Abstract
Endospanin-1 is a negative regulator of the cell surface expression of leptin receptor (OB-R), and endospanin-2 is a homologue of unknown function. We investigated the mechanism for endospanin-1 action in regulating OB-R cell surface expression. Here we show that endospanin-1 and -2 are small integral membrane proteins that localize in endosomes and the trans-Golgi network. Antibody uptake experiments showed that both endospanins are transported to the plasma membrane and then internalized into early endosomes but do not recycle back to the trans-Golgi network. Overexpression of endospanin-1 or endospanin-2 led to a decrease of OB-R cell surface expression, whereas shRNA-mediated depletion of each protein increased OB-R cell surface expression. This increased cell surface expression was not observed with OB-Ra mutants defective in endocytosis or with transferrin and EGF receptors. Endospanin-1 or endospanin-2 depletion did not change the internalization rate of OB-Ra but slowed down its lysosomal degradation. Thus, both endospanins are regulators of postinternalization membrane traffic of the endocytic pathway of OB-R.
Keywords: Cell Surface Receptor, Endocytosis, Endosomes, Lysosomes, Membrane Trafficking, Protein Sorting
Introduction
Leptin and the leptin receptor are key proteins in pathways regulating energy balance and body weight in both humans and rodents (1–3). The leptin receptor is a single membrane-spanning protein of the class I cytokine receptor family. The leptin receptor gene is transcribed in a number of different messenger RNAs by alternative promoter usage and mRNA splicing (4). Most of these transcripts are splice variants that encode receptor isoforms with different cytoplasmic tails, which are referred to as OB-Ra, OB-Rc, OB-Rd (short isoforms), and OB-Rb (long isoform). The long splice variant OB-Rb mediates the majority of the effects of leptin, mainly via the JAK-STAT signaling pathway (1, 5, 6). The exact function of the short isoforms is still unclear. Short and long isoforms follow very similar intracellular membrane trafficking pathways (7–9).
Recently, we identified a negative regulator of leptin receptor function (10). This protein, originally named leptin receptor gene-related protein, or OB-RGRP, is encoded by a transcript expressed from the leptin receptor gene (11). The OB-RGRP mRNA shares two exons that are untranslated in leptin receptor transcripts and contains two additional exons that are absent in leptin receptor transcripts. This transcript is named LEPROT (leptin receptor overlapping transcript). This overlapping gene structure is conserved in humans and rodents (11, 12). We named this protein endospanin-1, due to its topology and intracellular localization. We have shown that endospanin-1 down-regulates leptin signaling by lowering the expression of the receptor at the cell surface and thus the sensitivity of the cell to leptin. Furthermore, the physiological relevance of endospanin-1 as a regulator of OB-R function was evaluated in mice. The expression of a lentivirus-delivered short hairpin RNA (shRNA) directed against endospanin-1 in the arcuate nucleus of the hypothalamus prevented the development of high fat diet-induced obesity (10). These data suggest that endospanin-1 may be a new potential target for obesity treatment. However, the cellular mechanisms leading to the up-regulation of OB-R cell surface expression in endospanin-1-depleted cells are still unknown.
Endospanin-1 has no amino acid sequence in common with any leptin receptor isoform. Endospanin-1 is a small protein of 131 residues and is the type member of a new family of proteins conserved from yeast to humans. A first clue to the function of endospanin family members at the cellular level was provided by a study that identified the yeast endospanin-1 homologue Vps55p as a protein involved in the transport of proteins to the vacuole (13). Disruption of the VPS55 gene led to a phenotype of abnormal secretion of the vacuolar carboxypeptidase Y and of blocking of a late step of the endocytic pathway, which indicated that Vps55p functions as a regulator of membrane traffic between endosomes and the vacuole, the yeast degradative organelle equivalent of the mammalian lysosome. Moreover, human endospanin-1 expressed in a vps55Δ yeast strain complements these phenotypic defects, suggesting that the function of these proteins has been conserved throughout evolution.
We set out to study the cellular mechanisms underlying the regulation of OB-R cell surface expression by endospanin-1. In parallel, we studied a human endospanin-1 homologue of unknown function, which is encoded by the gene LEPROTL-1 (leptin receptor overlapping transcript-like 1) (14). We named this protein endospanin-2. We show that both endospanins modulate the expression of leptin receptors at the cell surface. Furthermore, we provide evidence that they regulate OB-R endocytosis and degradation at a postinternalization step.
EXPERIMENTAL PROCEDURES
Plasmids
Human endospanin-2 cDNA was obtained by PCR from a HeLa cell cDNA library. Both endospanin-1 (11) and endospanin-2 cDNAs were subcloned into the expression vector pCIneo (Promega). To study the transmembrane topology, a tag, MYPYDVPDYADIS, with a single influenza virus hemagglutinin (HA) epitope (underlined sequence) was inserted at the N terminus of endospanin-2, or a single vesicular stomatitis virus G protein (VSV-G)3 epitope tag, PYTDIEMNRLGK, was introduced in its second putative hydrophilic loop between the Asp60 and Ala61 residues or at the C terminus. To obtain endospanin-1-HA-L1, a tag, DSAYPYDVPDYAASE, with a single HA epitope was introduced between Glu28 and Asp29 residues of endospanin-1 in the first putative loop located between membrane-spanning domains 1 and 2. Similarly, endospanin-2-HA-L1 was constructed by inserting a tag, GASYPYDVPDYAGA, between Tyr30 and Asn31 residues. To generate green fluorescent protein (GFP)-tagged versions of both proteins, the C-terminal VSV-G tag was excised and was replaced by the coding sequence of EGFP. To construct endospanin-1–3HA and endospanin-2–3HA, a C-terminal extension, RIPGLINIFYPYDVPDYAGYPYDVPDYAGSYPYDVPDYAAQC, containing three HA epitopes was added to each protein. All constructs were generated by PCR-based mutagenesis, as described previously (8). The pcDNA3-OB-Ra-Luc and pcDNA3-endospanin-1-YFP constructs that were used for BRET experiments have been described previously (10, 15). To generate the pcDNA3-endospanin-2-YFP vector, endospanin-2 cDNA was PCR-amplified to introduce N- and C-terminal BamHI sites and used to replace endospanin-1 coding sequence by endospanin-2 coding sequence in pcDNA3-endospanin-1-YFP plasmid. All constructs were verified by DNA sequencing.
The oligonucleotides encoding the endospanin-1 shRNA were 5′-GATCCCCCTGGCATATTTCTTCACTATTCAAGAGATAGTGAAGAAATATGCCAGTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAACTGGCATATTTCTTCACTATCTCTTGAATAGTGAAGAAATATGCCAGGGG-3′; the oligonucleotides encoding the endospanin-2 shRNA were 5′-GATCCCCTACTGGCCCCTCTTTGTTCTTCAAGAGAGAACAAAGAGGGGCCATATTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAATACTGGCCCCTCTTTGTTCTCTCTTGAAGAACAAAGAGGGGCCAGTAGGG-3′; the oligonucleotides encoding the control shRNA (targeting firefly luciferase) were 5′-GATCCCCGGATTCCAATTCAGCGGGAGCCACCTGATGAAGCTTGATCAGGTGGCTCCCGCTGAATTGGAATCCATTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAATGGATTCCAATTCAGCGGGAGCCACCTGATCAAGCTTCATCAGGTGGCTCCCGCTGAATTGGAATCCGGG. These oligonucleotides were annealed and subcloned downstream of the H1 promoter in pSUPER.retro.puro (OligoEngine, Seattle, WA) by using BglII and HindIII. The GFP gene was added by inserting a CMV-GFP fragment into the XhoI restriction site of pSUPER.retro.puro. These plasmids were named pSRP/Luc-shRNA, pSRP/endo1-shRNA, and pSRP/endo2-shRNA. They were used for the production of retroviral vectors.
The plasmids encoding HA-tagged OB-Ra and mutants OB-RΔct (previously referred to as OB-RΔ1 (8)), OB-Ra/tm, and 2K2R have been described (8, 16). The plasmid pCIneoVSVG-OBRa was constructed by replacing the HA tag of pCIneoHA-OBRa by a VSV-G tag epitope, using an overlapping PCR method. The expression vectors for Rab5-GFP and Rab7-GFP were kindly provided by Dr. M. Zerial (Max Planck Institute, Dresden, Germany).
Adenovirus Vectors
Recombinant defective adenovirus vectors expressing endospanin-1, endospanin-2, or GFP were generated by homologous recombination in Escherichia coli, as described (17). Viral stocks were generated, amplified in HER-911 cells (18), and titered in the same cells by the TCID50 method. Adenoviruses expressing HA-tagged OB-Ra or OB-Rb have been described elsewhere (8).
Cell Culture
Normal rat kidney (NRK), 293T human embryo kidney cell, and human cervix carcinoma (HeLa) cell lines were obtained from the American Type Tissue Culture Collection (Manassas, VA). NRK cells were grown on Petri dishes in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (50 units/ml), and streptomycin (50 μg/ml) at 37 °C in an atmosphere of 5% CO2. HeLa cells were grown on Petri dishes in α-minimal Eagle's medium supplemented with 10% fetal bovine serum.
Retrovirus Generation and Infection
The retroviral packaging vectors were a generous gift from Dr. Jean Dubuisson. Retrovirus production was conducted by co-transfection of HEK-293T cells with three plasmids: a plasmid (300 ng) expressing the murine leukemia virus gag and pol genes; a plasmid (300 ng) expressing a heterologous envelope protein, VSV-G (19); and a transfer vector harboring a specific shRNA sequence under control of the H1 promoter (pSRP/Luc-shRNA, pSRP/endo1-shRNA, or pSRP/endo2-shRNA; 400 ng). Briefly, 4 × 106 HEK-293T cells were seeded on 6-well, poly-d-lysine-coated plates 24 h before transfection. For each shRNA, 4 μl of ExGen 500 reagent (Euromedex, Strasbourg, France) was combined with the three plasmids in 100 μl of a 9 g/liter NaCl solution and added onto the cells in 1 ml of Opti-MEM (Invitrogen). After a 6-h incubation, 1.5 ml of DMEM supplemented with 10% fetal bovine serum was added. The cells were examined for GFP expression by fluorescence microscopy 24 h after transfection. Virus-containing cell supernatants were collected, centrifuged, filtered through 0.45-μm pore-sized membranes, and concentrated 50 times by centrifugation at 300 × g for 10 min using an Amicon Centricon Plus-20 (Millipore). HeLa cells were infected by adding 300 μl of viral concentrated supernatant and 300 μl of medium supplemented with 8 μg/ml Polybrene to 3 × 104 cells in 12-well plates. One ml of medium was added 4 h after infection. Infection efficiency was monitored by GFP expression after 72 h. For single clone selection, cells were diluted 100 times in 10-cm plates and incubated 10 days with αMEM supplemented with 2 μg/ml of puromycin at 37 °C. GFP-positive clones were isolated using cloning cylinders and grown in a 96-well plate in αMEM medium at 37 °C. siRNA efficiency was tested by quantitative RT-PCR.
Antibodies
Polyclonal antisera were prepared by immunizing rabbits with synthetic dodecapeptides corresponding to the C termini of endospanin-1 or endospanin-2 coupled to keyhole limpet hemocyanin via an additional N-terminal Cys residue. Affinity columns were produced by coupling the same peptides to sulfolink beads (Pierce). After an initial step of ammonium sulfate precipitation, the immunoglobulin fractions were run through the affinity columns, eluted with 4 m MgCl2, 10 mm Na2PO4, pH 7.4, and dialyzed against PBS (138 mm NaCl, 2.7 mm KCl, 1.2 mm KH2PO4, 8.1 mm Na2HPO4, pH 7.0).
Polyclonal antibodies to internal epitopes of endospanin-1 or endospanin-2 were produced and affinity-purified by Eurogenetec. Peptides used for the immunization of rabbits and for the purification of the antibodies from the immune sera are located between residues Ile51 and Cys66 for endospanin-1 and between Cys50 and Ala65 for endospanin-2.
Mouse monoclonal antibody (mAb) 16B12, and rat mAb 3F10 (anti-HA) were from BabCo and Roche Applied Science, respectively. Mouse mAb P5D4 (anti-VSV-G) was from Roche Applied Science. Mouse mAb 2F7 (anti-rat TGN38) was a kind gift from Dr. G. Banting (University of Bristol, UK) (20). Mouse mAb to the transferrin receptor was from Zymed Laboratories Inc.. Mouse mAbs to EEA1 (early endosome antigen 1) and to LAMP-1 (lysosome-associated membrane protein 1) were from BD Biosciences. Rabbit polyclonal anti-EGFR antibody (sc-03) and goat polyclonal anti-actin were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Sheep polyclonal anti-human TGN46 was from Serotec. Alexa488-conjugated goat anti-rat or anti-mouse IgG and Alexa488-conjugated donkey anti-sheep IgG were purchased from Molecular Probes. Cy3-conjugated goat anti-mouse, anti-rat, or anti-rabbit IgG and horseradish peroxidase-conjugated secondary antibodies were from Jackson Immunoresearch.
Immunoblotting
Samples were diluted with an equal volume of 2 times concentrated SDS sample buffer (21), incubated for about 10 min at 95 °C, and run in SDS-PAGE. For endospanin detection, samples were incubated at 37 °C in order to avoid insolubilization of these highly hydrophobic proteins at 95 °C. Proteins were transferred to nitrocellulose (0.45-μm; Schleicher and Schuell or Invitrogen). Blocking and antibody incubations were carried out in TBS (137 mm NaCl, 20 mm Tris, pH 7.4) containing 5% dried skim milk and 0.1% Nonidet P-40. Immunoreactive bands were identified using peroxidase-conjugated anti-rabbit or anti-mouse antibodies, and detection was carried out using chemiluminescence (Renaissance kit from PerkinElmer Life Sciences or ECL+ kit from GE Healthcare).
Immunofluorescence Microscopy
Indirect immunofluorescence localization was performed on NRK and HeLa cells grown onto glass coverslips. To express HA-tagged or GFP-tagged constructs, HeLa cells were transfected with FuGene-6 reagent (Roche Applied Science) according to the manufacturer's instructions. Cells were fixed with 3% paraformaldehyde in PBS and permeabilized by freeze-thawing and/or by a 5-min incubation in 0.1% Triton X-100 in PBS at room temperature. For freeze-thaw permeabilization, cells were quickly frozen at −80 °C in a minimal volume of PBS and then quickly thawed at room temperature by the addition of a larger volume of PBS. Both primary and secondary antibody incubations were carried out in PBS containing 10% goat or horse serum for 30 min at room temperature. The coverslips were mounted on slides by using Mowiol 4-88-containing mounting medium (Calbiochem). Endospanin-1 and -2 were detected using affinity-purified rabbit polyclonal antibodies diluted 1:500. Anti-TGN38 mAb was diluted 10 times; anti-LAMP-1 was diluted 100 times; and anti-TGN46, anti-transferrin receptor monoclonal, anti-VSV-G epitope mAb P5D4 (Roche Applied Science), and anti-EEA1 mAb were used at a 1:500 dilution. Alexa488- and/or Cy3-labeled-conjugated secondary antibodies were used for visualization. Images were obtained with an Axiophot 2 microscope (Zeiss) equipped with a cooled charge-coupled device Micromax (Princeton). Confocal microscopy was performed with an SP2 confocal laser-scanning microscope (Leica) using a ×100/1.4 numerical aperture oil immersion lens. Double label immunofluorescence signals were sequentially collected using single fluorescence excitation and acquisition settings to avoid cross-over. Images were processed using Adobe Photoshop software.
Membrane Extraction Studies
HeLa cells were transfected by electroporation with endospanin-1–3HA or endospanin-2–3HA constructs. Cells from one subconfluent 10-cm dish were trypsinized, resuspended in 0.8 ml of HEBS (20 mm Hepes, 137 mm NaCl, 5 mm KCl, 0.7 mm Na2PO4, and 6 mm dextrose, pH 7.05), and transfected by electroporation with 18 μg of plasmid DNA in a cell pulser (Bio-Rad) with the following settings: 975 microfarads, 224 V (22). 24 h later, cells were rinsed in cold PBS, scraped off the dish, and pelleted. Cells were resuspended in 20 mm Tris-Cl, pH 7.4, complemented with protease inhibitors, and lysed by passing the cell suspension through a 22-gauge syringe. Nuclei and unbroken cells were pelleted at 900 × g for 5 min. Postnuclear supernatant was aliquoted and centrifuged at 100,000 × g for 30 min to pellet crude membranes. Pellets were resuspended with the following extraction solutions: homogenization buffer only; 1.5 m NaCl; 0.1 m Na2CO3, pH 11.4; 2 m urea; or 1% Triton X-100. The membranes were incubated at 4 °C for 30 min with occasional mixing and then centrifuged at 100,000 × g for 30 min. The supernatant was removed, and the pellet was resuspended in an equal volume of homogenization buffer. Aliquots of supernatants and pellets were analyzed by SDS-PAGE and immunoblotting.
Antibody Uptake
Cells were rinsed with prewarmed α-minimal Eagle's medium and overlaid with 0.25 ml of α-minimal Eagle's medium, containing the mAb 3F10 (anti-HA, rat IgG, 0.2 μg/ml) and/or P5D4 (anti-VSV-G, mouse IgG, 1 μg/ml). At the end of the incubation, the medium was removed, and the cells were immediately rinsed with PBS, fixed, and processed for immunofluorescence. Internalized antibodies were revealed with a Cy3-conjugated goat anti-rat IgG and/or an Alexa488-conjugated goat anti-mouse IgG antibody, after permeabilization with Triton X-100.
Quantitative RT-PCR
Cell lines expressing shRNA were grown in 6-well plates, and total RNA was extracted using a Nucleospin RNA II extraction kit (Macherey-Nagel), which includes a DNase I treatment. cDNA was obtained from 1 μg of RNA using the HighCap cDNA Archive kit (Applied Biosystems) in a final volume of 100 μl. Quantitative RT-PCR analysis was performed using the Taqman probe approach (Applied Biosystems, Foster City, CA), using 4 ng of reverse transcribed RNA and premade probes designed by the manufacturer. The ratio of the mRNA level of each gene to that of 18 S ribosomal RNA was calculated by the ΔΔCt method (23), and a value of 100 was assigned to the control HeLa cells. Each experiment was performed in triplicate and repeated twice.
Cell Surface Biotinylation Assays
Stable cell lines expressing shRNA were seeded in 6-well plates and transfected with 1 μg of plasmid expressing HA-tagged OB-Ra, OB-RaΔct, OB-Ra/tm, OB-Ra2K2R, or OB-Rb using FuGene-6 reagent (Roche Applied Science) according to the manufacturer's instructions. 24 h post-transfection, cells were put on ice and rinsed twice with cold PBS. To label cell surface proteins, the cells were incubated with the cleavable, EZ-Link Sulfo-NHS-SS-Biotin (0.3 mg/ml) (Pierce) in PBS for 15 min at 4 °C, and the biotinylation solution was renewed for another 15-min incubation. Unreacted biotin was quenched twice with 50 mm glycine in PBS for 10 min. Cells were lysed in 500 μl of lysis buffer (50 mm Tris, pH 7.4, 100 mm NaCl, 1% Triton X-100, 0.1% SDS, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science)) for 20 min on ice. Cell lysates were centrifuged for 5 min at 5,000 rpm, and biotinylated proteins were recovered by overnight incubation using streptavidin-agarose beads (Amersham Biosciences) at 4 °C. Beads were washed three times with lysis buffer; twice with 50 mm Tris, pH 7.4, 100 mm NaCl, 0.5% SDS, 0.5% sodium deoxycholate, 0.1% Triton X-100, 2 mm EDTA; twice with 50 mm Tris, pH 7.4, 500 mm NaCl, 0.1% Triton X-100, 2 mm EDTA; and once in 50 mm Tris, pH 7.4, 100 mm NaCl, 2 mm EDTA. Beads were then resuspended in Laemmli sample buffer containing 50 mm dithiothreitol and incubated at 95 °C for 10 min. The bound proteins were detected by immunoblotting.
Endocytosed Receptor Degradation Assay
Cells were surface-biotinylated on ice, as described before. After the treatment with glycine, the cells were incubated at 37 °C with prewarmed α-minimal Eagle's medium for the indicated times (up to 8 h). Cells were lysed, and biotinylated leptin, transferrin, or EGF receptors were revealed by immunoblotting after purification with streptavidin-agarose beads. Films were scanned, and amounts of biotinylated OB-R were measured using the ImageJ software (National Institutes of Health).
Endocytosis Assay Based on Cell Surface Biotinylation
Cell surface biotinylation and resistance to the cleavage by reduced glutathione after internalization were as described previously (8).
Pulse-Chase
Cells expressing endospanin-2-3HA were labeled with [35S]methionine/cysteine labeling mix (100 μCi/ml; Promix, PerkinElmer Life Sciences) for 30 min and then chased in a medium containing 1 mm methionine and 1 mm cysteine. Cell lysis and immunoprecipitation with anti-HA antibody were as described previously (8). Immunoprecipitated proteins were separated by SDS-PAGE followed by fluorography and quantified with a PhosphorImager.
Northern Blots
Total RNA from mouse tissues were isolated using RNAplus (Quantum Bioprobe). RNA was resolved in 1% agarose/formaldehyde, transferred to Hybond-N membranes (Amersham Biosciences), and hybridized according to the manufacturer's instructions. cDNA probes were made using polymerase chain reaction (PCR) products with primers as follows: 5′-TCGAAGCTTTACTGGCCCCTGTTCG-3′ as a forward primer and 5′-GGAGGATCCGTAGAGGGACATT-3′ as a reverse primer for mouse endospanin-1; 5′-ACGAATTCACCGCCATGGCAGGCATC-3′ as a forward primer and 5′-ACTCTAGACCACTGCTGCCAGCTGAAG-3′ as a reverse primer for mouse endospanin-2.
BRET Assay
BRET experiments, luminescence, and fluorescence measurements were performed as described (24, 25), using lumino/fluorometers FusionTM (Packard Instrument Co.) and Mithras (Berthold Technologies GmbH). Four independent experiments were performed, and results were expressed in milli-BRET units as a function of donor/acceptor relative ratio (total fluorescence over total luminescence measured). 1 milli-BRET unit corresponds to the BRET ratio values multiplied by 1000.
siRNA Transfection
Smart pool siRNA (Dharmacon 032248-01, 020301-00, and D-001810-10) were obtained from Thermo Fisher Scientific. siRNA transfections were performed as described previously (16).
RESULTS
Phylogenetic Analysis of Endospanins
Sequences homologous to human endospanin-1 were found in all sequenced eukaryote genomes but not in prokaryotes. All of these genes encode for putative proteins of 125–140 residues (supplemental Fig. S1). In the human genome, the LEPROTL-1 gene encodes for an uncharacterized protein related to endospanin-1 (14). We designated this protein endospanin-2. In vertebrates, two genes are present in each genome, which encode for orthologs of human endospanin-1 and endospanin-2 (supplementary Fig. S1). The predicted human and mouse endospanin-2 sequences are shown in Fig. 1A, along with the endospanin-1 sequences of both species and Vps55p, the member of this family of proteins found in the yeast Saccharomyces cerevisiae (13). Human and mouse endospanin-2 show only two conservative changes (Lys/Gln at position 32 and Ile/Val at position 86) among 131 residues but are less conserved with respect to endospanin-1 (67 and 72% identity for human and mouse proteins, respectively). Interestingly, when a larger number of endospanin family members were aligned, most of the conserved residues appeared clustered in regions that are predicted to form transmembrane domains (supplemental Fig. S1).
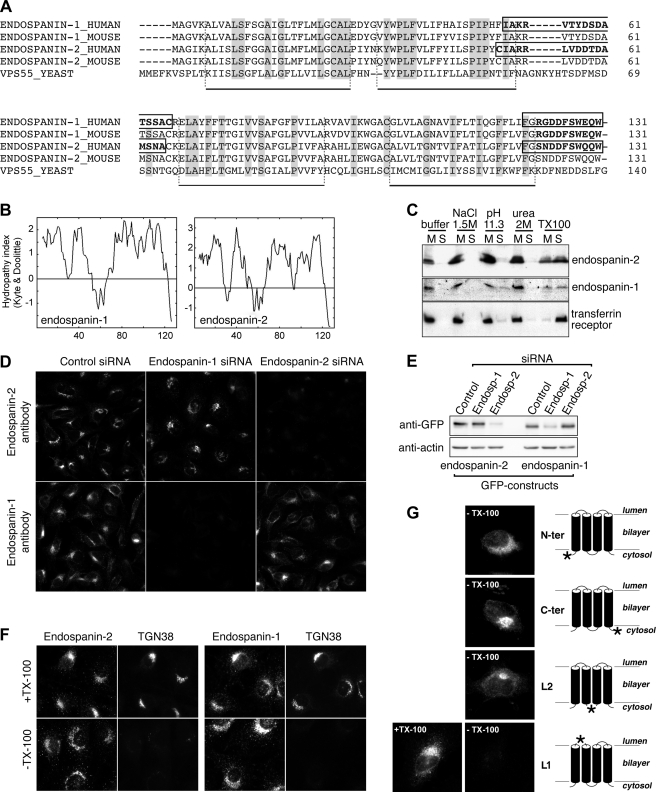
FIGURE 1.
Membrane association and transmembrane topology of endospanins. A, protein alignment. Predicted amino acid sequences of human and murine endospanin-1 and -2 and of yeast Vps55p were aligned. Conserved residues are highlighted in gray. Putative transmembrane domains are indicated under the alignment. Peptides used for antibody production are outlined in the protein sequences of human endospanins. B, hydropathy plots of human endospanins. C, membrane association of endospanins. Postnuclear membrane pellets of HeLa cells transfected with 3HA-tagged endospanin-1 or endospanin-2 were extracted with various ice-cold disruptive agents and centrifuged at 100,000 × g, and the resulting supernatants (S) and membrane pellets (M) were analyzed by SDS-PAGE and anti-HA and anti-transferrin receptor immunoblot. Membrane pellets were resuspended in solutions containing the following final concentrations of extracting agents: homogenization buffer only; 1.5 m NaCl; 0.2 m Na2CO3, pH 11.3; 2 m urea; and 1% Triton X-100. D, characterization of anti-endospanin antibodies. HeLa cells were transfected with siRNA to endospanin-1 or to endospanin-2 or with control siRNA and analyzed by immunofluorescence using anti-endospanin-1 antibody or anti-endospanin-2 antibody. All images were recorded with the same settings. E, efficiency and specificity of siRNA-mediated depletions were verified by immunoblot analysis after transfection of GFP-tagged endospanin-1 or endospanin-2 constructs. Immunoblot analyses were performed using anti-GFP and anti-actin antibodies. F, transmembrane topology of endospanins. NRK cells were fixed with 3% paraformaldehyde, freeze-thawed to rupture the plasma membrane, and subsequently treated with detergent to permeabilize internal membranes (top panels) or left untreated (bottom panels). Cells were analyzed by double label immunofluorescence using antibodies to endospanin-1 or endospanin-2 (left panels) and to TGN38 (right panels). Note that the C termini of endospanins were accessible to antibodies with or without detergent treatment, whereas the lumenal epitope recognized by the anti-TGN38 antibody could be detected only after detergent treatment. G, HeLa cells were transiently transfected to express endospanin-2 tagged at positions indicated by a star in the schemes in the right panels (N-ter, C-ter, L1, and L2 indicate the positions of the tags in the constructs). 24 h later, cells were fixed and freeze-thawed to selectively rupture the plasma membrane in order to determine by immunofluorescence the cytoplasmic orientation of the tag. The L1 construct was co-transfected with GFP in order to visualize transfected cells and analyzed in parallel after Triton X-100 permeabilization (+TX-100).
Endospanins Are Tetraspanning Membrane Proteins
Hydropathy plot analysis indicated that both endospanins contain four hydrophobic stretches of sufficient length (20–21 residues) to be transmembrane domains (Fig. 1B). Therefore, we investigated their membrane association. After transfection of 3HA-tagged constructs of these proteins in HeLa cells, both endospanins could be detected by immunoblotting with an apparent size of ∼16 kDa, consistent with the molecular mass of proteins tagged with three HA epitopes. Both proteins efficiently pelleted from postnuclear supernatants of cell homogenates by centrifugation at 100,000 × g. They could not be extracted from these pellets by sodium chloride, urea, or sodium carbonate but were solubilized by detergent, confirming that they are integral membrane proteins, as expected for proteins with four transmembrane domains (Fig. 1C).
We raised polyclonal antibodies to the C-terminal dodecapeptide or to an internal epitope located in the loop between putative trans-membrane domains 2 and 3 (Fig. 1A). All the antibodies detected overexpressed proteins but failed to detect endogenous ones in immunoblot experiments (data not shown). Despite their lack of reactivity against endogenous endospanins in immunoblot experiments, siRNA-mediated depletion of endospanins confirmed that antibodies to C termini specifically detect endogenous proteins in immunofluorescence microscopy (Fig. 1, D and E). Therefore, we used an assay based on immunofluorescence to study the topology of the proteins. We used NRK cells to compare the accessibility to antibodies of the C termini of endospanins with that of an antibody to a lumenal epitope in rat TGN38 (20) in selectively permeabilized cells. When the plasma membrane of NRK cells was permeabilized by freeze-thawing without detergent, the C termini of endospanins were accessible to antibodies. A perinuclear compartment was decorated, together with a cytosolic punctuate compartment, with both antibodies. In contrast, a monoclonal antibody recognizing a lumenal epitope of rat TGN38 did not decorate the same cells (Fig. 1F). After detergent permeabilization, both TGN38 and endospanin perinuclear immunoreactivities were observed (Fig. 1F). This indicates that the C termini of endogenous endospanin-1 and endospanin-2 were exposed to the cytoplasmic side of membranes.
To further investigate the topology of these proteins across cellular membranes, we then used a similar approach in HeLa cells expressing epitope-tagged endospanin-2. The plasma membrane was selectively permeabilized by freeze-thawing, and the accessibility of antibodies to the tag was monitored. HA or VSV-G tags fused to the N or C terminus or in the hydrophilic region located between the second and third transmembrane domains (L2) were readily accessible to antibodies (Fig. 1G). In contrast, HA epitopes inserted in the first or the third hydrophilic loops were not detected in freeze-thawed cells, although they were detected after detergent treatment (Fig. 1G) (data not shown), suggesting an extracytoplasmic orientation for both of these loops. Similarly, tags inserted at the N or C termini of endospanin-1 were accessible to antibodies in freeze-thawed cells without detergent (data not shown). This result is consistent with results of a previous BRET study, which indicated that both N and C termini of endospanin-1 are exposed on the same side of the membrane (10). Based on these results and the hydropathy profiles, we conclude that endospanins are likely to be tetraspanning membrane proteins, with their N and C termini on the cytoplasmic side of membranes.
Ubiquitous Expression of Endospanins
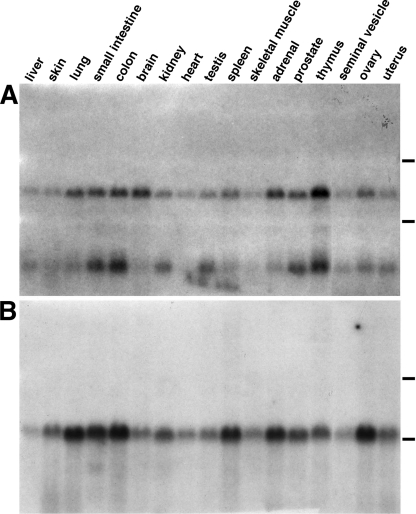
Endospanin-2 expression was examined by Northern blotting of total RNA from mouse tissues with a 32P-labeled cDNA probe encompassing the entire open reading frame. Two mRNAs were detected in each tissue (Fig. 2A). The ubiquitous expression of two endospanin-2 mRNAs was also observed in human tissues (data not shown).
FIGURE 2.
Tissue distribution of endospanins. Multiple mouse tissue total RNA blot was hybridized with a mouse endospanin-2 cDNA probe (A) and then with a mouse endospanin-1-specific cDNA probe (B). The positions of 18 and 28 S ribosomal RNA are indicated by lines on the right. Notice the presence of two bands of ∼2.7 and ∼0.7-kb endospanin-2 mRNA, probably resulting from alternative polyadenylation site usage.
The transcripts probably result from an alternative polyadenylation site usage. Examination of the 3′ end of human and mouse endospanin-2 sequences found in EST databases revealed two types of endospanin-2 transcripts of ∼0.7 and 2.7 kb, consistent with their observed migration in Northern blot. They are identical to each other over 619 nucleotides, encompassing the entire coding sequence, and differ by a ∼2-kb extension in the 3′-untranslated region of the larger transcript. Both in mouse and human cDNAs, a putative polyadenylation site, ATTAAAA, is found 170–180 nucleotides downstream from the stop codon, which is included in a larger highly conserved region of 3′-UTR of the larger cDNA (80% identity over 576 nucleotides between human and mouse sequences). Polyadenylation of endospanin-2 mRNA at this site would give a transcript of ∼0.7 kb, a size consistent with the observed migration of the shorter transcript. It is thus very likely that both endospanin-2 transcripts detected in Northern blots originate from a single gene and that they encode the same protein because the alternative polyadenylation would only result in a shorter 3′-UTR and would not affect the coding sequence.
For comparison, the same blot of mouse tissues was rehybridized with a probe containing the sequences of exons 3 and 4 of the mouse endospanin-1 gene. This probe is specific for the endospanin-1 transcript and will not detect any of the OB-R transcripts (11). As expected, a single ubiquitous ∼2-kb signal was detected in all the mouse tissues examined (Fig. 2B), as already detected in human tissues (11). Small variations in the relative levels of expression of endospanin-1 and endospanin-2 mRNAs appeared to be parallel from one tissue to another, suggesting a possible regulated co-expression of both proteins, although their genes were found on different chromosomes.
Intracellular Localization of Endospanins
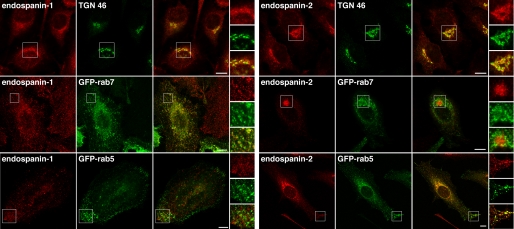
The intracellular localization of endogenous endospanins was examined in HeLa cells by confocal microscopy. As already shown in NRK cells (Fig. 1F), the anti-endospanin-1 and anti-endospanin-2 antibodies revealed a perinuclear staining pattern in immunofluorescence. The perinuclear compartment labeled with anti-endospanin-2 also contained the TGN marker TGN46 (Fig. 3). In contrast, endospanin-1 perinuclear staining overlapped only partially with TGN46. Most of the endospanin-1 staining overlapped with Rab7, indicating a localization of endospanin-1 in late endosomes and to a lesser extent in the TGN. The anti-endospanin-2 antibody also revealed a weaker peripheral punctate staining pattern, which partially co-localized with Rab5 rather than Rab7. In contrast, endospanin-1 did not co-localize with Rab5 in most of the cells (Fig. 3). Altogether these data indicate that both endospanins are membrane proteins of the TGN-endosomal system and that they have different steady-state distributions in HeLa cells.
FIGURE 3.
Intracellular localization of endospanins. HeLa cells were transfected with either GFP-Rab7 or GFP-Rab5 constructs as fluorescent endosomal markers or left untransfected, fixed, and processed for immunofluorescent detection of TGN46 (untransfected cells) and/or endogenous endospanin-1 or endospanin-2. Bars, 10 μm.
Intracellular Trafficking of Endospanins
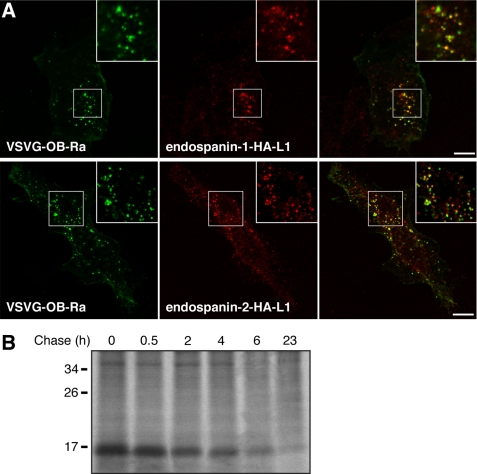
To gain insights into the function of endospanins, we sought to determine the intracellular trafficking of these proteins, more precisely their ability to traffic to the cell surface. We transfected HeLa cells with plasmids expressing an endospanin-1 or an endospanin-2 construct that were HA-tagged in the first hydrophilic loop and carried out antibody uptake experiments using an anti-HA antibody. We observed a punctate staining of anti-HA antibodies in transfected cells, indicating that endospanins had been expressed at the cell surface and then endocytosed with the antibodies (Fig. 4A). As expected, the antibody uptake was not observed with cells expressing C-terminally HA-tagged endospanin (data not shown). Anti-HA antibodies could be observed in endosomes but were never detected in the Golgi/TGN area, even after longer uptakes (data not shown). In co-uptake experiments, internalized HA-tagged endospanin-1 and endospanin-2 were found partially localized in endosomes that also contained internalized VSV-G-tagged OB-Ra (Fig. 4A). When cells expressing HA-tagged endospanin-2 were chased with no antibody, after an initial antibody uptake, the signal became undetectable after 4–6 h, meaning that the tagged proteins had probably been transported to lysosomes, where the bound antibody had been degraded.
FIGURE 4.
Intracellular traffic of endospanins. A, antibody uptake. Endospanin-1 (endospanin-1-HA-L1) or endospanin-2 (endospanin-2-HA-L1) tagged in their first hydrophilic loop and VSV-G-tagged OB-Ra were co-expressed in HeLa cells. Cells were incubated in the presence of anti-HA mAb 3F10 and anti-VSV-G mAb P5D4 for 1 h, rinsed, fixed, and processed for immunofluorescent detection of anti-HA (red) and anti-VSV-G (green) internalized antibodies. Bars, 10 μm. B, pulse-chase analysis of endospanin-2 turnover. HeLa cells transfected with 3HA-tagged endospanin-2 were pulse-labeled for 30 min and chased for the indicated times. Cell lysates were immunoprecipitated with an anti-HA antibody. Samples were separated by SDS-PAGE, and immunoprecipitated proteins were visualized by fluorography.
To verify whether lysosomes were the final destination of these proteins or if they were recycled from a late endosomal compartment after dissociation from the antibody, we assessed the turnover of endospanin-2 with pulse-chase experiments. HeLa cells expressing 3HA-tagged endospanin-2 were labeled for 30 min with [35S]methionine/cysteine and chased over a 23-h time course. Endospanin-2 was found to turn over with a half-life of about 2 h (Fig. 4B). This result reveals a turnover rate of endospanin-2, which is entirely compatible with a lysosomal degradation after endocytosis and transport to lysosomes. Similar results were obtained with the endospanin-2 construct HA-tagged in the first hydrophilic loop, which was used in the antibody uptake experiments described before (data not shown). Taken together, the results of antibody uptake and pulse-chase experiments suggest that endospanins traffic to the cell surface, and do not recycle back to the TGN after endocytosis but are rather transported to endosomes together with endocytosed leptin receptors and finally to lysosomes, where they are probably degraded.
Interaction of Endospanins with Leptin Receptor
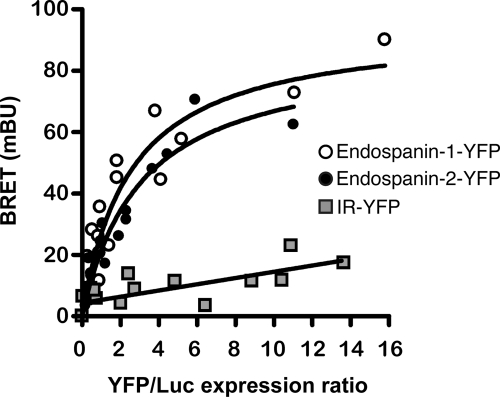
We have previously shown that endospanin-1 interacts with the leptin receptor OB-Ra and that its overexpression down-regulates the cell surface expression of OB-Ra and OB-Rb (10). To determine if endospanin-2 also could interact with the leptin receptor, we used a BRET donor saturation assay that we previously developed to study OB-R/endospanin-1 interaction. As already reported (10), a significant and saturable energy transfer was observed in cells co-expressing OB-Ra fused to luciferase (OB-Ra-Luc) and various quantities of endospanin-1-YFP fusion protein. Very similar results were obtained in cells co-expressing OB-Ra-Luc and endospanin-2-YFP fusion proteins (Fig. 5). The specificity of these interactions was confirmed by the absence of any significant transfer between OB-Ra-Luc and YFP-tagged insulin receptor. These data indicate that OB-Ra interacts with both endospanins in intact cells.
FIGURE 5.
Interaction of OB-R with endospanins. Interaction of OB-Ra and endospanin-1 or endospanin-2 was studied by BRET in intact cells. BRET donor saturation curves were generated by expressing constant amounts of OB-Ra-Luc and increasing quantities of the indicated YFP-tagged proteins. The BRET, total luminescence, and total fluorescence were measured in three independent experiments. Data obtained for the BRET acceptors endospanin-1-YFP and endospanin-2-YFP were best fitted with a nonlinear regression equation assuming a single binding site, and those obtained for insulin receptor (IR)-YFP were best fitted with a linear regression equation. mBU, milli-BRET units.
Endospanins Regulate Leptin Receptor Cell Surface Expression
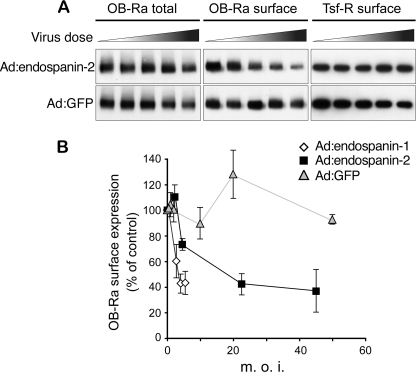
We next tested whether endospanin-2 overexpression has any impact on OB-R cell surface expression. HeLa cells were infected with recombinant OB-Ra adenovirus and increasing doses of endospanin-1, endospanin-2, or control GFP-expressing recombinant adenoviruses, and OB-R cell surface expression was quantified using a surface protein biotinylation assay. As observed previously for endospanin-1 (10), a dose-dependent decrease of OB-Ra cell surface expression was observed with endospanin-2 (Fig. 6A) but not with the control. In contrast, the total amount of receptor was not affected, except for the higher dose of endospanin-2 virus. This slight decrease of OB-Ra expression was not linked to the overexpression of endospanin-2 because a similar decrease was observed with the higher dose of the control virus (Fig. 6A). To observe a similar down-regulation of OB-Ra cell surface expression, a dose of endospanin-2 adenovirus about 5 times higher than that of endospanin-1 adenovirus was required (Fig. 6B). This difference could result from a lower efficacy of endospanin-2 adenovirus compared with endospanin-1 adenovirus to overexpress the transgenic protein or could reflect higher endogenous expression levels of endospanin-2 than endospanin-1. The inability of our antibodies to detect endogenous proteins in Western blot did not allow us to test these possibilities. As observed previously for endospanin-1, endospanin-2 overexpression had no impact on transferrin receptor cell surface expression. These data suggest that endospanin-2, as previously shown for endospanin-1 (10), is able to regulate OB-Ra cell surface expression.
FIGURE 6.
Endospanin-2 overexpression reduces OB-Ra cell surface expression. A, HeLa cells were transduced with a fixed dose of Ad:OB-Ra and increasing doses of Ad:endospanin-2 or Ad:GFP as a control. Cell surface proteins were biotinylated on ice. Biotinylated material was isolated with streptavidin-Sepharose beads and revealed by immunoblot analysis using an anti-HA antibody to detect OB-Ra (OB-Ra surface) or an anti-transferrin receptor (Tsf-R surface) antibody. The relative amounts of OB-Ra in cell lysates were analyzed in parallel (OB-Ra total). B, to compare endospanin-1 and endospanin-2, HeLa cells were transduced with Ad:OB-Ra and increasing doses of Ad:endospanin-1, Ad:endospanin-2, or Ad:GFP. Cell surface proteins were isolated as described above. The relative ratio of biotinylated (surface) over total protein amounts was plotted as a function of the multiplicity of infection of the adenoviruses. Data are means ± S.D. (error bars) of four independent experiments. m.o.i., multiplicity of infection.
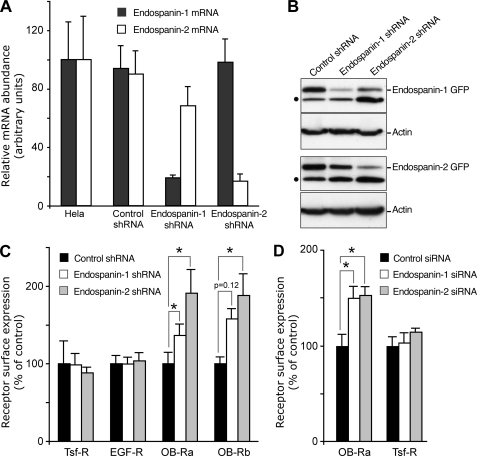
In order to confirm the data obtained using adenovirus-mediated overexpression, we then knocked down endospanin expression by RNA interference. Stable cell lines expressing shRNA directed against each transcript were obtained; independent clones were isolated, and endospanin-1 and -2 mRNA levels were quantified using real-time PCR (Fig. 7A). In cells expressing endospanin-1 or endospanin-2 shRNA, mRNA levels of the corresponding transcript were decreased by ∼80% as compared with HeLa cells (Fig. 7A). As expected, no effect on mRNA abundance was observed in cells expressing a control shRNA. The effect of shRNAs on protein expression was confirmed using GFP-tagged endospanins transiently expressed in stable cell lines (Fig. 7B). Lower levels of endospanin-1 or endospanin-2 proteins were observed in the stable cell line expressing the corresponding shRNA.
FIGURE 7.
shRNA-mediated suppression of endospanins increases leptin receptor cell surface expression. A, effect of shRNA on endospanin-1 and endospanin-2 mRNA levels. Total RNA was extracted from HeLa cells and from the different cell lines expressing either control, endospanin-1, or endospanin-2 shRNA. The levels of endospanin-1 and endospanin-2 mRNA were monitored by real-time quantitative PCR. The data are the mean ± S.D. (error bars) of triplicate experiments and are presented as percentage of the control (HeLa), to which the 100% value was arbitrary attributed. B, effect of shRNA on protein levels. shRNA-expressing cells were transfected with GFP-tagged endospanin-1 or endospanin-2. Immunoblot analysis was performed using anti-GFP antibodies, which detected both endospanin-GFP constructs and GFP (dot) that is constitutively expressed in the cell lines. The same blot was probed with an anti-actin antibody to ensure equal loading of the different samples. C, effect of shRNA on leptin receptor cell surface expression. Cell lines expressing control, endospanin-1, or endospanin-2 shRNAs were transfected with plasmids expressing either short (OB-Ra) or long (OB-Rb) leptin receptor isoforms. 48 h post-transfection, cell surface expression was measured as described in the legend to Fig. 6B, using anti-HA antibody (for OB-Ra and OB-Rb) or antibodies to transferrin receptor (Tsf-R) or to EGF receptor (EGF-R). Data are means ± S.E. (error bars) of triplicate experiments (*, p < 0.05, Wilcoxon test). D, effect of siRNA on leptin receptor cell surface expression. HeLa cells were transfected twice with control (black), endospanin-1 (white), or endospanin-2 (gray) siRNA pools and transduced with Ad:OB-Ra 48 h after the second siRNA transfection. 24 h later, cell surface expression was measured as described in the legend to Fig. 6B, using anti-HA antibody (for OB-Ra) or an antibody to transferrin receptor. Data are mean ± S.D. (error bars) of at least three experiments.
Stable cell lines expressing either endospanin-1 or endospanin-2 shRNA were used to assess the role of these proteins in the modulation of OB-R cell surface expression. We observed a 1.5–2-fold increase in cell surface expression of both OB-Ra and OB-Rb in cells expressing either endospanin-1 or endospanin-2 shRNA as compared with control cells (Fig. 7C). To determine if the knockdown of endospanin-1 or endospanin-2 could affect other receptors expressed at the cell surface, we quantified transferrin and EGF receptors and showed that neither endospanin-1 nor endospanin-2 shRNA had any effect on their expression at the cell surface (Fig. 7C).
To verify the specificity of each endospanin depletion, we monitored the cell surface expression of OB-Ra in HeLa cells transfected with siRNA. An increase of about 1.5-fold of OB-Ra cell surface expression was observed both in cells transfected with endospanin-1 siRNA and in cells transfected with endospanin-2 siRNA (Fig. 7D). In contrast, the same cells did not show any significant variation of the cell surface expression of transferrin receptor. We also verified that the transfection of an antisense oligonucleotide to endospanin-1, an experimental system that we previously used to down-regulate endospanin-1 and increase OB-R cell surface expression (10), had no effect on endospanin-2 mRNA (data not shown). These results confirm that the depletion of each endospanin specifically leads to an increased OB-R cell surface expression.
Taken together, the results of RNAi experiments are consistent with those obtained using adenovirus-mediated overexpression and suggest that both endospanins function as negative regulators of OB-R cell surface expression.
Endospanins Regulate OB-R Trafficking in the Endocytic Pathway
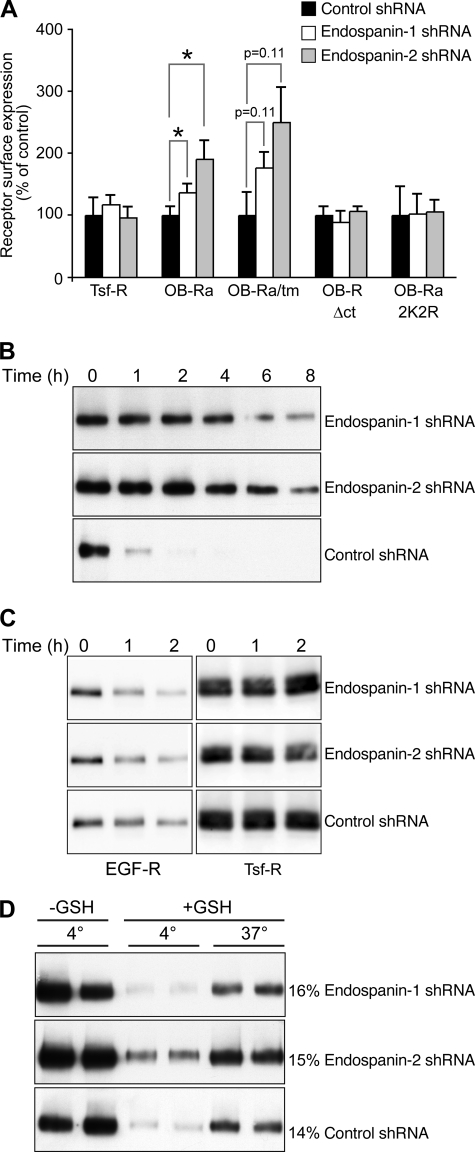
To assess whether endospanins regulate OB-R expression in endocytic or secretory pathways, we expressed OB-Ra mutants impaired at different steps of their intracellular traffic. Three constructs with increased cell surface expression were used: OB-Ra/tm, a construct with increased transport to the cell surface from the biosynthetic pathway, and OB-RΔct and 2K2R, two constructs with reduced internalization from the cell surface due to the lack of cytosolic tail or lack of ubiquitin acceptor sites, respectively (8, 16). Our results suggest that OB-Ra/tm cell surface expression was up-regulated in endospanin-1 and in endospanin-2 shRNA-expressing cells in a way similar to OB-Ra (Fig. 8A). On the other hand, the cell surface expression of both endocytosis-defective mutants was not raised in cells depleted of endospanin-1 or endospanin-2. These results suggest that endospanins regulate OB-R trafficking in the endocytic pathway.
FIGURE 8.
Effect of endospanin depletion on OB-Ra endocytosis and degradation. A, cell lines expressing control, endospanin-1, or endospanin-2 shRNAs were transfected with plasmids expressing either OB-Ra or trafficking mutants OB-Ra/tm, OB-RΔct, and 2K2R. Cell surface expression was measured as described in the legend to Fig. 6B. Data are means ± S.E. of triplicate experiments. B, degradation of endocytosed OB-Ra. Cell lines expressing control, endospanin-1, or endospanin-2 shRNAs were transduced with OB-Ra-expressing adenovirus and submitted to biotinylation of cell surface proteins at 4 °C. Cells were then incubated at 37 °C for the indicated times, and biotinylated OB-Ra was analyzed by immunoblot. C, degradation of biotinylated EGF receptor (EGF-R) and transferrin receptor (Tsf-R). Cells were surface-biotinylated and incubated with 0.25 μg/ml EGF for the indicated times. Biotinylated EGF and transferrin receptors were analyzed by immunoblot. D, effect of endospanin shRNAs on OB-Ra internalization. Cell lines expressing control, endospanin-1, or endospanin-2 shRNAs were transduced with OB-Ra-expressing adenovirus, biotinylated at 4 °C, and allowed to internalize for 15 min at 37 °C or kept on ice (4 °C). Endocytosed OB-Ra protected from glutathione reduction (+GSH) was isolated using streptavidin beads and quantified by immunoblot using anti-HA antibody, together with 40% of the proteins initially biotinylated and not submitted to glutathione reduction (−GSH). The numbers to the right indicate the percentage of internalized protein.
To confirm these results, we monitored the degradation rate of endocytosed OB-Ra. Stable cell lines expressing shRNA were infected with OB-Ra-expressing adenovirus and subjected to biotinylation of cell surface proteins at 4 °C. Cells were then incubated at 37 °C to allow endocytosis and lysosomal degradation of plasma membrane proteins. Our results show that in the presence of endospanin-1 or endospanin-2 shRNA, OB-Ra was significantly stabilized (Fig. 8B). For comparison, we also monitored the degradation of EGFR and transferrin receptor, two receptors whose cell surface expression was not altered in endospanin-1 or endospanin-2 shRNA cell lines. As expected, EGFR was degraded after endocytosis, whereas transferrin receptor was not (Fig. 8C). For EGFR, such a major inhibitory effect on receptor degradation was not observed. These results indicate that endospanins regulate OB-R trafficking but not EGFR trafficking in the endocytic pathway.
We next measured OB-Ra internalization rate in the three cell lines, using an assay based on cell surface biotinylation and glutathione cleavage resistance. The levels of protected OB-Ra were similar in all cell lines (Fig. 8D). This indicates that the increased OB-Ra expression at the cell surface observed in cells deprived of endospanin-1 or endospanin-2 is not due to a blockade of the internalization step. Thus, it is likely that endospanins regulate the traffic of the leptin receptor at a postinternalization step of the endocytic pathway.
Endospanins Regulate OB-R Transport to Lysosomes
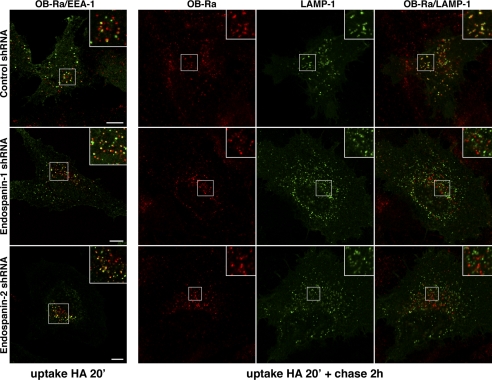
Because the degradation of endocytosed leptin receptors was reduced in shRNA-expressing cells with no change in their internalization from the cell surface, we analyzed their intracellular localization by confocal microscopy. OB-R-expressing cells were incubated for 20 min at 37 °C with anti-HA mAb to label internalized receptors. The cells were either fixed after the antibody uptake or incubated for an additional chase of 2 h with no antibody before fixation. Cells were then processed for immunofluorescent detection of internalized anti-HA antibody and double-labeled with an anti-EEA1 or anti-LAMP-1 antibody. As previously observed in HeLa and COS cells (16), internalized OB-Ra was partially colocalized with EEA1 in early endosomal membranes after a 20-min uptake (Fig. 9). A very similar pattern was observed for OB-Rb (data not shown). In cells depleted of endospanin-1 or endospanin-2, we did not observe any major changes in intracellular localization of internalized OB-Ra or OB-Rb at the end of the uptake (Fig. 9) (data not shown).
FIGURE 9.
Effect of endospanin depletion on OB-Ra endosomal transport. Cells expressing control, endospanin-1, or endospanin-2 shRNAs were transfected with an expression plasmid for HA-tagged OB-Ra, allowed to internalize anti-HA antibody for 20 min, and processed for double label immunofluorescent detection of internalized antibody together with early EEA1 or chased for 2 h with no antibody in the presence of leupeptin and processed for double label immunofluorescent detection of internalized antibody together with LAMP-1. Bars, 10 μm.
To be able to observe endocytosed receptors in LAMP-1-positive compartments after 2 h of chase, we had to perform the experiment in the presence of leupeptin, an inhibitor of lysosomal degradation. Leupeptin treatment had no detectable impact on the co-localization of internalized OB-Ra with EEA1 following the antibody uptake (data not shown) but probably prevented the degradation of endocytosed antibodies and/or receptors in late endocytic compartments. Endocytosed antibodies could be observed in LAMP-1-positive structures in control cells after 2 h of chase (Fig. 9). In contrast, the colocalization with LAMP-1 was dramatically reduced in cells depleted of endospanin-1 or endospanin-2 (Fig. 9). These results suggest that endospanins regulate OB-R trafficking in the late endocytic pathway.
Taken together, our results show that the internalization of leptin receptors from the cell surface occurs normally and that their lysosomal degradation is reduced in cells depleted of endospanin-1 or endospanin-2. Internalized leptin receptors do not accumulate in endocytic compartments of these cells, suggesting that they are probably recycled back to the plasma membrane, resulting in their increased expression at the cell surface. Based on these results, we conclude that endospanins are probably involved in the regulation of postinternalization steps of OB-R trafficking in the endocytic pathway.
DISCUSSION
In this study, we characterized endospanin-1, a protein originally identified as an open reading frame in a splice variant transcript from the OB-R gene (11), as well as endospanin-2, a recently identified endospanin-1 homologue (14). The mRNAs of both proteins were detected in all tissues examined, suggesting ubiquitous expression. At the tissue level, a regulated co-expression of both proteins was suggested by Northern blot experiments. Endospanins were found to be integral membrane proteins with four transmembrane domains of 20 or 21 residues. This means that about two-thirds of these small proteins (131 residues) are probably embedded within the membrane bilayer, leaving only very short connecting loops at the lumenal side of the membranes and a not much longer hydrophilic loop, together with the N- and C- termini on the cytoplasmic side.
Homologues of endospanins were found in animals, plants, and fungi but not in prokaryotes, and all of these homologues have four putative transmembrane domains. No sequence similarity was found to any members of other families of proteins with four transmembrane domains. Together with the conservation of some intron positions in their genes, this indicates that endospanin-1 and endospanin-2 define a family of evolutionarily conserved tetraspanning membrane proteins.
Our results suggest that endospanins regulate membrane traffic and/or cargo sorting in the endocytic pathway. At steady state, endospanin-1 is mainly localized in endosomes, whereas endospanin-2 is mostly localized in the TGN and also to a lesser extent in endosomes. Both endospanins traffic to the plasma membrane, where they are internalized in the same endocytic compartments as leptin receptors. siRNA-mediated depletion of endospanin-1 or endospanin-2 slowed down the transport to lysosomes and the degradation of endocytosed OB-R by interfering with a postinternalization step, and this in turn increased their cell surface expression, probably by increasing or inducing their recycling to the plasma membrane. Because the principles of membrane trafficking are conserved from yeast to neurons, results obtained with HeLa cells are likely to be valid for other cell types. These features are quite similar to those of Vps55p, a member of this family expressed in S. cerevisiae, which is implicated in late endosome-to-vacuole transport (13, 26). This suggests that the members of this family provide a function in membrane traffic that is common to phylogenetically distant eukaryotes. The membrane-spanning domains seem to be of special importance for this function because they are the most conserved.
Most membrane traffic regulatory proteins identified up to now are either soluble or single spanning transmembrane proteins. There are also examples of multispanning proteins, beside Vps55p, that have been implicated in the regulation of membrane traffic. Some of them, like the PMP22 (peripheral myelin protein 22) or stargazin, for instance, are small proteins with four transmembrane domains, which function in membrane traffic as escort proteins (27, 28). Each of them specifically interacts with neosynthetized proteins (P1 myelin protein and glutamate receptors, respectively) and is essential to direct these proteins to their final destination. Another interesting example is the yeast membrane protein Rer1p that cycles between the ER and Golgi complex. Rer1p interacts with the transmembrane domain of various endoplasmic reticulum membrane proteins and is required for their retrieval (29). In this context, it is noteworthy that both endospanins interact with leptin receptors in transfected cells. It will be interesting to determine if this interaction is also found between endogenous proteins and if it is necessary for the modulation of the surface expression of leptin receptors.
Other tetraspanning proteins do not function as escort proteins but rather as regulators of a specific step in membrane traffic. For instance, Got1p and Sft2p are two small tetraspanning proteins that control a fusion step in the transport of proteins to the Golgi complex in S. cerevisiae (30). MAL/VIP17, a tetraspanning membrane protein unrelated to endospanins, is another example. It functions as a regulator of TGN to apical plasma membrane traffic in the secretory pathway of polarized cells (31). Vps55p appears to be such a general regulator of the membrane traffic between endosomes and the vacuole (13, 26). Our results indicate that endospanins function in endosomes. However, we cannot exclude an additional function of these proteins in the biosynthetic pathway, given their dual distribution in the trans-Golgi network and endosomes.
Our results suggest that endospanin-1 and endospanin-2 fulfill very similar functions. One possibility would be that both proteins function as parts of the same complex. However, there are no data supporting this idea. On the contrary, their different intracellular localization at steady state rather suggests that both proteins could fulfill related functions in separate protein complexes. In yeast, there is evidence that Vps55p is part of a functional protein complex that also includes Vps68p, another small membrane protein with four putative transmembrane domains (26). Two genes for Vps68p-like proteins are also found in the human genome,4 suggesting that the whole complex has been duplicated in humans. Whether these two Vps68p homologues also interact with endospanins or function in the regulation of OB-R intracellular trafficking remains an open question.
The function of endospanins is apparently restricted to a surprisingly small set of cargoes. Initially, we screened endospanin-1 specificity by assaying cell surface expression of selected receptors. The silencing of endospanin-1 only resulted in the up-regulation of cell surface expression of leptin receptors OB-Ra and OB-Rb, among the few receptors that have been tested so far. Recently, it has been reported that both endospanins also regulate cell surface expression of the growth hormone receptor (GHR) (32). Although cellular mechanisms underlying this modulation of GHR expression were not investigated, similarities with OB-R can be found. Much like with leptin receptors, endospanin-1 and endospanin-2 overexpression decreased GHR cell surface expression, their silencing increased GHR cell surface expression, and this modulation of GHR expression appeared to be at a post-transcriptional level. This indicates that the function of endospanins is not restricted to leptin receptors. For EGFR, a receptor whose surface expression was not altered by endospanin-1 or endospanin-2 depletion, the transport to lysosomes was not affected, confirming the lack of involvement of endospanins in the regulation of its trafficking in the endocytic pathway. However, we cannot exclude the possibility that the endocytic traffic of other receptors and membrane proteins may be controlled by endospanin-1 and/or endospanin-2 with a minimal impact on their steady state cell surface expression, depending on other properties of the proteins. It would be interesting to investigate other receptors in order to further probe the specificity of endospanin-1 and endospanin-2 function.
In obese patients, increased circulating leptin levels are associated with reduced leptin-induced signaling. The mechanisms underlying this leptin resistance state are still poorly understood. We have previously shown that targeting endospanin-1 in the arcuate nucleus of the hypothalamus prevents diet-induced obesity in mice by increasing OB-R cell surface expression (10). This suggests that altered regulation of OB-R cell surface expression contributes to the onset of leptin resistance. In vitro studies have shown that OB-R is endocytosed in a constitutive and ligand-independent manner (7–9), indicating that the down-regulation of this receptor is not mediated by ligand-induced internalization. We now provide evidence that endospanin-1 functions by facilitating OB-R degradation. Therefore, we can hypothesize that an increase in endospanin-1 expression levels might be associated with leptin resistance, as it was recently suggested for GH resistance under conditions of fasting and type 1 diabetes mellitus (32). It would be of interest to determine if endospanin-1 is actually overexpressed in the arcuate nucleus of mice fed a high fat diet. In addition, we now show that endospanin-2 has a similar function in vitro. Whether endospanin-2 is also required for OB-R degradation in vivo is a critical question that remains to be addressed. If it is required, then conceivably endospanin-2 could be targeted pharmacologically, in addition to endospanin-1, so as to increase OB-R levels and enhance leptin sensitivity.
Supplementary Material
Acknowledgments
We kindly acknowledge the technical assistance of Delphine Delcroix and Frédérique Dewitte. We thank Jean Dubuisson for support and for the retroviral packaging vectors, Marino Zerial for the GFP-tagged Rab constructs, Georges Banting for TGN38 antibody, and Sophana Ung for assistance in the preparation of the figures. We thank Francis Vasseur, Rosine Haguenauer-Tsapis, Naima Belgareh-Touzé, and Christiane Volland for helpful discussions and critical reading of the manuscript.
This research was supported in part by grants from CNRS (Programme Biologie Cellulaire), INSERM (Programme National de Recherche sur le Diabète), and the Région Nord-Pas de Calais.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
K. Séron, C. Couturier, S. Belouzard, J. Bacart, D. Monté, L. Corset, O. Bocquet, J. Dam, V. Vauthier, C. Lecœur, B. Bailleul, B. Hoflack, P. Froguel, R. Jockers, and Y. Rouillé, unpublished observation.
- VSV-G
- vesicular stomatitis virus G protein
- BRET
- bioluminescence resonance energy transfer
- EGFR
- epidermal growth factor receptor
- TGN
- trans-Golgi network
- NRK
- normal rat kidney
- GHR
- growth hormone receptor.
REFERENCES
- 1. Friedman J. M., Halaas J. L. (1998) Nature 395, 763–770 [DOI] [PubMed] [Google Scholar]
- 2. Schwartz M. W., Woods S. C., Porte D., Jr., Seeley R. J., Baskin D. G. (2000) Nature 404, 661–671 [DOI] [PubMed] [Google Scholar]
- 3. Bates S. H., Myers M. G., Jr. (2003) Trends Endocrinol. Metab. 14, 447–452 [DOI] [PubMed] [Google Scholar]
- 4. Tartaglia L. A. (1997) J. Biol. Chem. 272, 6093–6096 [DOI] [PubMed] [Google Scholar]
- 5. Frühbeck G. (2006) Biochem. J. 393, 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zabeau L., Lavens D., Peelman F., Eyckerman S., Vandekerckhove J., Tavernier J. (2003) FEBS Lett. 546, 45–50 [DOI] [PubMed] [Google Scholar]
- 7. Barr V. A., Lane K., Taylor S. I. (1999) J. Biol. Chem. 274, 21416–21424 [DOI] [PubMed] [Google Scholar]
- 8. Belouzard S., Delcroix D., Rouillé Y. (2004) J. Biol. Chem. 279, 28499–28508 [DOI] [PubMed] [Google Scholar]
- 9. Uotani S., Bjørbaek C., Tornøe J., Flier J. S. (1999) Diabetes 48, 279–286 [DOI] [PubMed] [Google Scholar]
- 10. Couturier C., Sarkis C., Séron K., Belouzard S., Chen P., Lenain A., Corset L., Dam J., Vauthier V., Dubart A., Mallet J., Froguel P., Rouillé Y., Jockers R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19476–19481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bailleul B., Akerblom I., Strosberg A. D. (1997) Nucleic Acids Res. 25, 2752–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindell K., Bennett P. A., Itoh Y., Robinson I. C., Carlsson L. M., Carlsson B. (2001) Mol. Cell. Endocrinol. 172, 37–45 [DOI] [PubMed] [Google Scholar]
- 13. Belgareh-Touzé N., Avaro S., Rouillé Y., Hoflack B., Haguenauer-Tsapis R. (2002) Mol. Biol. Cell 13, 1694–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y., Ying K., Xie Y., Zhou Z., Wang W., Tang R., Zhao W., Zhao S., Wu H., Gu S., Mao Y. (2001) Biochim. Biophys. Acta 1517, 327–331 [DOI] [PubMed] [Google Scholar]
- 15. Couturier C., Jockers R. (2003) J. Biol. Chem. 278, 26604–26611 [DOI] [PubMed] [Google Scholar]
- 16. Belouzard S., Rouillé Y. (2006) EMBO J. 25, 932–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chartier C., Degryse E., Gantzer M., Dieterle A., Pavirani A., Mehtali M. (1996) J. Virol. 70, 4805–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fallaux F. J., Kranenburg O., Cramer S. J., Houweling A., Van Ormondt H., Hoeben R. C., Van Der Eb A. J. (1996) Hum. Gene Ther. 7, 215–222 [DOI] [PubMed] [Google Scholar]
- 19. Bartosch B., Dubuisson J., Cosset F. L. (2003) J. Exp. Med. 197, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horn M., Banting G. (1994) Biochem. J. 301, 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 22. Chu G., Hayakawa H., Berg P. (1987) Nucleic Acids Res. 15, 1311–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Ayoub M. A., Couturier C., Lucas-Meunier E., Angers S., Fossier P., Bouvier M., Jockers R. (2002) J. Biol. Chem. 277, 21522–21528 [DOI] [PubMed] [Google Scholar]
- 25. Ayoub M. A., Levoye A., Delagrange P., Jockers R. (2004) Mol. Pharmacol. 66, 312–321 [DOI] [PubMed] [Google Scholar]
- 26. Schluter C., Lam K. K., Brumm J., Wu B. W., Saunders M., Stevens T. H., Bryan J., Conibear E. (2008) Mol. Biol. Cell 19, 1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen L., Chetkovich D. M., Petralia R. S., Sweeney N. T., Kawasaki Y., Wenthold R. J., Bredt D. S., Nicoll R. A. (2000) Nature 408, 936–943 [DOI] [PubMed] [Google Scholar]
- 28. Sanders C. R., Ismail-Beigi F., McEnery M. W. (2001) Biochemistry 40, 9453–9459 [DOI] [PubMed] [Google Scholar]
- 29. Sato K., Sato M., Nakano A. (2003) Mol. Biol. Cell 14, 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conchon S., Cao X., Barlowe C., Pelham H. R. (1999) EMBO J. 18, 3934–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puertollano R., Martín-Belmonte F., Millán J., de Marco M. C., Albar J. P., Kremer L., Alonso M. A. (1999) J. Cell Biol. 145, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Touvier T., Conte-Auriol F., Briand O., Cudejko C., Paumelle R., Caron S., Baugé E., Rouillé Y., Salles J. P., Staels B., Bailleul B. (2009) J. Clin. Invest. 119, 3830–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.