereas another study indicated that ongoing transcription moves cohesin from its initial loading sites where Scc2 resides to converging intergenic sites (34). Furthermore, cohesin relocalization associated with transcriptional changes appears to be Scc2-independent (35). In mammalian cells, although Nipbl was found to colocalize with a subset of cohesin binding sites in mouse embryonic stem cells (13), it was unclear how Nipbl binding is altered in different cell types and differentiation stages and how it relates to changes in cohesin binding. Our results show that Nipbl binding is induced at cohesin binding sites and is required for cohesin binding during gene activation, which is clearly distinct from what was observed in yeast, indicating that Nipbl plays an important role in differentiation-induced cohesin binding in mammals. Importantly, our results provide the first evidence that partial reduction of Nipbl by heterozygous mutation is sufficient to cause alteration of cohesin-mediated chromatin interactions. This may provide important mechanistic insight into Cornelia de Lange syndrome, a human developmental disorder linked to Nipbl haploinsufficiency (36).
Role of Cohesin in β-globin Gene Expression
Although the β-globin locus was shown to undergo dynamic chromatin interaction changes during differentiation, the molecular mechanism and significance of these chromatin interactions were unclear. Although CTCF mediates the HS5 and 3′HS1 insulator interaction, deletion of CTCF sites failed to affect β-globin gene expression, and other HS sites in the LCR are required for proper β-globin expression (2, 37–40). Our results indicate that cohesin binds not only to CTCF sites but also to other HS sites in the LCR and indeed mediates chromatin interactions both at the CTCF insulator sites and at enhancer regions. This role of cohesin is conserved in mouse and human cells. Our comparison of CTCF and cohesin depletion in human K562 cells strongly suggests that cohesin promotes β-globin gene expression by mediating the interaction between non-CTCF sites in the LCR and specific globin target genes. Interestingly, a recent study using the same cell line showed that CTCF depletion has an effect on Gγ expression, although the effect on chromatin interactions was not examined inside of the β-globin locus (31). This apparent discrepancy may be due to different methods and/or durations of CTCF depletion, which was shorter in our study. Nevertheless, under our CTCF depletion conditions that affected cohesin binding at the insulator sites and the insulator interaction, there was no effect on the enhancer-target gene interaction and Gγ expression. Thus, our results highlight the distinction between CTCF insulator-dependent and independent functions of cohesin at this locus.
Multiple Effects of Cohesin in Gene Expression
Cohesin and cohesin-associated factors affect gene expression in different contexts in multiple organisms. It was first reported in S. cerevisiae that cohesin affects boundary function at the HMR locus (41). Similarly, cohesin interferes with the distal enhancer-promoter interactions for the cut and ultrabithorax genes in Drosophila (42). In mammalian cells, RNA polymerase II transcription has a tendency to stall at cohesin/CTCF binding sites (43), suggesting that the presence of cohesin may serve as a roadblock for transcription. Perhaps to negate this interfering effect of cohesin, yeast cohesins move out of gene regions as transcription is activated and accumulate in regions of transcriptional convergence (34, 35). In addition, cohesin/Nipbl may play an active role in gene silencing. A recent study reported the interaction of Nipbl with histone deacetylases, indicative of its role in promoting deacetylation and transcriptional silencing (44). We previously found the binding of cohesin to heterochromatic repeat regions in human cells, which is Nipbl- dependent but CTCF-independent, also suggesting its link to repressive chromatin organization and gene silencing (15).
In contrast, our current results demonstrate that cohesin recruitment is part of a differentiation-induced gene activation process in which cohesin mediates distal enhancer-target gene interactions. In this context, cohesin does not appear to interfere with transcription. This is in agreement with other studies that suggest that cohesin binding promotes gene expression. Many cohesin binding sites coincide with RNA polymerase II in transcriptionally active gene regions in Drosophila (45), and genetic analysis suggested that the cohesin subunit Rad21 and Nipped-B (Drosophila Nipbl homolog) have trithorax (trxG) function important for hedgehog gene expression (46). A significant overlap between the binding of the mediator complex and cohesin in the enhancer and promoter regions of active genes was observed in mouse embryonic stem cells (13). A similar overlap was found between tissue-specific transcription factors and cohesin at non-CTCF sites in human cancer cells (14). These studies collectively suggest that there are different modes by which cohesin exerts its effect on transcription, resulting in gene insulation, repression, or activation.
Specificity of Cohesin Function in Gene Regulation
How are the functional specificities of cohesin determined? This may be dictated by chromatin context and how cohesin is recruited. Although CTCF recruits cohesin to its binding sites, cohesin recruitment to D4Z4 heterochromatin requires H3K9me3 and HP1γ (15). The cohesin loading factor Nipbl, rather than cohesin itself, interacts with HP1 (15, 47). Nipbl was also shown to interact with the mediator complex, although whether mediator is required for Nipbl and cohesin recruitment has not been examined (13). In contrast, cohesin, and not Nipbl, binds to CTCF (15, 48, 49). At the β-globin locus, it was previously found that the NF-E2 subunit p18 interacts with the cohesin components SMC1 and Rad21 (50). Thus, cohesin interaction with NF-E2 may contribute to the differentiation-induced cohesin binding to the HS2 and/or globin gene promoter. In addition, crucial β-globin gene activators such as erythroid kruppel-like factor, GATA-binding factor 1, and LIM domain binding 1, which also affect chromatin interactions (5, 51, 52), may contribute to the recruitment of cohesin.
We hypothesize that through different mechanisms, cohesin is recruited to certain chromatin regions to stabilize and/or facilitate their interaction to ensure proper gene expression during development. In that sense, cohesin may contribute to the “epigenetic memory” of chromatin interactions in each cell type and differentiation stage. Further studies will be necessary to address how binding and functional specificities of cohesin are determined and to characterize the gene regulatory networks involving cohesin.
Supplementary Material
Acknowledgments
We thank Drs. Ann Dean, Paolo Sassone-Corsi, and Peter Verrijzer for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HD052860 (to A. D. L. and A. L. C.), DK044746 (to M. T. G.), HL065440 (to M. T. G.), AR058548 (to K. Y.), HD062951 (to K. Y.), and T32 CA113265 (to R. C.). This work was also supported by FSH Society Helen Younger and David Younger Fellowship Grant FSHS-DHY-001 (to K. Y.) and FSHS-DHY-002 (to W. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S2, and additional references.
- CTCF
- CCCTC-binding factor
- LCR
- locus control region
- Nipbl
- Nipped-B-like
- DMSO
- dimethyl sulfoxide
- E11.5
- embryonic day 11.5
- ChIP
- chromatin immunoprecipitation
- IP
- immunoprecipitation
- Q-PCR
- quantitative PCR
- 3C
- chromosome conformation capture
- Ct
- threshold cycles
- MEL
- mouse erythroleukemia
- NF-E2
- NF-erythroid-derived 2
- HS
- DNase I hypersensitive site
- SMC1
- structural maintenance of chromosomes 1.
REFERENCES
- 1. Schneider R., Grosschedl R. (2007) Genes Dev. 21, 3027–3043 [DOI] [PubMed] [Google Scholar]
- 2. Splinter E., Heath H., Kooren J., Palstra R. J., Klous P., Grosveld F., Galjart N., de Laat W. (2006) Genes Dev. 20, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palstra R. J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. (2003) Nat. Genet. 35, 190–194 [DOI] [PubMed] [Google Scholar]
- 4. Carter D., Chakalova L., Osborne C. S., Dai Y. F., Fraser P. (2002) Nat. Genet. 32, 623–626 [DOI] [PubMed] [Google Scholar]
- 5. Song S. H., Hou C., Dean A. (2007) Mol. Cell 28, 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bulger M., Groudine M. (2010) Dev. Biol. 339, 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wood A. J., Severson A. F., Meyer B. J. (2010) Nat. Rev. Genet. 11, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wendt K. S., Peters J. M. (2009) Chromosome Res. 17, 201–214 [DOI] [PubMed] [Google Scholar]
- 9. Zlatanova J., Caiafa P. (2009) J. Cell Sci. 122, 1275–1284 [DOI] [PubMed] [Google Scholar]
- 10. Hadjur S., Williams L. M., Ryan N. K., Cobb B. S., Sexton T., Fraser P., Fisher A. G., Merkenschlager M. (2009) Nature 460, 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mishiro T., Ishihara K., Hino S., Tsutsumi S., Aburatani H., Shirahige K., Kinoshita Y., Nakao M. (2009) EMBO J. 28, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nativio R., Wendt K. S., Ito Y., Huddleston J. E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J. M., Murrell A. (2009) PLoS Genet. 5, e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., Taatjes D. J., Dekker J., Young R. A. (2010) Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt D., Schwalie P. C., Ross-Innes C. S., Hurtado A., Brown G. D., Carroll J. S., Flicek P., Odom D. T. (2010) Genome Res. 20, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng W., de Greef J. C., Chen Y. Y., Chien R., Kong X., Gregson H. C., Winokur S. T., Pyle A., Robertson K. D., Schmiesing J. A., Kimonis V. E., Balog J., Frants R. R., Ball A. R., Jr., Lock L. F., Donovan P. J., van der Maarel S. M., Yokomori K. (2009) PLoS Genet. 5, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawauchi S., Calof A. L., Santos R., Lopez-Burks M. E., Young C. M., Hoang M. P., Chua A., Lao T., Lechner M. S., Daniel J. A., Nussenzweig A., Kitzes L., Yokomori K., Hallgrimsson B., Lander A. D. (2009) PLoS Genet. 5, e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., Cobb B. S., Yokomori K., Dillon N., Aragon L., Fisher A. G., Merkenschlager M. (2008) Cell 132, 422–433 [DOI] [PubMed] [Google Scholar]
- 18. Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. (2007) Nat. Protoc. 2, 1722–1733 [DOI] [PubMed] [Google Scholar]
- 19. Kooren J., Palstra R. J., Klous P., Splinter E., von Lindern M., Grosveld F., de Laat W. (2007) J. Biol. Chem. 282, 16544–16552 [DOI] [PubMed] [Google Scholar]
- 20. Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 21. Cai S., Lee C. C., Kohwi-Shigematsu T. (2006) Nat. Genet. 38, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 22. Kong X., Mohanty S. K., Stephens J., Heale J. T., Gomez-Godinez V., Shi L. Z., Kim J. S., Yokomori K., Berns M. W. (2009) Nucleic Acids Res. 37, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 24. Friend C., Scher W., Holland J. G., Sato T. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 378–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farrell C. M., West A. G., Felsenfeld G. (2002) Mol. Cell. Biol. 22, 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andrews N. C., Erdjument-Bromage H., Davidson M. B., Tempst P., Orkin S. H. (1993) Nature 362, 722–728 [DOI] [PubMed] [Google Scholar]
- 27. Kovach J. S., Marks P. A., Russell E. S., Epler H. (1967) J. Mol. Biol. 25, 131–142 [DOI] [PubMed] [Google Scholar]
- 28. Dostie J., Richmond T. A., Arnaout R. A., Selzer R. R., Lee W. L., Honan T. A., Rubio E. D., Krumm A., Lamb J., Nusbaum C., Green R. D., Dekker J. (2006) Genome Res. 16, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q., Peterson K. R., Fang X., Stamatoyannopoulos G. (2002) Blood 100, 3077–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benz E. J., Jr., Murnane M. J., Tonkonow B. L., Berman B. W., Mazur E. M., Cavallesco C., Jenko T., Snyder E. L., Forget B. G., Hoffman R. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 3509–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hou C., Dale R., Dean A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douay L., Giarratana M. C. (2009) Methods Mol. Biol. 482, 127–140 [DOI] [PubMed] [Google Scholar]
- 33. Kogut I., Wang J., Guacci V., Mistry R. K., Megee P. C. (2009) Genes Dev. 23, 2345–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. (2004) Nature 430, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bausch C., Noone S., Henry J. M., Gaudenz K., Sanderson B., Seidel C., Gerton J. L. (2007) Mol. Cell. Biol. 27, 8522–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu J., Krantz I. D. (2009) Clin. Genet. 76, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alami R., Bender M. A., Feng Y. Q., Fiering S. N., Hug B. A., Ley T. J., Groudine M., Bouhassira E. E. (2000) Genomics 63, 417–424 [DOI] [PubMed] [Google Scholar]
- 38. Bender M. A., Byron R., Ragoczy T., Telling A., Bulger M., Groudine M. (2006) Blood 108, 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bender M. A., Reik A., Close J., Telling A., Epner E., Fiering S., Hardison R., Groudine M. (1998) Blood 92, 4394–4403 [PubMed] [Google Scholar]
- 40. Farrell C. M., Grinberg A., Huang S. P., Chen D., Pichel J. G., Westphal H., Felsenfeld G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14554–14559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Donze D., Adams C. R., Rine J., Kamakaka R. T. (1999) Genes Dev. 13, 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rollins R. A., Korom M., Aulner N., Martens A., Dorsett D. (2004) Mol. Cell. Biol. 24, 3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wada Y., Ohta Y., Xu M., Tsutsumi S., Minami T., Inoue K., Komura D., Kitakami J., Oshida N., Papantonis A., Izumi A., Kobayashi M., Meguro H., Kanki Y., Mimura I., Yamamoto K., Mataki C., Hamakubo T., Shirahige K., Aburatani H., Kimura H., Kodama T., Cook P. R., Ihara S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jahnke P., Xu W., Wülling M., Albrecht M., Gabriel H., Gillessen-Kaesbach G., Kaiser F. J. (2008) Nucleic Acids Res. 36, 6450–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Misulovin Z., Schwartz Y. B., Li X. Y., Kahn T. G., Gause M., MacArthur S., Fay J. C., Eisen M. B., Pirrotta V., Biggin M. D., Dorsett D. (2008) Chromosoma 117, 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hallson G., Syrzycka M., Beck S. A., Kennison J. A., Dorsett D., Page S. L., Hunter S. M., Keall R., Warren W. D., Brock H. W., Sinclair D. A., Honda B. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lechner M. S., Schultz D. C., Negorev D., Maul G. G., Rauscher F. J., 3rd (2005) Biochem. Biophys. Res. Commun. 331, 929–937 [DOI] [PubMed] [Google Scholar]
- 48. Stedman W., Kang H., Lin S., Kissil J. L., Bartolomei M. S., Lieberman P. M. (2008) EMBO J. 27, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rubio E. D., Reiss D. J., Welcsh P. L., Disteche C. M., Filippova G. N., Baliga N. S., Aebersold R., Ranish J. A., Krumm A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brand M., Ranish J. A., Kummer N. T., Hamilton J., Igarashi K., Francastel C., Chi T. H., Crabtree G. R., Aebersold R., Groudine M. (2004) Nat. Struct. Mol. Biol. 11, 73–80 [DOI] [PubMed] [Google Scholar]
- 51. Drissen R., Palstra R. J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. (2004) Genes Dev. 18, 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vakoc C. R., Letting D. L., Gheldof N., Sawado T., Bender M. A., Groudine M., Weiss M. J., Dekker J., Blobel G. A. (2005) Mol. Cell 17, 453–462 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.