Abstract
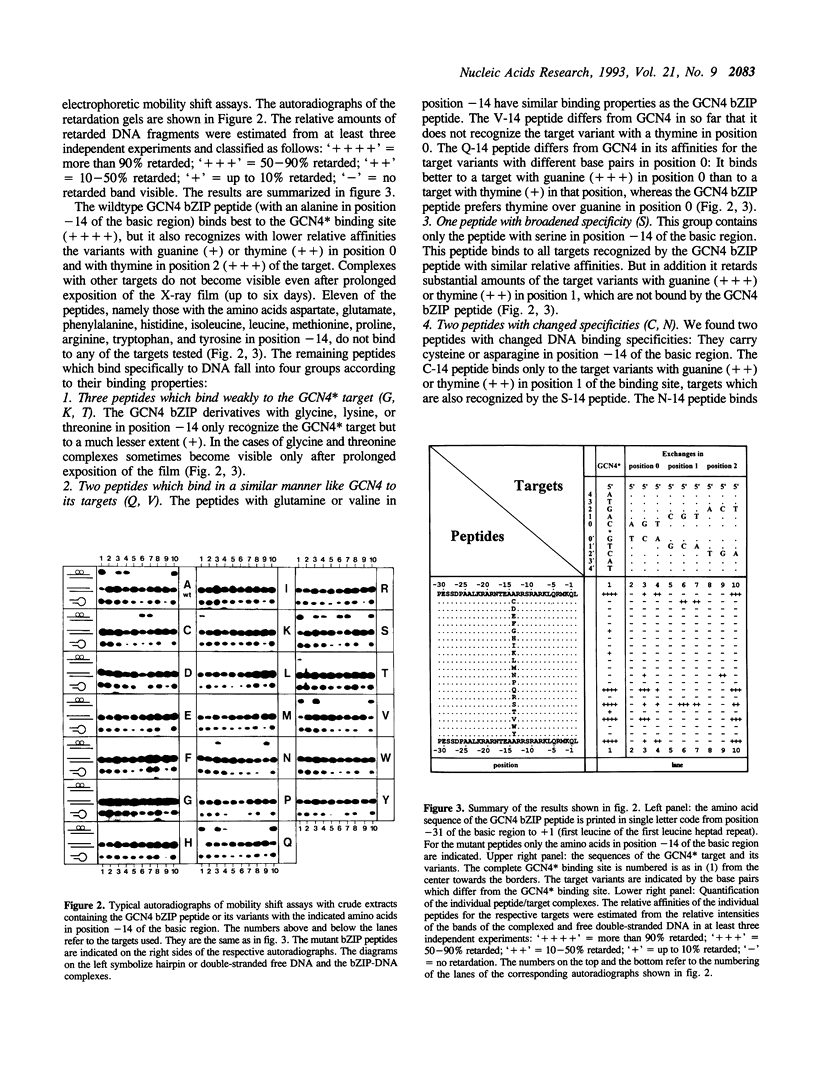
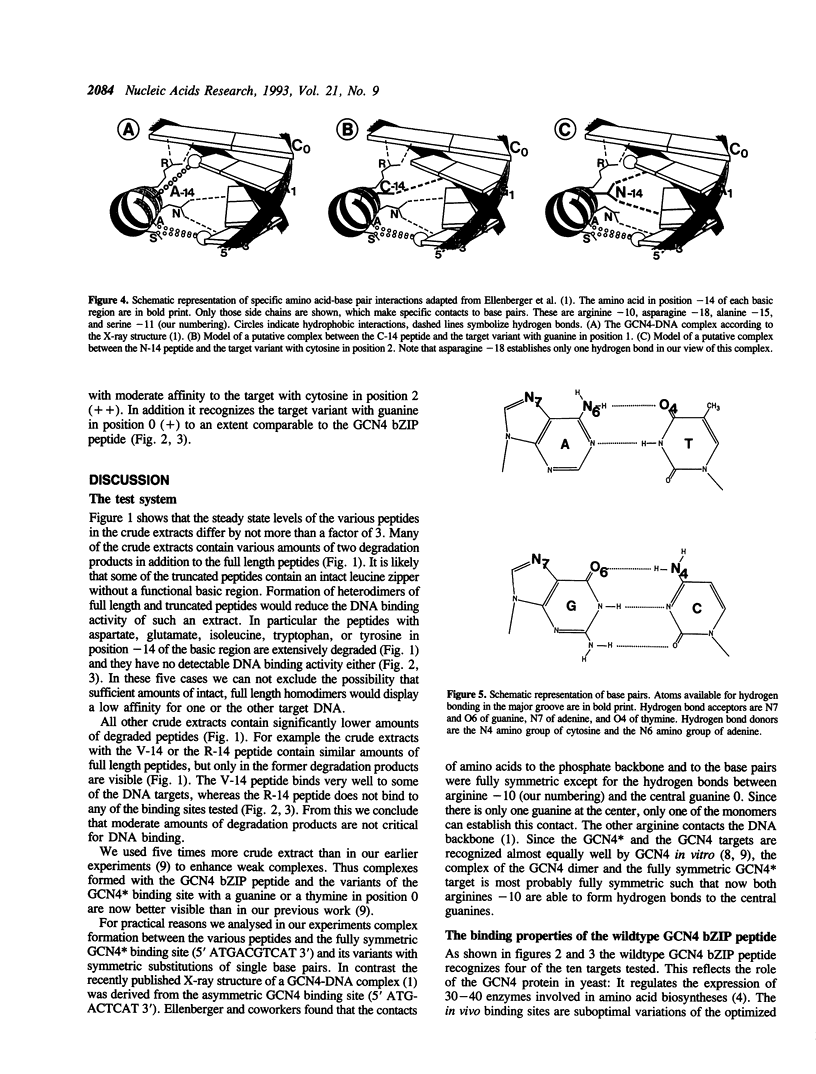
The X-ray structure of a GCN4 DNA complex (1) shows, that specific DNA binding of the GCN4 basic region is mediated by a complicated network of base pair and DNA backbone contacts. According to the X-ray structure, alanine -14 of the basic region of GCN4 (we define the first leucine of the leucine zipper as +1) makes a hydrophobic contact to the methyl group of the thymine next to the center of the GCN4 binding site 5' ATGACTCAT 3'. We tested the DNA binding properties of the nineteen derivatives of GCN4, which carry all possible amino acids in position -14 of the basic region. Substitution of alanine -14 of GCN4 by either asparagine or cysteine changes the DNA binding specificity. Serine in this position broadens the specificity for position 1 of the target, whereas other amino acids either retain or decrease GCN4 specificity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agre P., Johnson P. F., McKnight S. L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989 Nov 17;246(4932):922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- Busch S. J., Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990 Feb;6(2):36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Involvement of an initiation factor and protein phosphorylation in translational control of GCN4 mRNA. Trends Biochem Sci. 1990 Apr;15(4):148–152. doi: 10.1016/0968-0004(90)90215-w. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Kanaan M. N., Marzluf G. A. Mutational analysis of the DNA-binding domain of the CYS3 regulatory protein of Neurospora crassa. Mol Cell Biol. 1991 Sep;11(9):4356–4362. doi: 10.1128/mcb.11.9.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature. 1989 Aug 17;340(6234):568–571. doi: 10.1038/340568a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley W. S., Harshman K. D., Parker C. S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989 Mar;3(3):283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- Oas T. G., McIntosh L. P., O'Shea E. K., Dahlquist F. W., Kim P. S. Secondary structure of a leucine zipper determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1990 Mar 27;29(12):2891–2894. doi: 10.1021/bi00464a001. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Brandl C. J., Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989 Jul;9(7):2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sellers J. W., Vincent A. C., Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990 Oct;10(10):5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]




