Abstract
Genetic studies involving zebrafish and mice have demonstrated that the protein Gon4l (Gon4-like) is essential for hematopoiesis. These studies also suggested that Gon4l regulates gene expression during hematopoietic development, yet the biochemical function of Gon4l has not been defined. Here, we describe the identification of factors that interact with Gon4l and may cooperate with this protein to regulate gene expression. As predicted by polypeptide sequence conservation, Gon4l interacted and co-localized with the DNA-binding protein YY1 (Yin Yang 1). Density gradient sedimentation analysis of protein lysates from mouse M12 B cells showed that Gon4l and YY1 co-sediment with the transcriptional co-repressor Sin3a and its functional partner histone deacetylase (HDAC) 1. Consistent with these results, immunoprecipitation studies showed that Gon4l associates with Sin3a, HDAC1, and YY1 as a part of complexes that form in M12 cells. Sequential immunoprecipitation studies demonstrated that Gon4l, YY1, Sin3a, and HDAC1 could all associate as components of a single complex and that a conserved domain spanning the central portion of Gon4l was required for formation of this complex. When targeted to DNA, Gon4l repressed the activity of a nearby promoter, which correlated with the ability to interact with Sin3a and HDAC1. Our data suggest that Sin3a, HDAC1, and YY1 are co-factors for Gon4l and that Gon4l may function as a platform for the assembly of complexes that regulate gene expression.
Keywords: Cell Differentiation, Co-repressor Transcription, Gene Regulation, Histone Deacetylase, Transcription Factors, B Cell Development
Introduction
Antibody-secreting B cells are generated throughout life via a developmental pathway called B lymphopoiesis (1). At an early stage in this process, B cell progenitors undergo a developmental transition that results in suppression of residual multi-lineage potentiality and commitment to a B cell fate. This transition coincides with dramatic changes in gene expression, resulting in the up-regulation of B cell-specific genes and the down-regulation of genes that promote alternative lineage fates (2–5). In addition, B cell progenitors must express functional immunoglobulin heavy and light chain proteins to develop into mature B cells. Expression of these proteins requires rearrangement of the encoding loci as controlled by transcriptional and epigenetic mechanisms (6, 7). Thus, B lymphopoiesis relies on elaborate mechanisms of gene regulation to generate B cells that can respond to antigens and make antibodies.
In our previous study (8), we described a novel mouse strain named Justy (for just T cells). This strain carries a chemically induced point mutation that, in the homozygous state, causes a profound arrest in B lymphopoiesis but does not affect other major aspects of mouse physiology. The identified developmental defect coincides with sustained expression of genes normally targeted for repression during the early stages of B lymphopoiesis. These data suggest that the mutation impairs mechanisms required for gene repression in developing B cells. The lesion responsible was shown to be a point mutation that disrupts splicing of RNA transcribed from the Gon4l (Gon4-like) gene, resulting in dramatically reduced expression of the encoded protein. Collectively, these data demonstrate that Gon4l is required for B lymphopoiesis and suggest that this protein is important for the regulation of gene expression during this process.
Studies of Caenorhabditis elegans, Drosophila melanogaster, and zebrafish have also provided evidence that Gon4l is important for the regulation of gene expression in developmental pathways (9–13). For example, a null mutation in the zebrafish Gon4l ortholog disrupts the expression of critical erythroid genes and severely impairs primitive erythropoiesis (11). Also, studies of mutant zebrafish have suggested that the absence of Gon4l results in cell cycle arrest and increased cellular apoptosis in developing embryos (11, 12). The mechanisms underlying these latter effects are not known, but they could reflect disruption of gene regulation during development.
Despite its apparent biological importance, the function of Gon4l is essentially unknown. In this study, we used biochemical methods to demonstrate that Gon4l can support the formation of complexes that contain the DNA-binding protein YY1, the co-repressor Sin3a, and the histone-modifying enzyme histone deacetylase 1 (HDAC1).2 Using a reporter gene assay, we established that Gon4l can inhibit the activity of a target promoter. Moreover, mutational analysis demonstrated that a highly conserved region in the center of Gon4l was required for both interaction with YY1, Sin3a, and HDAC1 and inhibition of promoter activity. Our data support a model in which Gon4l functions together with YY1 and/or Sin3a/HDAC-containing complexes to regulate gene expression, which may represent a mechanism important for B lymphopoiesis and other developmental pathways.
EXPERIMENTAL PROCEDURES
Analysis of Gon4l Protein Homologies
Putative protein structural domains were identified using the SMART (14, 15) and NCBI BLAST databases. Protein sequence conservation was determined using Vector-NTI software (Invitrogen). Accession numbers for the analyzed protein sequences are: NM_139118 (human YY1AP), NP_001032622 (human GON4L), Q9DB00 (mouse Gon4l), ABP65284 (Danio rerio Gon4l), NP_001097352 (D. melanogaster Cdp1), and NM_001129314 (C. elegans GON-4).
Cell Culture
The B cell lines M12.4.1 (M12) (16), HAFTL1 (17), 38B9 (18), and 18.81A20 (19) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, and 50 μm 2-mercaptoethanol. Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm sodium pyruvate, and 2 mm l-glutamine.
Generation of M12 Cells Expressing FLAG-Gon4l
A DNA fragment encoding FLAG-Gon4l was ligated into the pEFOP.neo expression vector. pEFOP.neo was generated by replacing the Rous sarcoma virus promotor fragment in pOPRSV5.neo (20) with the EF1α promotor fragment from pEF-BOS (21). M12 cells were electroporated with pEFOP-FLAG-Gon4l or empty pEFOP.neo as described previously (22). Clonal cell lines were selected for and maintained in the medium described in the preceding section but containing 400 μg/ml G418 sulfate.
Plasmids and Transient Transfections
pcDNA3.1 (Invitrogen) was used as the backbone for all Gon4l expression constructs. A synthetic oligonucleotide duplex encoding the FLAG epitope tag was fused in-frame at the 5′ end of Gon4l coding sequences to generate plasmids that express FLAG-Gon4l proteins. A DNA fragment from pCMV-BD (Stratagene) encoding the GAL4 DNA-binding domain (DBD) was used to generate plasmids that express GAL4-DBD-Gon4l fusion proteins. pSPORT-YY1 and pCMV-SPORT6-HDAC1 were purchased from Open Biosystems. The latter plasmid was used to PCR amplify HDAC1 cDNA, which was then inserted into pCMV-HA (Clontech) to generate pCMV-HA-HDAC1. The expression plasmid CS2+MT-Sin3a (23) was provided by Dr. Donald Ayer (University of Utah). All of the transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitations (IPs) Using Whole Cell Lysates
The cells were suspended in a lysis buffer containing 50 mm Tris-HCl, pH 8.0, 120 mm NaCl, 0.5% Nonidet P-40, 0.2 mm sodium orthovanadate, 100 mm NaF, 50 μg/ml PMSF, and protease inhibitor mixture (Roche Applied Sciences). The lysates were gently rotated at 4 °C for 15 min and centrifuged at 16,100 × g for 10 min, after which supernatants were collected. To immunoprecipitate YY1 from M12 cell lysates, anti-YY1 antibody (Santa Cruz; sc-1703) and protein G-agarose beads (Santa Cruz; sc-2002) were used. For all other IP experiments, Dynal protein G beads (Invitrogen) coated with the appropriate antibodies were used as described (20). For sequential IPs, material recovered in the first IP was incubated with 100 μg/ml FLAG peptide (Sigma) in lysis buffer, and eluted material was used for the second IP. For single-round IPs or the second round of sequential IPs, immunoprecipitates were eluted with 1× NuPAGE sample buffer (Invitrogen). Antibodies used for IPs were: anti-FLAG (Sigma; clone M2), anti-mSin3a (Santa Cruz; sc-994), or anti-HDAC1 (Santa Cruz; sc-7872). Control IPs were performed using nonspecific mouse or rabbit IgG (Santa Cruz).
Nuclear Extract Preparation and IP
The nuclear extracts were prepared using previously described buffers (24). All of the procedures were done on ice or at 4 °C. 3 × 108 cells were washed with PBS and resuspended in three packed cell volumes of Buffer A. The cells were incubated for 10 min, after which Nonidet P-40 was added to a final concentration of 0.25%. After incubation for 10 min, the cells were transferred to a Dounce homogenizer (Kontes) and processed through the homogenizer 50 times with pestle B. The nuclei were pelleted by centrifugation at 700 × g for 10 min and resuspended in three packed cell volumes of Buffer B. The nuclei were transferred to a Dounce homogenizer, processed through the homogenizer 50 times with pestle B, rotated at 4 °C for 1 h, and then centrifuged at 16,100 × g for 10 min. Supernatants were collected and dialyzed against Buffer D for 4 h. Recovered material was centrifuged at 16,100 × g for 10 min, and supernatant was collected as nuclear extract. IPs were performed as described in the preceding section except that Buffer D lacking or containing 0.5% Nonidet P-40 was used as reaction and wash buffer.
Sucrose Gradient Centrifugation
M12 cells were lysed in PBS containing 0.5% Triton X-100 and 1 mm EDTA (25). Cell lysates were separated by centrifugation for 16 h at 41,000 rpm in a swinging bucket rotor (Beckman SW41Ti). Sedimentation standards (Bio-Rad precision plus standards mixed with Sigma thyroglobulin) were separated through gradients run in parallel with those used to separate cell lysates. The gradients were collected in 500-μl fractions and analyzed by immunoblot.
SDS-PAGE and Immunoblot Analysis
The proteins were separated by electrophoresis through 7% or 3–8% NuPAGE gels (Invitrogen) and transferred to PVDF (Millipore). The membranes were probed with the following antibodies: affinity-purified polyclonal anti-Gon4l antibodies (8) (supplemental Fig. S2), anti-FLAG (Sigma; clone M2), anti-YY1, anti-Myc, anti-HDAC1, anti-mSin3a, anti-GAL4-DBD (Santa Cruz; sc-1703, -40, -7872, -994, and -510, respectively), or anti-HA (Roche Applied Sciences; clone 3F10). Donkey anti-mouse or anti-rabbit IgG HRP (Santa Cruz; sc-2314 and -2313) was used as secondary antibody. Membrane-bound antibody complexes were visualized by chemiluminescence (SuperSignal West Pico, Thermo Scientific).
Immunofluorescent Confocal Microscopy
293T cells were transfected, replated 24 h later onto poly-l-lysine-coated coverslips, and cultured overnight. The cells were fixed with ice-cold methanol and permeabilized with 0.2% Triton X-100 in PBS. After incubation with 5% normal goat serum (Sigma), the cells were incubated overnight at 4 °C with primary antibodies diluted in blocking solution. The cells were washed and incubated with goat anti-rabbit antibodies conjugated to Alexa Fluor 568 and/or goat anti-mouse antibodies conjugated to Alexa Fluor 488, washed, and stained with TO-PRO-3 to visualize nuclei. The images were acquired using a Zeiss 510 confocal microscope attached to a digital camera. Primary antibodies used were: anti-FLAG (Sigma; clone M2 or polyclonal F7425), anti-YY1 (Santa Cruz; sc-281), or anti-Myc (Santa Cruz; sc-40, clone 9E10).
Luciferase Reporter Assays
293T cells cultured in 24-well plates were transfected with 100 ng of the plasmid pUAS-TK-Luc (provided by Dr. Christopher Glass, UCSD) (26, 27), 2 ng of the plasmid pRL-CMV (Renilla luciferase, Promega), and increasing amounts of expression plasmids encoding FLAG-Gon4l, GAL4-Gon4l fusions, or the GAL4-DBD. The total amount of DNA transfected was adjusted to 0.8 μg by adding pcDNA3.1 lacking a cDNA insert. 24 h after DNA was added, the cell lysates were prepared, and firefly and Renilla luciferase activities were determined using the dual luciferase reporter assay system (Promega) following the manufacturer's instructions. All of the samples were analyzed in triplicate, and the values obtained were used to calculate averages and standard deviations.
RESULTS
Gon4l Contains Homology to Transcriptional Regulators
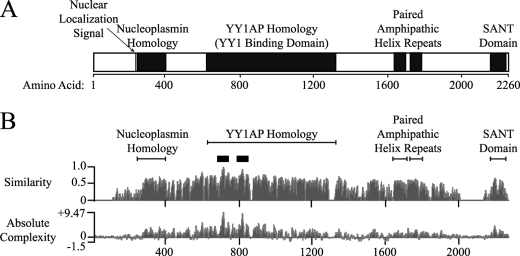
We and others (8, 11, 28) have used bioinformatic tools to identify structural motifs that may be formed by Gon4l (Fig. 1A). The N-terminal portion of mouse Gon4l (amino acids 1–610) is highly acidic (pI of 4.1) and contains a putative nuclear localization sequence (NLS) that is conserved among vertebrate orthologs. Adjacent to the NLS is a region with weak homology to nucleoplasmins, which have key roles in the regulation of chromatin structure (29). The central domain of Gon4l (amino acids 611–1364) is 78% similar to the human protein YY1AP (YY1-associated protein), which can interact with DNA-binding protein YY1 and influence YY1-mediated transcriptional regulation (30). The gene encoding YY1AP is unique to primates and was apparently generated by partial duplication of GON4L (31). Amino acids 1600–1850 of Gon4l span two separate regions that each have strong homology to the consensus paired amphipathic helix (PAH) repeat sequence. The mammalian transcriptional co-repressors Sin3a and Sin3b each contain four PAH repeats, which mediate protein-protein interactions important for transcriptional regulation (e.g. binding to HDACs) (32). Lastly, NMR analysis (Protein Data Bank accession code 1UG2) indicates that sequences near the C terminus of mouse Gon4l (amino acids 2171–2222) form a helix-loop-helix structure called a SANT domain (SWI3, ADA2, N-CoR, and TFIIIB) (33). This motif can mediate protein-protein interactions involved in transcriptional regulation (for example, see Ref. 34).
FIGURE 1.
Putative domains and amino acid sequence conservation in Gon4l. A, schematic of mouse Gon4l showing the locations of structural domains predicted to be formed based on bioinformatic analysis of the primary amino acid sequence. Descriptions of the indicated domains are provided in the text. B, similarity (top plot) and absolute complexity (bottom plot) plots generated using the ClustalW program to compare the amino acid sequences of human YY1AP and Gon4l proteins from human, mouse, zebrafish, D. melanogaster, and C. elegans. The similarity plot was generated by assigning values between 0 and 1 to each of the amino acids in the different Gon4l proteins, with values being directly proportional to the similarity of each residue to the corresponding amino acid in the consensus sequence. The values shown represent the sums of values for each amino acid position divided by the number of sequences analyzed. The absolute complexity plot was generated by performing pair-wise substitutions between all of the sequences analyzed. Scores were assigned to each pair-wise substitution that are directly proportional to the similarity between the amino acids involved. The values shown are the sums of all scores generated by pair-wise comparison divided by the number of pairs analyzed. The black boxes above the similarity plot span the most highly conserved regions of Gon4l.
We used the algorithm ClustalW to identify the most highly conserved amino acids among Gon4l proteins from invertebrate and vertebrate species (Fig. 1B). Consistent with roles in Gon4l function, sequences spanning the putative domains described above were all highly conserved. The most conserved stretches (>90% similarity) were within two segments near the N terminus of the YY1AP homology region (amino acids 700–730 and 799–829; supplemental Fig. S1). Based on studies of YY1AP, these residues may be important for interaction with YY1 (30).
Gon4l Interacts with the Transcription Factor YY1
To detect Gon4l expression in cells, we generated two different anti-Gon4l polyclonal antibodies as described previously (8). One antibody preparation is specific for the C-terminal region of Gon4l, and the other is specific for a fragment within the YY1AP homology region (supplemental Fig. S2A). The C-terminal antibodies detected full-length Gon4l and a slightly smaller protein in whole cell lysates (WCLs) from a set of mouse transformed B cell lines (supplemental Fig. S2B, top panel). Both of these proteins were detected in WCL from primary pro-B cells (8), suggesting that they are characteristically expressed in B lineage cells. Antibodies specific for the YY1AP homology region also detected full-length Gon4l but not the smaller species (supplemental Fig. S2C), suggesting the latter is either a Gon4l isoform that lacks the YY1AP homology region or a protein encoded by another gene that cross-reacts with the C-terminal Gon4l antibodies.
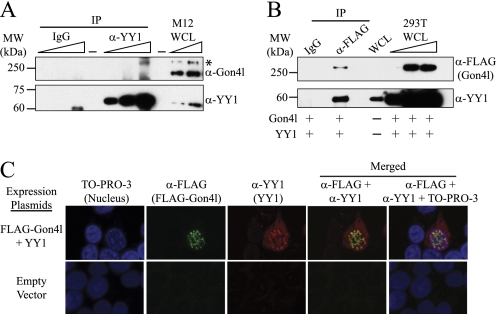
Given the homology to YY1AP, we determined whether mouse Gon4l and YY1 could interact. WCLs were prepared from M12 B cells and used to perform IPs with control IgG and YY1 antibodies. Immunoblot analysis of the recovered material using the C-terminal Gon4l antibodies showed that Gon4l co-immunoprecipitated with YY1 (Fig. 2A). The same result was obtained when Gon4l antibodies raised against the YY1AP homology domain were used to immunoblot material recovered by IP (data not shown). Notably, the recovered material did not contain the smaller protein detected in M12 WCL by the C-terminal Gon4l antibodies. This result is consistent with the apparent absence of the YY1AP homology in this protein as shown by immunoblot analysis (supplemental Fig. S2, B and C).
FIGURE 2.
Mouse Gon4l interacts with the zinc finger DNA-binding protein YY1. A, co-IP of YY1 and Gon4l from M12 WCLs. Anti-YY1 antibodies were used to IP YY1 from WCL. Nonspecific IgG was used to perform control IPs in parallel. Recovered material was immunoblotted with antibodies specific for the C terminus of Gon4l (top panel) or YY1 (bottom panel). The wedges at the top of the blots signify that serially increased amounts of sample were loaded. The asterisk in the top panel marks the position of Gon4l. B, co-IP of YY1 and FLAG-Gon4l expressed in transfected 293T cells. Anti-FLAG and nonspecific IgG were used to IP proteins from 293T WCLs. Recovered material was immunoblotted with antibodies specific for the FLAG epitope to detect Gon4l (top panel) or those specific for YY1 (bottom panel). C, co-localization of Gon4l and YY1 in the nucleus. The images shown were obtained by immunofluorescent confocal microscopy of 293T cells expressing FLAG-Gon4l and YY1 from transfected plasmids as noted at the left. 293T cells transfected with expression plasmid lacking a cDNA insert were used as a control. The cells were stained with anti-FLAG and anti-YY1 antibodies and then with the nuclear stain TO-PRO-3. The reagents used for staining are noted at the top. Yellow color in the merged images signifies co-localization of FLAG-Gon4l and YY1. All of the data shown are representative of at least three independent experiments. MW, molecular mass.
We next determined whether YY1 and Gon4l expressed from plasmids could interact, thus allowing us to define parts of Gon4l required for interaction with YY1 (see Fig. 8). A plasmid encoding FLAG epitope-tagged Gon4l (FLAG-Gon4l) was transiently co-transfected into 293T cells together with a plasmid encoding mouse YY1. Lysates from transfected cells were used to perform IPs with control IgG and FLAG antibodies. Immunoblot analysis demonstrated that YY1 specifically co-immunoprecipitated with FLAG-Gon4l (Fig. 2B). We also determined whether FLAG-Gon4l and YY1 co-localized in cells. The expression plasmids described above were co-transfected into 293T cells, which were then stained with the appropriate antibodies and analyzed by immunofluorescent confocal microscopy (Fig. 2C). FLAG-Gon4l was detected in the nucleus, whereas YY1 was seen in both the cytoplasm and the nucleus as previously reported (35). Among 60 cells staining positive for both proteins, 57 (∼95%) showed overlapping nuclear localization of YY1 and Gon4l. These data demonstrate that Gon4l and YY1 interact in cells and co-localize within the nucleus.
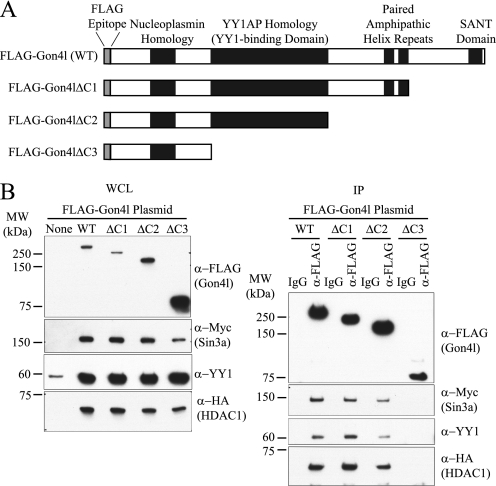
FIGURE 8.
The YY1AP homology domain of Gon4l is required for association with YY1 and Sin3a/HDAC1. A, schematic showing the structures of FLAG-Gon4l C-terminal truncation mutants used for the experiments shown in B. B, 293T cells were transfected with expression plasmid lacking a cDNA insert or with the indicated FLAG-Gon4l expression plasmid together with expression plasmids encoding YY1, Myc-Sin3a, and HA-HDAC1. Left panel, immunoblot analysis of WCLs probed with the indicated antibodies. Right panel, FLAG antibodies were used to IP FLAG-Gon4l proteins from WCLs. Nonspecific IgG was used to perform control IPs in parallel. Recovered material was immunoblotted with the indicated antibodies. The data shown are representative of at least two independent experiments. MW, molecular mass.
Gon4l Co-sediments with YY1, Sin3a, and HDAC1
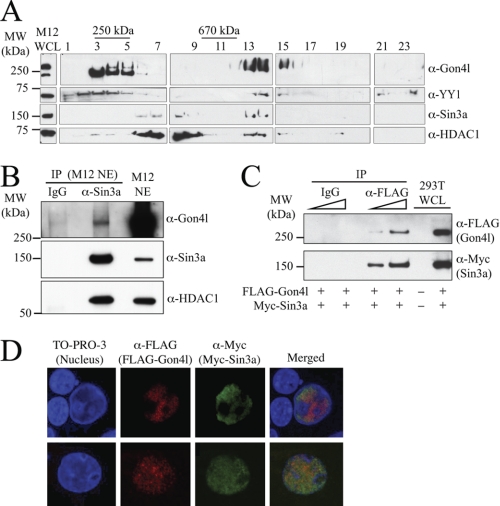
To identify other proteins that associate with Gon4l, density gradient sedimentation analysis was performed. WCLs from M12 cells were separated in 20–50% linear sucrose gradients by centrifugation. Protein standards were separated in identical gradients run in parallel so that molecular masses could be assigned to regions of the gradients. After centrifugation, gradient fractions were collected, and those containing M12 WCL were analyzed by immunoblot (Fig. 3A). A portion of the Gon4l protein expressed in M12 cells was detected in fractions containing species with molecular masses of ∼250 kDa, which is roughly the mass of monomeric Gon4l. However, the majority of Gon4l expressed in M12 cells was found in gradient fractions containing species with molecular masses greater than 670 kDa, suggesting that Gon4l within these fractions was stably associated with other factors. Further, a substantial amount of YY1 was detected in the same high molecular mass fractions.
FIGURE 3.
Gon4l interacts with the transcriptional co-repressor Sin3a and its functional partner HDAC1. A, co-sedimentation of Gon4l, YY1, Sin3a, and HDAC1 in sucrose density gradients. WCLs from M12 cells were loaded onto the top of a 20–50% sucrose gradient. After centrifugation, the fractions indicated across the top of the panel (odd numbers only are shown) were collected and analyzed by immunoblot using the antibodies listed on the right side of the panel. M12 WCL was used as a positive control for immunoblots (left side of panel). Protein standards were separated in parallel gradients, and fractions containing these are indicated at the top of the panel. The data shown are representative of at least three independent experiments. B, co-immunoprecipitation of Gon4l, Sin3a, and HDAC1 from M12 cell nuclear extracts (NE). Sin3a antibodies and nonspecific IgG were used to IP proteins from M12 NE. Recovered material was immunoblotted with antibodies specific for Gon4l (top panel), Sin3a (middle panel), or HDAC1 (bottom panel). M12 NE was used as a positive control for immunoblots. C, co-immunoprecipitation of FLAG-Gon4l and Myc-tagged Sin3a (Myc-Sin3a) expressed in transfected 293T cells. Anti-FLAG and nonspecific IgG were used to immunoprecipitate proteins from 293T WCLs. Recovered material was immunoblotted with antibodies specific for the FLAG or Myc epitopes to detect FLAG-Gon4l (top panel) and Myc-Sin3a (bottom panel), respectively. The wedges at the top of the blots signify that serially increased amounts of sample were loaded. 293T WCLs were used as control for immunoblots. D, co-localization of Gon4l and Sin3a in the nucleus. The images shown were obtained by immunofluorescent confocal microscopy of 293T cells expressing FLAG-Gon4l and Myc-Sin3a from transfected plasmids. 293T cells transfected with expression plasmid lacking a cDNA insert were used as a control (data not shown). The cells were stained with anti-FLAG and anti-Myc antibodies and then with the nuclear stain TO-PRO-3 as noted at the top of the panel. Yellow color in the merged images signifies co-localization of FLAG-Gon4l and Myc-Sin3a. The upper and lower sets of panels are each from an independent experiment. All of the data shown are representative of at least three independent experiments. MW, molecular mass.
Our previous study showed that the lack of Gon4l impairs gene repression in B cell progenitors (8). Because protein complexes containing HDACs are critical for gene repression, we determined whether Gon4l co-sedimented with HDACs and their functional partners. Immunoblot analysis revealed that HDAC1 and its associated co-repressor, Sin3a, were contained within the same high molecular mass fractions as Gon4l and YY1 (Fig. 3A). These results suggested that Sin3a and HDAC1 interact with Gon4l.
Gon4l Interacts with the Co-repressor Sin3a and Its Functional Partner HDAC1
We next determined whether Gon4l, Sin3a, and HDAC1 expressed in M12 cells could interact as suggested by the gradient analysis. Nuclear extracts were prepared from M12 cells and used to perform IPs with control IgG and Sin3a antibodies (Fig. 3B). As expected, HDAC1 was specifically recovered with Sin3a. In addition, Gon4l was recovered with Sin3a, indicating the two proteins associate in M12 cells. We also determined whether YY1 expressed by M12 cells co-immunoprecipitated with Gon4l, Sin3a, and HDAC1. YY1 was recovered with the other three proteins when IPs were performed using Sin3a antibodies, but only when the reaction and wash buffers used lacked detergent (supplemental Fig. S3). These data suggest that YY1 associates with a subset, rather than most or all, of the complexes formed by Gon4l, Sin3a, and HDAC1 in M12 cells.
We next determined whether Sin3a and Gon4l could interact when expressed from plasmids in transfected cells. An expression plasmid encoding Myc-Sin3a was co-transfected into 293T cells together with that encoding FLAG-Gon4l. Lysates were prepared from transfected cells and used to perform IPs with control IgG and anti-FLAG antibodies. Immunoblot analysis of recovered material demonstrated that Sin3a was co-immunoprecipitated with Gon4l (Fig. 3C). We also determined whether Gon4l and Sin3a co-localize in cells. Expression plasmids encoding FLAG-Gon4l or Myc-Sin3a were transiently co-transfected into 293T cells, which were then stained with antibodies that recognize the FLAG or Myc epitope tags. Immunofluorescent confocal microscopy analysis showed that both Sin3a and Gon4l localized almost exclusively to the nucleus (Fig. 3D). Although the distribution of Sin3a was slightly broader than that of Gon4l, substantial overlap between the localization patterns of the two proteins was observed.
FLAG-Gon4l Expressed from a Transgene Associates with Endogenous Sin3a, HDAC1, and YY1
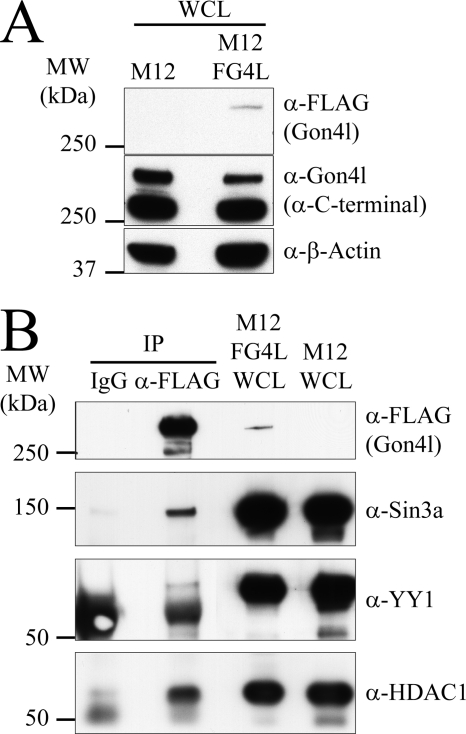
To confirm the findings described above, we created a system in which Gon4l expressed in M12 cells could be directly targeted for IP. Thus, we generated an M12 cell line (M12FG4L) that contains a stably integrated transgene encoding FLAG-Gon4l. Immunoblot analysis demonstrated that FLAG-Gon4l was expressed in M12FG4L cells and that the total amount of Gon4l protein present in these cells was similar to that maintained in parental M12 cells (Fig. 4A). Sucrose gradient sedimentation analysis of M12FG4L WCL demonstrated that FLAG-Gon4l distributed in the gradient in a manner essentially identical to that of native Gon4l, with the majority of FLAG-Gon4l being detected in fractions containing high molecular mass species (supplemental Fig. S4). As expected, these fractions also contained YY1. FLAG antibodies were used to immunoprecipitate FLAG-Gon4l from M12FG4L WCLs. Immunoblot analysis of recovered material demonstrated that Sin3a, HDAC1, and YY1 all co-immunoprecipitated with FLAG-Gon4l (Fig. 4B). As observed when Sin3a was targeted for IP, a limited amount of YY1 was recovered with FLAG-Gon4l, Sin3a, and HDAC1. This result again suggests that YY1 associates with only a subset of Gon4l-containing complexes formed in cells. These data confirm that Gon4l associates with Sin3a, HDAC1, and YY1 when all are expressed at physiologic levels within cells.
FIGURE 4.
Gon4l expressed at low levels from a stable transgene associates with endogenous Sin3a, HDAC1, and YY1. A, characterization of M12FG4L B cells, which express FLAG-Gon4l from a stably integrated transgene. M12 B cells were transfected with an expression plasmid encoding FLAG-Gon4l, and stable transfectants were obtained by drug selection, generating the M12FG4L line. M12FG4L WCLs were immunoblotted with antibodies specific for the FLAG epitope (top panel), the C terminus of Gon4l (middle panel), or β-actin (bottom panel), with the latter confirming equal protein loading in each lane. B, FLAG-Gon4l associates with endogenous Sin3a, HDAC1, and YY1. Anti-FLAG and nonspecific IgG were used to IP proteins from M12FG4L WCLs. Material recovered by IP was immunoblotted with the antibodies indicated on the right side of the panel to detect FLAG-Gon4l, Sin3a, YY1, and HDAC1. WCLs from M12FG4L and parental M12 cells were included in the immunoblot as controls. All of the data shown are representative of three independent experiments. MW, molecular mass.
Gon4l, YY1, Sin3a, and HDAC1 All Associate as Components of a Single Complex
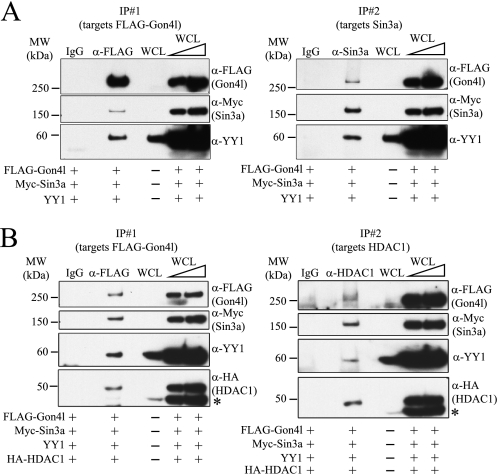
We next determined whether Gon4l could simultaneously interact with Sin3a and YY1. All three proteins were co-expressed in 293T cells, and IPs were performed using WCLs and anti-FLAG antibodies. FLAG-Gon4l, Sin3a, and YY1 were all detected in the recovered material (Fig. 5A, left panel). Sequential IP analysis was then performed: anti-FLAG antibodies were used for the first IP to capture Gon4l-containing complexes, and the recovered material was subjected to a second IP using anti-Sin3a antibodies, thus capturing Sin3a-containing complexes. Immunoblot analysis showed that Gon4l, Sin3a, and YY1 were all recovered by this procedure (Fig. 5A, right panel), demonstrating that these three proteins could interact as parts of a single complex.
FIGURE 5.
Gon4l, YY1, Sin3a, and HDAC1 can associate as parts of a single complex. 293T cells were co-transfected with the combination of expression plasmids indicated at the bottom of each panel. WCLs from transfected cells were subjected to sequential IPs. Nonspecific IgG was used to perform control IPs. Aliquots of material recovered from each IP were immunoblotted with antibodies that detect FLAG-Gon4l, Myc-Sin3a, YY1, or HA-HDAC1. The wedges at the top of the blots signify that serially increased amounts of WCLs were loaded. A, Gon4l, YY1, and Sin3a are all components of a single complex. Left panel, immunoblot analysis of proteins recovered by the first IP, which was performed using anti-FLAG antibodies. Right panel, immunoblot analysis of proteins recovered by the second IP, which was performed using anti-Sin3a antibodies. B, Gon4l, YY1, Sin3a, and HDAC1 are all components of a single complex. Left panel, immunoblot analysis of proteins recovered by the first IP, which was performed using anti-FLAG antibodies. Right panel, immunoblot analysis of proteins recovered by the second IP, which was performed using anti-HDAC1 antibodies. Immunoblotting for HA-HDAC1 was done using membranes already bound by YY1 antibodies, producing the signals marked with asterisks. All of the data shown are representative of at least three independent experiments. MW, molecular mass.
Sequential IP analysis was performed to determine whether HDAC1 could associate with the complex formed by Gon4l, YY1, and Sin3a. Expression plasmids encoding the four proteins were co-transfected into 293T cells, and sequential IPs were carried out to first isolate complexes containing Gon4l and then those containing HDAC1. All four proteins were recovered by both the first and second IPs (Fig. 5B). Similar results were obtained when anti-Sin3a antibodies were used for the second IP instead of those for HDAC1 (data not shown). These results demonstrate that HDAC1 binds to the complex formed by Gon4l, YY1, and Sin3a.
Gon4l Tethered to DNA Can Inhibit the Activity of a Nearby Promoter
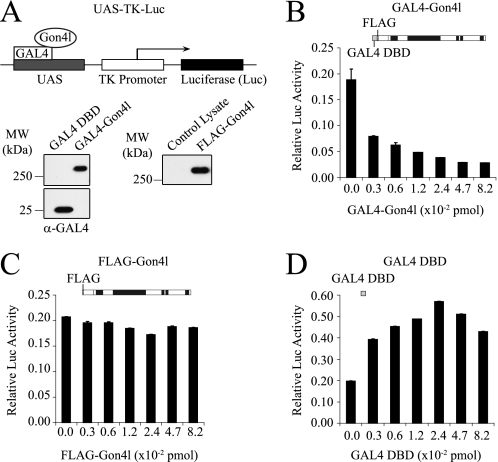
The finding that Gon4l associates with Sin3a and HDAC1 supports the conclusion that Gon4l functions in pathways that mediate gene repression. This idea is consistent with data from our previous study, which showed that the lack of Gon4l impairs repression of non-B-lineage genes in developing B cells (8). To determine whether Gon4l can mediate gene repression, we employed a reporter gene assay system in which Gon4l was directed to DNA-binding sites located upstream of a target promoter (Fig. 6A). An expression plasmid was constructed that encodes the DNA-binding domain of yeast GAL4 (GAL4-DBD) fused to the N terminus of Gon4l (GAL4-Gon4l). As controls, expression plasmids encoding the GAL4-DBD only or FLAG-Gon4l were used. Immunoblot analysis of WCLs from 293T cells transfected with these plasmids confirmed protein expression (Fig. 6A). Different amounts of each expression plasmid were transiently transfected into 293T cells together with a plasmid containing a reporter gene. This reporter gene consists of an upstream activating sequence (UAS) encoding five GAL4-binding sites upstream of a promoter element from the herpes simplex virus that drives expression of firefly luciferase (26, 27). As a control, empty expression plasmid was co-transfected with the reporter construct. A plasmid that expresses Renilla luciferase was included in all transfections. 24 h after the DNA was added, cell lysates were prepared and analyzed for firefly and Renilla luciferase activities, with the latter being used to normalize transfection efficiencies. Relative to the control, luciferase activity was decreased in a dose-dependent manner when the GAL4-Gon4l expression plasmid was co-transfected with the reporter plasmid (Fig. 6B). In contrast, co-transfection of the FLAG-Gon4l expression plasmid had essentially no effect on luciferase activity (Fig. 6C), whereas co-transfection of the GAL4-DBD only expression plasmid modestly increased luciferase activity (Fig. 6D). These data show that Gon4l bound to DNA can repress the activity of a nearby promoter.
FIGURE 6.
The Gon4l protein can repress transcription when bound to DNA near a promoter. A, schematic (upper panel) representing the luciferase reporter gene used and the GAL4-Gon4l fusion protein. FLAG-Gon4l is fused at the N terminus to the DBD of the yeast protein GAL4. Expression plasmids encoding FLAG-Gon4l or GAL4-DBD were used as controls. The UAS-TK-Luciferase reporter gene consists of a UAS encoding five tandem GAL4-binding sites upstream of the TK promoter region that drives luciferase expression. Immunoblots (lower panels) confirmed expression of the indicated proteins. B–D, 293T cells were co-transfected with the UAS-TK-Luciferase reporter construct and the indicated amounts of the relevant expression plasmids. A plasmid expressing Renilla luciferase was also co-transfected to provide a normalization control. 24 h after the addition of DNA, the cells were harvested, and the lysates were analyzed for firefly and Renilla luciferase activities. Graphs show the normalized luciferase activity (y axis) expressed from the reporter plasmid when co-transfected with the indicated amount of expression plasmid (x axis): GAL4-Gon4l (B), FLAG-Gon4l (C), or GAL4-DBD (D). The values shown are the averages and standard deviations of triplicate samples after normalization to Renilla luciferase activity. All of the plots are representative of data obtained from at least three independent experiments. MW, molecular mass.
The YY1AP Homology Region of Gon4l Mediates Both Transcriptional Repression and Interaction with YY1, Sin3a, and HDAC1
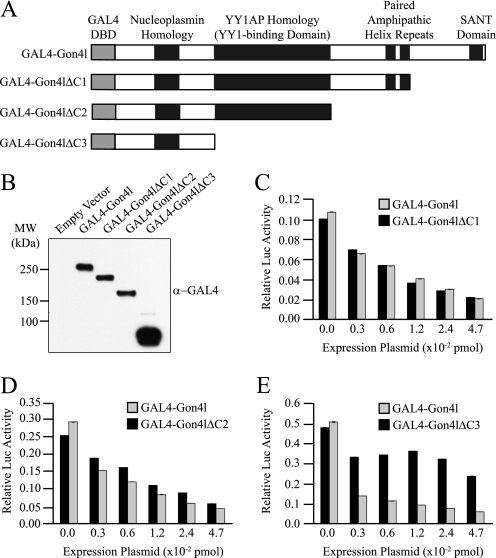
To determine the regions of Gon4l required to repress reporter gene activity, we constructed expression plasmids encoding Gon4l C-terminal truncation mutants fused to the GAL4-DBD (Fig. 7A). Expression of the encoded proteins in transiently transfected 293T cells was confirmed by immunoblot (Fig. 7B). The reporter gene assay showed that luciferase activity was decreased when the reporter construct was co-transfected with an expression plasmid encoding either a GAL4-Gon4l fusion protein lacking sequences encoding the SANT domain or one lacking those spanning both the SANT domain and PAH repeats (Fig. 7, C and D). In contrast, co-transfection of the reporter gene with an expression plasmid encoding a GAL4-Gon4l fusion lacking sequences spanning the YY1AP homology region did not reduce luciferase activity (Fig. 7E), despite greater expression of this protein relative to the other GAL4-Gon4l fusion proteins (Fig. 7B). These data indicate that transcriptional repression by Gon4l requires the region spanning the YY1AP homology.
FIGURE 7.
The YY1AP homology domain of Gon4l is required for transcriptional repression. A, schematic showing the structures of GAL4 DBD-Gon4l fusion proteins used for the experiments shown in B–E. B, immunoblot analysis of whole cell lysates from 293T cells transfected with the indicated GAL4 DBD-Gon4l expression plasmids. Anti-GAL4 antibodies were used for the immunoblot. C–E, 293T cells were co-transfected with the pUAS-TK-Luciferase reporter construct and the indicated amounts of GAL4 DBD-Gon4l expression plasmids. A plasmid expressing Renilla luciferase was also co-transfected to provide a normalization control. Graphs show the normalized luciferase activity (y axis) expressed from the reporter plasmid when co-transfected with the indicated amount of GAL4-Gon4l expression plasmid (x axis). All of the plots are representative of data obtained from at least three independent experiments. MW, molecular mass.
To correlate these results with the ability to interact with Sin3a and HDAC1, expression plasmids encoding the Gon4l C-terminal truncation mutants fused to the FLAG epitope tag were generated and used for IP studies (Fig. 8A). Each of these proteins was expressed in 293T cells together with Sin3a, HDAC1, and YY1 (Fig. 8B, left panel). WCLs from these cells were used to conduct IPs with control or FLAG antibodies. Recovered material was probed for the presence of Myc-Sin3a, YY1, and HA-HDAC1 (Fig. 8B, right panel). The lack of amino acid sequences spanning the SANT domain and the two PAH repeats had no obvious effect on interactions between Gon4l and the other proteins. However, the absence of the YY1AP homology region in Gon4l prevented association with Sin3a and HDAC1 as well as that with YY1. Thus, the YY1AP homology region in Gon4l is required for both transcriptional repression and association with Sin3a and HDAC1. These data support the conclusion that repression of promoter activity by Gon4l involves interaction with Sin3a and HDAC1.
DISCUSSION
Gon4l orthologs are found throughout the phylogeny of multicellular organisms (31). Studies of mutations in invertebrate and vertebrate species have demonstrated that Gon4l is important for several developmental processes, including somite formation, erythropoiesis, and B lymphopoiesis (8–13, 36). Further, these studies have provided evidence that Gon4l functions to regulate gene expression during these developmental processes, yet nothing is known about the molecular function of Gon4l. This report provides data supporting the conclusion that Gon4l regulates gene expression by associating with YY1, Sin3a, and HDAC1.
The central region of Gon4l bears strong homology to the human protein YY1AP, which interacts with the zinc finger transcription factor YY1 (30). Similar to what has been reported for YY1AP, we found Gon4l and YY1 co-immunoprecipitated and co-localized in the nucleus. These data suggest that interaction with YY1 is one mechanism by which Gon4l and associated factors are recruited to target genes. In addition, our data demonstrate that Gon4l, YY1, and Sin3a/HDAC1 form a complex, which supports the idea that Gon4l functions in pathways that mediate gene repression, as suggested by our previous study (8). As components of a Gon4l-containing complex, YY1 and Sin3a/HDAC1 (and perhaps other factors that interact with Sin3a) could serve DNA binding and catalytic functions, respectively, resulting in repression of gene expression. Consistent with this idea, our data demonstrate that Gon4l bound to DNA can repress the activity of a nearby promoter. Notably, we found that this activity and the ability to interact with YY1, Sin3a, and HDAC1 both require the highly conserved YY1AP homology in Gon4l. These data suggest that the YY1AP homology domain of Gon4l mediates the assembly of functionally important complexes that, at least in some cases, contain Sin3a and HDAC1.
Our data show that co-IP of YY1 with Gon4l is more difficult to detect than Sin3a/HDAC1. Although multiple explanations can be imagined, this result is nonetheless consistent with the possibility that the extent of constitutive interaction between YY1 and Gon4l is modest. This could reflect competition between YY1 and other DNA-binding proteins for interaction with Gon4l-containing complexes. This idea is appealing given the apparent central role of Gon4l in regulating B cell development (8). This scenario would also be consistent with studies showing that Sin3a, and thus likely complexes containing Sin3a, can interact with several different DNA-binding proteins (32).
Functional roles for the SANT domain or PAH repeats in Gon4l were not revealed by our study. However, it seems highly likely that these domains are important for Gon4l function. Indeed, it has been demonstrated that sequences spanning the SANT domain in Gon4l are required to rescue erythropoiesis in Gon4l mutant zebrafish (11). The PAH repeats in the Sin3 proteins mediate interaction with transcription factors or with non-DNA-binding co-factors (32, 37). Thus, the association of Gon4l-containing complexes with DNA may in some cases depend on interactions mediated by the PAH repeats (38, 39). The importance of such interactions would likely depend on the promoter structure of a given target gene. In addition, drawing from the examples of Sin3a and Sin3b, the presence of two PAH repeats in Gon4l suggests the ability to interact with other proteins in addition to YY1 and Sin3a/HDAC1. In support of this conclusion, Gon4l was detected in gradient fractions containing complexes of at least 670 kDa in mass, which is larger than the total molecular mass of Gon4l, YY1, and Sin3a/HDAC1 combined (∼520 kDa). Thus, it is likely that other co-factors for Gon4l remain to be identified. Of note in this regard, a recent report provided evidence that the PAH repeats and SANT domain of zebrafish Gon4l mediate interaction with the proteins MCM3 and MCM4 (12), which are important for DNA replication but likely function in additional pathways (40).
Our findings suggest that Gon4l functions by collaborating with YY1 and Sin3a/HDAC1. YY1 is a highly conserved member of the polycomb group protein family that mediates transcriptional activation and repression (41). Several studies have demonstrated that Sin3a functions as a transcriptional co-repressor (32). However, both of these proteins likely have roles in additional pathways, including those that control cell division, genome stability, and DNA repair (42–45). Further, analysis of mice engineered to lack YY1 in B cell progenitors has demonstrated that YY1 has a role in immunoglobulin gene rearrangement (36), a process that requires DNA repair. With regard to Gon4l, studies of zebrafish bearing null mutations in the Gon4l ortholog ugly duckling have provided evidence that this protein is important for cell division and genome stability in addition to the regulation of gene expression (11, 12). These findings, combined with our observations linking Gon4l to Sin3a and YY1, suggest that Gon4l may be important for pathways in addition to those that mediate gene repression. Thus, further characterization of the functional relationships between Gon4l, YY1, and Sin3a could not only increase our understanding of how gene repression is enforced but may also provide novel insights regarding mechanisms that regulate cell proliferation and genome integrity.
Supplementary Material
Acknowledgments
We thank Dr. Peng Jiang for assistance with bioinformatic analysis, Drs. Christopher Glass and Donald Ayer for providing plasmids, Dr. Peter Mohler for use of the sucrose gradient maker and helpful advice, Dr. Curt Sigmund for use of the luminometer, and the University of Iowa Central Microscope Facility for help with performing immunofluorescent confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI067489 (to J. D. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- HDAC
- histone deacetylase
- DBD
- DNA-binding domain
- IP
- immunoprecipitation
- PAH
- paired amphipathic helix
- WCL
- whole cell lysate
- UAS
- upstream activating sequence.
REFERENCES
- 1. Hardy R. R., Kincade P. W., Dorshkind K. (2007) Immunity 26, 703–714 [DOI] [PubMed] [Google Scholar]
- 2. Hoffmann R., Seidl T., Neeb M., Rolink A., Melchers F. (2002) Genome Res. 12, 98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rumfelt L. L., Zhou Y., Rowley B. M., Shinton S. A., Hardy R. R. (2006) J. Exp. Med. 203, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hystad M. E., Myklebust J. H., Bø T. H., Sivertsen E. A., Rian E., Forfang L., Munthe E., Rosenwald A., Chiorazzi M., Jonassen I., Staudt L. M., Smeland E. B. (2007) J. Immunol. 179, 3662–3671 [DOI] [PubMed] [Google Scholar]
- 5. Laiosa C. V., Stadtfeld M., Graf T. (2006) Annu. Rev. Immunol. 24, 705–738 [DOI] [PubMed] [Google Scholar]
- 6. Jung D., Giallourakis C., Mostoslavsky R., Alt F. W. (2006) Annu. Rev. Immunol. 24, 541–570 [DOI] [PubMed] [Google Scholar]
- 7. Sen R., Oltz E. (2006) Curr. Opin. Immunol. 18, 237–242 [DOI] [PubMed] [Google Scholar]
- 8. Lu P., Hankel I. L., Knisz J., Marquardt A., Chiang M. Y., Grosse J., Constien R., Meyer T., Schroeder A., Zeitlmann L., Al-Alem U., Friedman A. D., Elliott E. I., Meyerholz D. K., Waldschmidt T. J., Rothman P. B., Colgan J. D. (2010) J. Exp. Med. 207, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedman L., Santa Anna-Arriola S., Hodgkin J., Kimble J. (2000) Dev. Biol. 228, 350–362 [DOI] [PubMed] [Google Scholar]
- 10. Hammerschmidt M., Pelegri F., Mullins M. C., Kane D. A., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C. P., Jiang Y. J., Kelsh R. N., Odenthal J., Warga R. M., Nüsslein-Volhard C. (1996) Development 123, 143–151 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., Du L., Osato M., Teo E. H., Qian F., Jin H., Zhen F., Xu J., Guo L., Huang H., Chen J., Geisler R., Jiang Y. J., Peng J., Wen Z. (2007) Blood 110, 99–106 [DOI] [PubMed] [Google Scholar]
- 12. Lim C. H., Chong S. W., Jiang Y. J. (2009) Mol. Biol. Cell 20, 4183–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bulchand S., Menon S. D., George S. E., Chia W. (2010) J. Cell Sci. 123, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 14. Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Letunic I., Doerks T., Bork P. (2009) Nucleic Acids Res. 37, D229–D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamano T., Kim K. J., Leiserson W. M., Asofsky R. (1982) J. Immunol. 129, 1403–1406 [PubMed] [Google Scholar]
- 17. Davidson W. F., Pierce J. H., Rudikoff S., Morse H. C., 3rd. (1988) J. Exp. Med. 168, 389–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alt F., Rosenberg N., Lewis S., Thomas E., Baltimore D. (1981) Cell 27, 381–390 [DOI] [PubMed] [Google Scholar]
- 19. Siden E. J., Baltimore D., Clark D., Rosenberg N. E. (1979) Cell 16, 389–396 [DOI] [PubMed] [Google Scholar]
- 20. Rowland S. L., Tremblay M. M., Ellison J. M., Stunz L. L., Bishop G. A., Hostager B. S. (2007) J. Immunol. 179, 4645–4653 [DOI] [PubMed] [Google Scholar]
- 21. Mizushima S., Nagata S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hostager B. S., Haxhinasto S. A., Rowland S. L., Bishop G. A. (2003) J. Biol. Chem. 278, 45382–45390 [DOI] [PubMed] [Google Scholar]
- 23. Fleischer T. C., Yun U. J., Ayer D. E. (2003) Mol. Cell. Biol. 23, 3456–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lutterbach B., Westendorf J. J., Linggi B., Patten A., Moniwa M., Davie J. R., Huynh K. D., Bardwell V. J., Lavinsky R. M., Rosenfeld M. G., Glass C., Seto E., Hiebert S. W. (1998) Mol. Cell. Biol. 18, 7176–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinzel T., Lavinsky R. M., Mullen T. M., Söderstrom M., Laherty C. D., Torchia J., Yang W. M., Brard G., Ngo S. D., Davie J. R., Seto E., Eisenman R. N., Rose D. W., Glass C. K., Rosenfeld M. G. (1997) Nature 387, 43–48 [DOI] [PubMed] [Google Scholar]
- 27. Chen J. D., Evans R. M. (1995) Nature 377, 454–457 [DOI] [PubMed] [Google Scholar]
- 28. Ohtomo T., Horii T., Nomizu M., Suga T., Yamada J. (2007) Amino Acids 33, 645–652 [DOI] [PubMed] [Google Scholar]
- 29. Frehlick L. J., Eirín-López J. M., Ausió J. (2007) Bioessays 29, 49–59 [DOI] [PubMed] [Google Scholar]
- 30. Wang C. Y., Liang Y. J., Lin Y. S., Shih H. M., Jou Y. S., Yu W. C. (2004) J. Biol. Chem. 279, 17750–17755 [DOI] [PubMed] [Google Scholar]
- 31. Kuryshev V. Y., Vorobyov E., Zink D., Schmitz J., Rozhdestvensky T. S., Münstermann E., Ernst U., Wellenreuther R., Moosmayer P., Bechtel S., Schupp I., Horst J., Korn B., Poustka A., Wiemann S. (2006) Genomics 88, 143–151 [DOI] [PubMed] [Google Scholar]
- 32. Grzenda A., Lomberk G., Zhang J. S., Urrutia R. (2009) Biochim. Biophys. Acta 1789, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boyer L. A., Latek R. R., Peterson C. L. (2004) Nat. Rev. Mol. Cell Biol. 5, 158–163 [DOI] [PubMed] [Google Scholar]
- 34. Mo X., Kowenz-Leutz E., Laumonnier Y., Xu H., Leutz A. (2005) Genes Dev. 19, 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palko L., Bass H. W., Beyrouthy M. J., Hurt M. M. (2004) J. Cell Sci. 117, 465–476 [DOI] [PubMed] [Google Scholar]
- 36. Liu H., Schmidt-Supprian M., Shi Y., Hobeika E., Barteneva N., Jumaa H., Pelanda R., Reth M., Skok J., Rajewsky K., Shi Y. (2007) Genes Dev. 21, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Silverstein R. A., Ekwall K. (2005) Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 38. Guenther M. G., Barak O., Lazar M. A. (2001) Mol. Cell. Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de la Cruz X., Lois S., Sánchez-Molina S., Martínez-Balbás M. A. (2005) Bioessays 27, 164–175 [DOI] [PubMed] [Google Scholar]
- 40. Forsburg S. L. (2004) Microbiol. Mol. Biol. Rev. 68, 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gordon S., Akopyan G., Garban H., Bonavida B. (2006) Oncogene 25, 1125–1142 [DOI] [PubMed] [Google Scholar]
- 42. Wu S., Shi Y., Mulligan P., Gay F., Landry J., Liu H., Lu J., Qi H. H., Wang W., Nickoloff J. A., Wu C., Shi Y. (2007) Nat. Struct. Mol. Biol. 14, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu S., Hu Y. C., Liu H., Shi Y. (2009) Mol. Cell. Biol. 29, 6245–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jazayeri A., McAinsh A. D., Jackson S. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1644–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dannenberg J. H., David G., Zhong S., van der Torre J., Wong W. H., Depinho R. A. (2005) Genes Dev. 19, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.