Abstract
Receptor tyrosine kinases are critical targets for the regulation of cell survival. Cancer patients with abnormal receptor tyrosine kinases (RTK) tend to have more aggressive disease with poor clinical outcomes. As a result, human epidermal growth factor receptor kinases, such as EGFR (HER1), HER2, and HER3, represent important therapeutic targets. Several plant polyphenols including the type III polyketide synthase products (genistein, curcumin, resveratrol, and epigallocatechin-3-galate) possess chemopreventive activity, primarily as a result of RTK inhibition. However, only a small fraction of the polyphenolic structural universe has been evaluated. Along these lines, we have developed an in vitro route to the synthesis and subsequent screening of unnatural polyketide analogs with N-acetylcysteamine (SNAc) starter substrates and malonyl-coenzyme A (CoA) and methylmalonyl-CoA as extender substrates. The resulting polyketide analogs possessed a similar strucutral polyketide backbone (aromatic-2-pyrone) with variable side chains. Screening chalcone synthase (CHS) reaction products against BT-474 cells resulted in identification of several trifluoromethylcinnamoyl-based polyketides that showed strong suppression of the HER2-associated PI3K/AKT signaling pathway, yet did not inhibit the growth of nontransformed MCF-10A breast cells (IC50 > 100 µm). Specifically, 4-trifluoromethylcinnamoyl pyrone (compound 2e) was highly potent (IC50 < 200 nm) among the test compounds toward proliferation of several breast cancer cell lines. This breadth of activity likely stems from the ability of compound 2e to inhibit the phosphorylation of HER1, HER2, and HER3. Therefore, these polyketide analogs might prove to be useful drug candidates for potential breast cancer therapy.
Keywords: chalcone synthase, inhibitors, N-acetylcysteamine (SNAc) precursors, polyketides, tyrosine kinases
Introduction
The human epidermal growth factor receptor (HER) family, which includes EGFR (HER1), and HER2, and HER3 serve as important targets for selective cancer therapy.[1] Overexpression of HER2 is found in ~30% of human breast cancers and is associated with more aggressive and drug resistant tumors.[2] Dimerization of the HER receptors (HER1/HER2, HER2/HER2, and HER2/HER3) induces autophosphorylation of specific tyrosine residues within the cytoplasmic catalytic kinase domain of the activated receptor.[3] These tyrosine autophosphorylation residues activate downstream cell proliferation (mitogen-activated protein kinase, MAPK) and the survival (phosphatidylinnositol-3 kinase, PI3K/Akt[4]) signaling pathway. HER2 is also the preferred heterodimeric partner with other HER receptors. Thus, HER2 is an attractive target for cancer drug development.
Several small molecule tyrosine kinase (TK) inhibitors, such as lapatinib, gefitinib, and erlotinib, which target the intracellular kinase domain of the HER receptor, are clinically available.[5] However, these compounds have toxic side effects, including skin rash, diarrhea, and thrombocytopenia.[6] In addition, in some lung cancer patients treated with gefitinib and erlotinib, somatic mutations were observed in the EGFR kinase domain;[7] this resulted in altered drug sensitivity and increased drug resistance.[8] Thus, development of safer, yet highly potent and selective TK inhibitors is needed.
Products of the type III polyketide synthases, including polyphenols such as catechins, genistein, resveratrol, and curcumin, possess anticancer and antiangiogenic activity, are capable of stimulating apoptosis in cancer cells, and appear to serve as chemopreventive agents against oxidative damage in normal cells.[9] The diverse biological activity of polyphenolic-based polyketides relates to the ability of these structurally complex molecules to activate or inhibit one or more molecular targets that are critical in cancer cell proliferation and/or metastasis.[9] In addition, these natural products are generally nontoxic and have been used clinically for targeting HER2,[10] cyclooxygenase-2,[11] nuclear factor κB (NF-κB) transcription factor,[12] protein kinases (e.g., Akt and MAPKs),[13] Bcl-2,[14] among others.
Despite the large number of polyphenols that exist in nature, only a small fraction of the polyketide and polyphenolic structural universe can be tapped because of difficulties in isolation of these compounds. Diversity-oriented synthesis (DOS) of natural product-like polyphenolic compounds[15] and de novo polyphenol synthesis by plant-based type III polyketide synthases (PKS) are possible alternative routes to the generation of novel polyphenols. Although DOS has great potential to generate diverse natural product-like architectures, multiple steps are required for polyketide chain elongation and cyclization,[16] whereas, type III PKS alone is capable of performing both steps. Exploiting these PKS to synthesize natural and unnatural polyphenols is possible and might be expected to yield unexplored structurally diverse natural products.[17] Many secondary metabolism pathways serve as excellent candidates for this strategy, including polyketides, isoprenoids, lignans, etc., and rapid microarray-based screens can be developed based on key targets, e.g., caspase family enzymes, kinases, phosphatases, histone deacetylases, as well as cell-based assays.[18] The polyketide synthases are particularly well-suited for in vitro manipulation. The type III PKS are fundamentally the simplest among the three classes of PKS, and are comprised of homodimeric enzymes that catalyze decarboxylative condensation of malonyl-CoA or methylmalonyl-CoA onto a wide range of starter-CoA substrates. In nature, these enzymes catalyze the synthesis of a diverse array of flavonoids, chalcones, stilbenes, and together with other enzymes, a large number of hybrid molecules with activities as varied as anti-infectives to anticancer agents.[19] Moreover, examples of natural post-PKS tailoring reactions are known,[20] and our previous efforts have led to hybrid natural-unnatural polyketide synthesis through post-PKS tailoring reactions.[18a]
Chalcone synthase (CHS) is the key enzyme involved in polyphenol biosynthesis and possesses a very wide substrate specificity,[21] and utilizes both aromatic and aliphatic CoA esters as substrates.[22] The natural function of CHS is to initiate polyphenol biosynthesis through sequential Claisen condensation of coumaroyl-CoA as a starter substrate with malonyl-CoA as a chain extender substrate. After three rounds of chain elongation, the resulting linear tetraketide intermediate is cyclized to form the chalcone naringenin. In some cases, immature polyphenols are produced through truncated condensation reactions to yield triketide and tetraketide products that are then cyclized to produce coumaroyl-4-hydroxy-2-pyrones.[20] CHS possesses a high degree of substrate promiscuity, including both aromatic and aliphatic CoA esters as substrates,[22] albeit far less promiscuity is evident for the extender substrate.[21, 23] We reasoned that such broad specificity could be used to facilitate the discovery of new molecular entities with potent biological activities.
In the current work, we used an in vitro precursor-directed synthesis strategy employing CHS with a range of structurally diverse aromatic starter substrates to generate compounds that possessed nanomolar activity against proliferation of several HER2 overexpressing breast cancer cell lines. Importantly, these compounds were inactive against a nontumorigenic breast cell line (MCF-10A), thereby providing highly selective and potent biological activity with potential therapeutic applications.
Results and Discussion
In Vitro Synthesis of Unnatural Polyketides
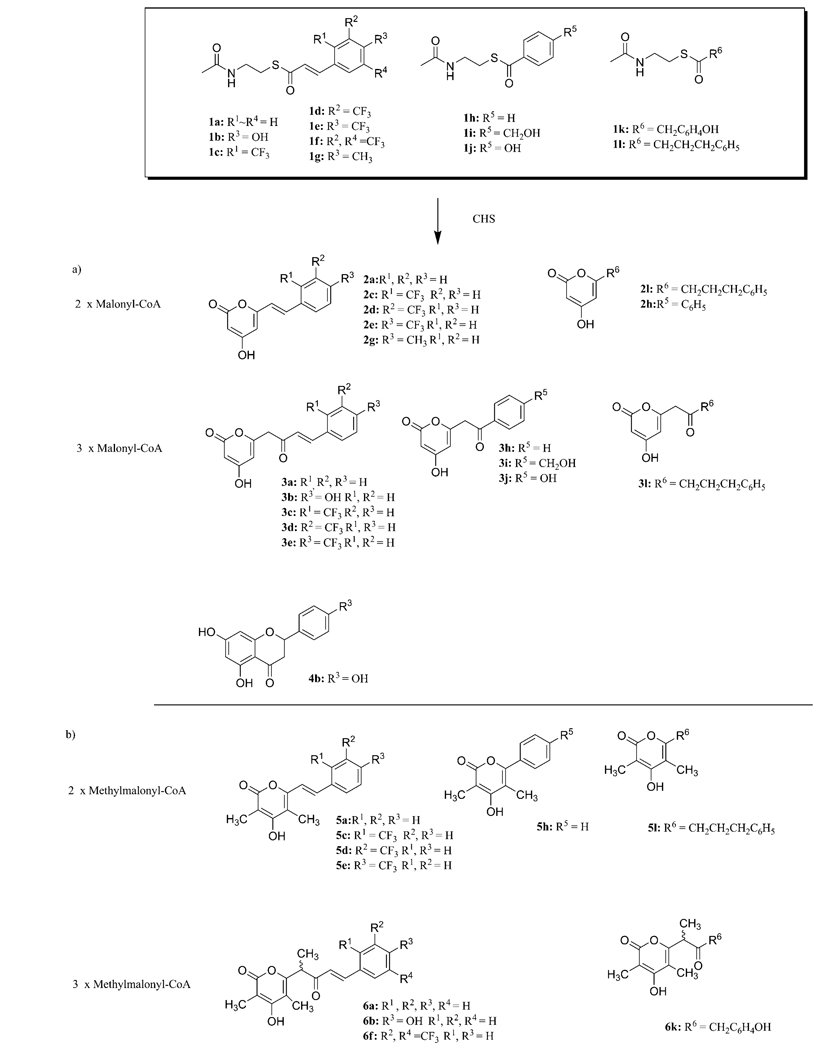
To obtain novel (unnatural) polyphenols derived from Type III PKS, we performed in vitro precursor-directed synthesis with CHS from alfalfa (Medico sativa) and various N-acetylcysteamine (SNAc) thioester starter substrates (Figure 1S), with malonyl-CoA or methylmalonyl-CoA serving as extender substrates (Scheme 1). To ensure that CHS reacted with SNAc esters in a manner similar to the respective CoA esters, a standard reaction was performed by using p-coumaryl-SNAc and malonyl-CoA as starter and extender substrates, respectively. HPLC-MS/MS measurement of the reaction mixture resulted in two peaks with parent ions [M+H]+ at m/z 273 and [M−H]− at m/z 271, indicating the successive condensation of three units of malonyl-CoA. Based on MS/MS-ESI fragmentation analysis, one fragment at m/z 227 corresponded to [M−H−CO2]−, and this indicates the presence of an α-pyrone ring (3b), which was terminated without aromatic ring formation. The second peak gave a similar UV spectrum (λmax 296 nm) and mass spectrum identical to the expected product naringenin (4b). Thus, the SNAc starter substrate was effectively used by CHS to generate the natural product of the reaction.
Scheme 1.
CHS-catalyzed synthesis of unnatural polyphenols from cinnamoyl-SNAc starter substrates with a) malonyl-CoA and b) methylmalonyl-CoA as extender units. Isolated yields of compounds 2–6 ranged from 5–17 %.
Encouraged by this result, we proceeded to perform CHS catalysis on a range of starter substrates with both malonyl-CoA and methylmalonyl-CoA as extender substrates (Scheme 1). The products from 21 enzymatic reactions were analyzed by HPLC-MS, and this resulted in the identification of a large number of structurally diverse products, including tri- and tetraketide pyrones and fully formed flavonoids. 1H and 13C NMR analyses were performed on seven of these products, thus confirming the MS identification.
Inhibitory Effect of CHS Reaction Products on the HER2-PI3K-Akt Signaling Pathway
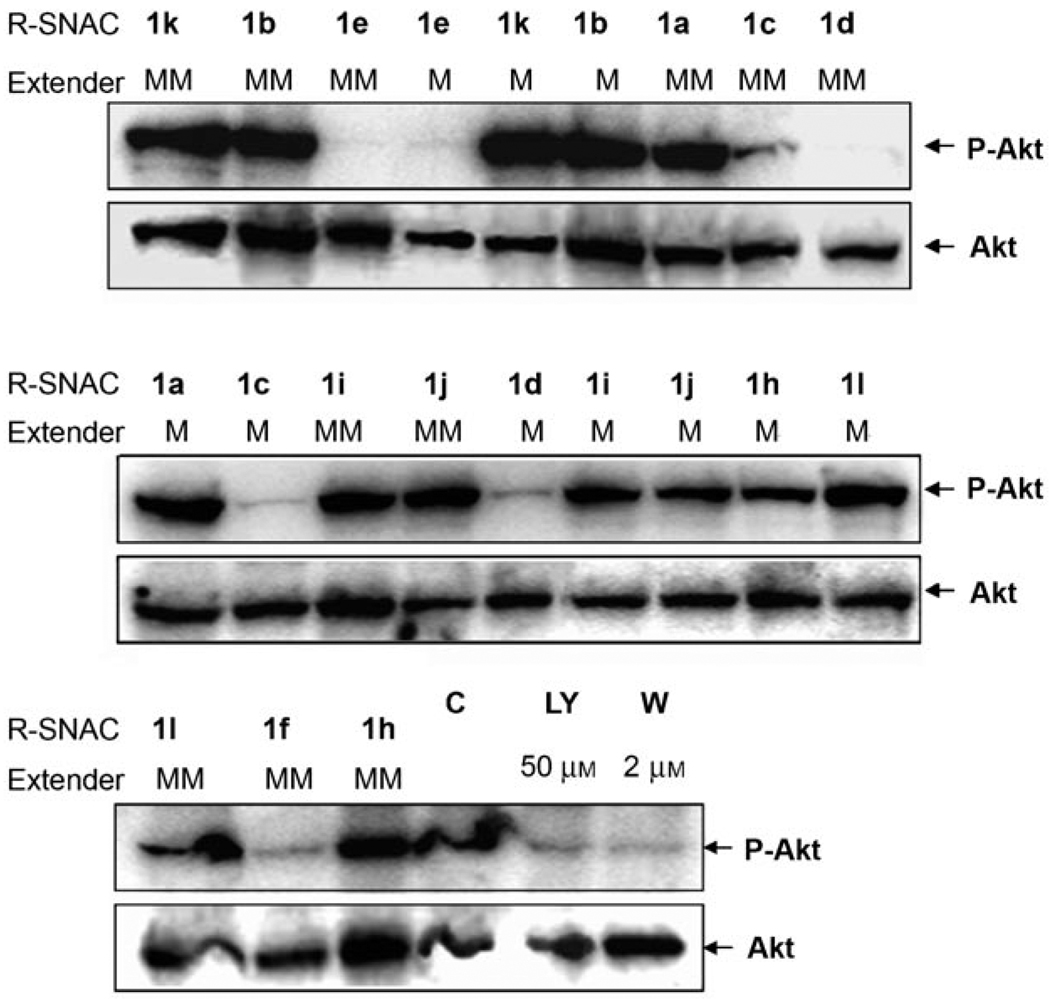
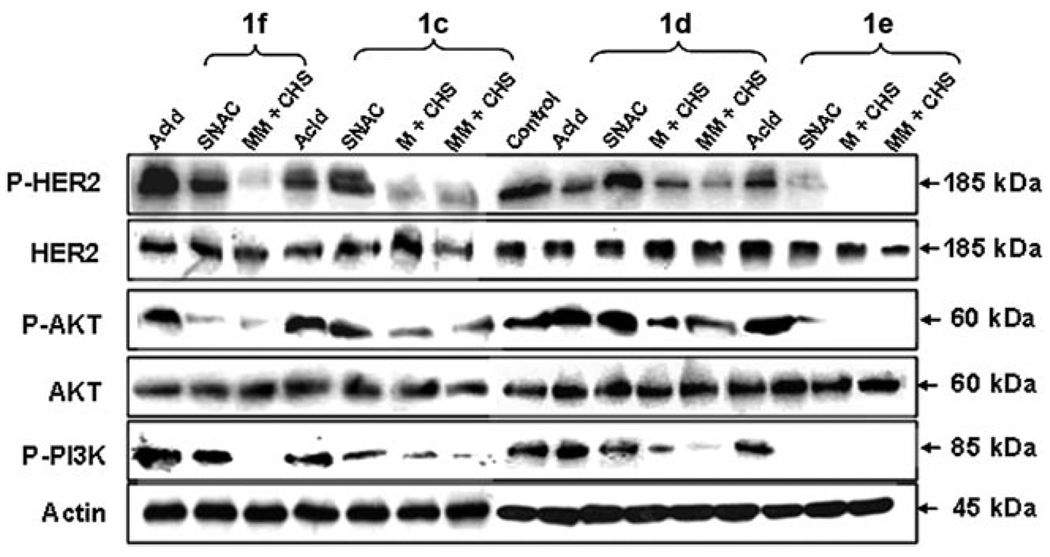
CHS reaction products were then extracted from the reaction mixtures into ethyl acetate and evaluated as inhibitors of the HER2-associated PI3K/Akt signaling pathway. HER2-overexpressing BT-474 cells[24] were then used to investigate whether the polyketide products suppressed HER2 signaling. The extracted reaction mixtures were redissolved in DMSO. The resulting product mixtures were used in 2D BT-474 cell culture assays in 6-well plates. The inhibitory activity of these product mixtures were assessed against the HER2-PI3K-Akt signaling pathway (Figure 1 and 2). As a negative control, the vehicle containing 1% (v/v) DMSO was used and it showed the expected high levels of phospho-Akt (p-Akt), which is the downstream output of HER2 activation; thus, this demonstrated the active PI3K-Akt signaling cascade. Positive controls consisted of 2 µm wortmannin and 50 µm LY-294002, which are known irreversible PI3K inhibitors.[25] The reaction mixtures derived from 2-, 3-, and 4-trifluoromethylcinnamoyl-SNAc starter substrates (1c–1f) appeared to strongly inhibit formation of phospho-HER2 (p-HER2), which shuts down the downstream kinase signaling pathway such that negligible phosphorylation of PI3K and Akt occur (Figure 3). This inhibition did not take place with 50 µm of the free cinnamic acid derivatives, although some activity was observed with 50 µm of the 3,5-bis(trifluoromethyl)cinnamoyl-SNAc (compound 1f) and 2-trifluoromethylcinnamoyl-SNAc (compound 1c) starter substrates (reduced levels of p-Akt generated). Therefore, only in the presence of the full CHS product mixtures was there significant inhibitory activity. These results were not influenced by the effects of cell concentration, as the actin signal remained relatively constant (Figure 3).
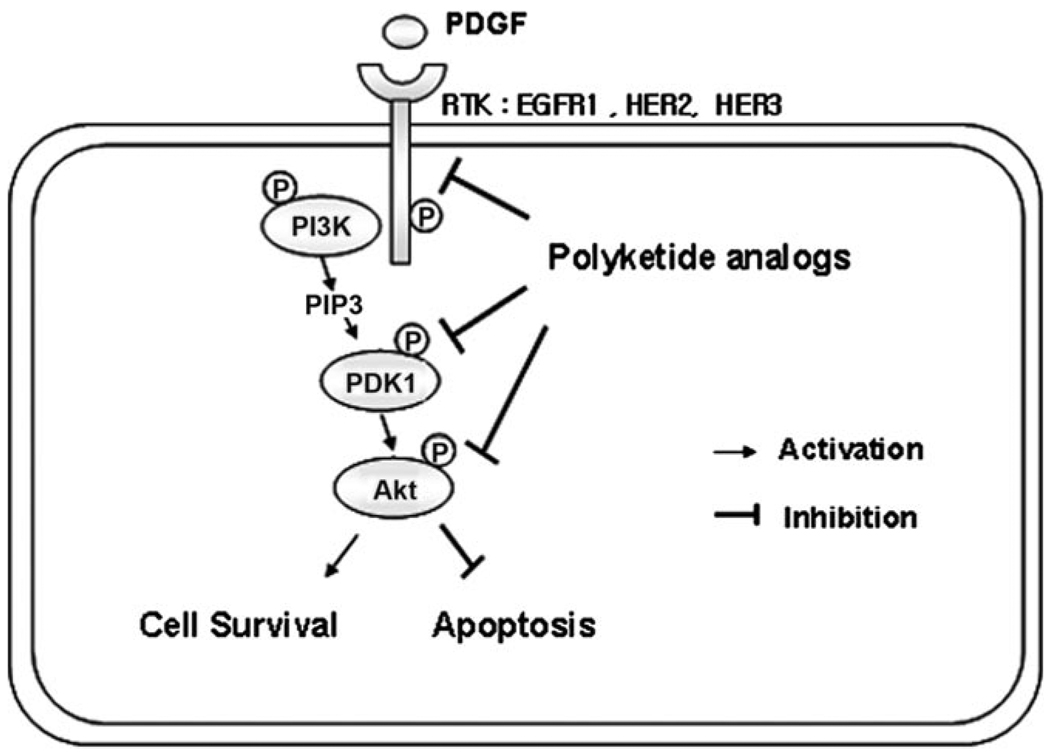
Figure 1.
Schematic representation of HER2-mediated PI3K-Akt signaling pathway. Activation of HER2 initiates the activation of the cytoplasmic tyrosine kinase domain and subsequent events to activate downstream of PI3K and Akt. The activation of Akt suppresses apoptosis and stimulates the cell survival pathway.
Figure 2.
Effect of precursor-directed CHS reaction extracts on the level of phospho-Akt in HER2-overexpressing human breast cancer BT-474 cells in 6-well plate 2D cultures. Abbreviation: C, control (without treatment); LY, LY294001; W, wortmannin; M, malonyl-CoA; MM, methylmalonyl-CoA. Whole-cell lysates were subjected to SDS-PAGE analysis and probed with anti-phospho-Akt (Ser473) and anti-Akt antibody, respectively.
Figure 3.
Effect of trifluoromethylcinnamic acid-based compounds (corresponding acids, SNAc esters, and their CHS reaction extracts) on HER2-mediated PI3K-Akt signaling pathway. The BT-474 cells, which are confluent in 6-well plates (3 mL), were treated for 3 h with different trifluoromethylcinnamic acids (50 µm), trifluoromethylcinnamoyl-SNAc esters (50 µm), and their CHS reaction extracts (10 µL). Whole-cell lysates were subjected to SDS-PAGE analysis and probed with anti-phospho-HER2 (Tyr1221/1222), anti-HER2, anti-phospho-Akt (Ser473), anti-Akt antibody, anti-phospho-PI3K (Tyr508), and anti-actin, respectively.
High-Yield Production of Bioactive Polyketide Analogs using Immobilized CHS
The reaction mixtures from the trifluoromethylcinnamoyl-SNAc starter substrates (with either malonyl-CoA or methylmalonyl-CoA as extender substrates) possessed the highest biological activity. However, these mixtures were highly impure and the product concentrations were low (< 3–5% conversion), as a result of the instability of CHS under the in vitro reaction conditions (half-life of 5 h). Following our previous work,[26] we immobilized CHS onto Ni-NTA agarose beads; this also served as a method to purify the enzyme through its N-terminal His8-tagged construction. This approach resulted in improved CHS stabilization (half-life of ca. 7 days, data not shown) and ~20% conversion (Figure 2S in the Supporting Information) for both 2- and 4-trifluoromethylcinnamoyl-SNAc starter substrates, by using malonyl-CoA as extender, which enabled us to isolate the triketide pyrone products (compounds 2c and 2e, respectively, structures confirmed by NMR, see Supporting Information) after scale-up (50 mL reaction volume).
Cell Specific Growth Inhibition Assays by Bioactive Polyketide Analogs
Shutting down the HER2 signaling pathway was expected to activate apoptosis, which would lead to growth inhibition[1] at increased doses of the CHS products. This was tested by using purified compounds 2c and 2e for dose response assays against BT-474 cells. As shown in Figure 3S (Supporting Information), both compounds showed strong growth inhibition activity against BT-474 cells, with IC50 values of 120 and 30 nm, respectively (Table 1). Interestingly, neither compound was active against nontransformed MCF10A breast cells (IC50 > 100 µm, Table 1).
Table 1.
Cell growth inhibition by 2- and 4-trifluoromethylcinnamoyl-SNAc (1c and 1e), their corresponding pyrones (2c and 2e), and 4-methylcinnamoyl pyrone (2g) on human normal (MCF-10A) and tumor-derived cell lines (MCF-7 and BT-474).
|
Cell line Compounds |
BT-474 |
IC50 (µm) MCF-7 |
MCF-10A |
|---|---|---|---|
| Compound 2e | 0.03 ± 0.001 | 0.3 ± 0.02 | > 100 |
| Compound 1e | 1.0 ± 0.1 | 1.5 ± 0.2 | > 100 |
| Compound 2c | 0.12 ± 0.02 | 3.1 ± 0.1 | > 100 |
| Compound 1c | 5.5 ± 0.2 | 15 ± 0.2 | > 100 |
| Compound 2g | 55 ± 1.5 | > 100 | > 100 |
The trifluoromethyl group is critical for inhibitory activity. When the CF3 group of compound 2e is replaced with the CH3 group of compound 2g, an 1800-fold loss in inhibitory activity is obtained (Table 1). The pyrone component is also important for inhibitory activity; the SNAc starter substrates were less inhibitory than the pyrone products (ca. 30-fold lower activity for compound 1e vs. compound 2e).
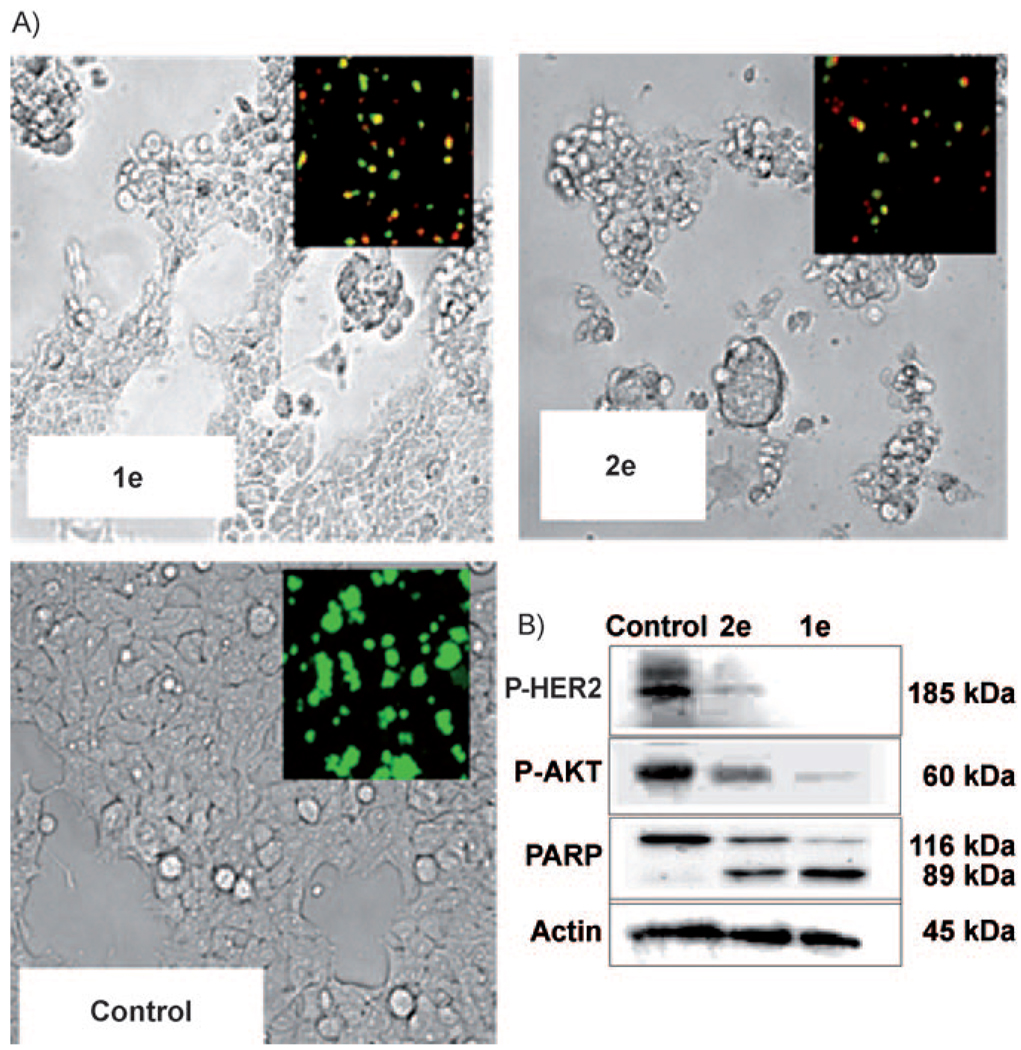
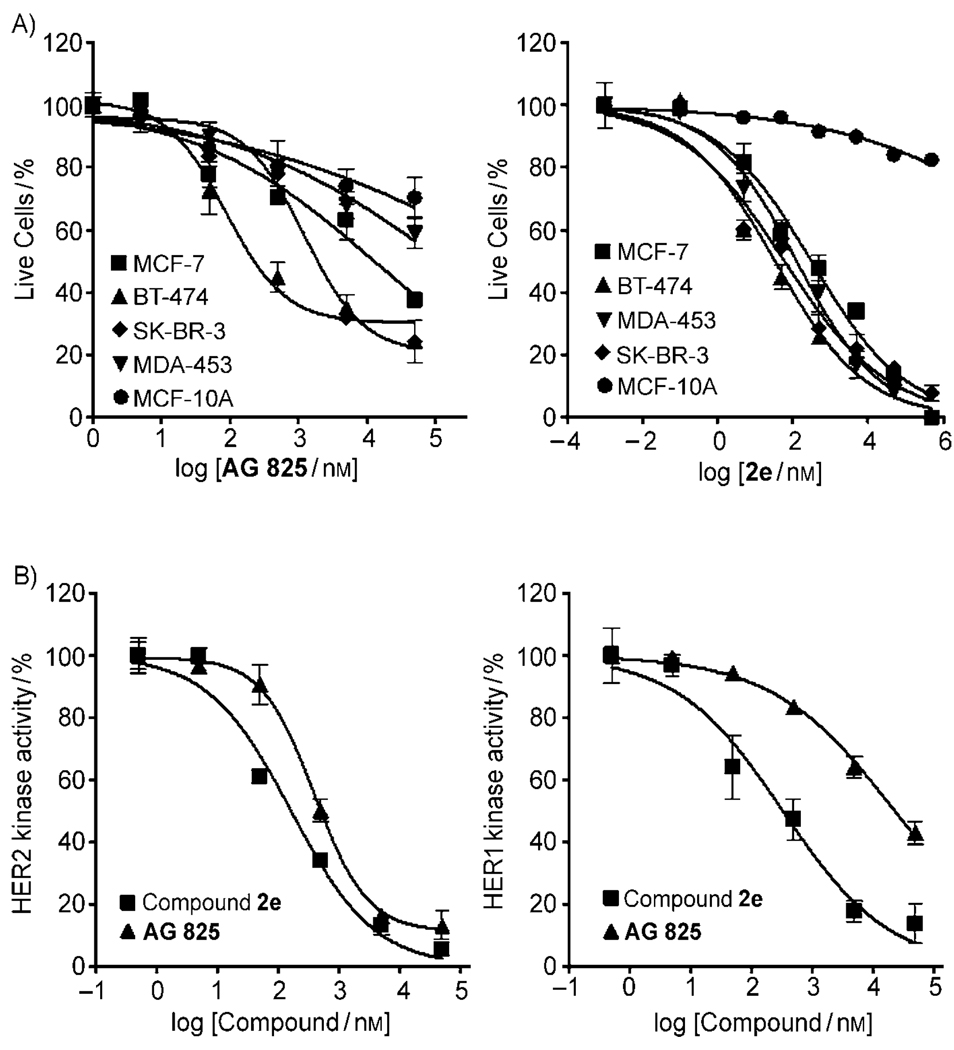
In addition to dramatic growth inhibition, compound 2e strongly activated apoptosis in BT-474 cells, as demonstrated by severe cellular morphological changes and disruption of the cell membrane. This was reflected in increasing red-colored cells (dead cells) by labeling nuclei with membrane-impermeable ethidium homodimer-1 (Figure 4a, insets). Poly (ADP-ribose) polymerase (PARP) cleavage to an 89 KDa fragment, the hallmark of apoptosis, was also shown to be strongly activated by compound 2e (Figure 4b). No evidence of apoptosis (as indicated by lack of PARP cleavage) was obtained with the CH3 analog (compound 2g) and much lower apoptosis and PARP cleavage was evident with the SNAc starter substrate (compound 1e); this is consistent with the trifluoromethyl-containing pyrone as being a highly active HER2-pathway inhibitor. Due to the highly potent growth inhibition activity of compound 2e towards BT-474 cells, we performed similar experiments for both compound 2e and a known HER2 specific inhibitor, AG-825,[27] against two other HER2-overexpressing human breast cancer cell lines, including SKBR-3[28a] and trastuzumab (Herceptin)-resistant MDA-MB-453 cells[28] (Figure 5a). Compound 2e was highly potent against both cell lines with IC50 values of 60 and 130 nm against SKBR-3 and MDA-MB-453, respectively, whereas AG-825 was less potent against these cell lines (IC50 values of BT-474, SKBR-3 and MDA-MB-453 were 0.84, 2.7 and 132 µm, respectively). Trastuzumab-resistant MDA-MB-474 cells showed the lowest sensitivity against AG-825. When we compared the growth inhibition of compound 2e with commercially available multiple-kinase inhibitor drug, lapatinib (Tykerb), Lapatinib had IC50 values against SKBR-3 cells of 37–76 nm[29] and IC50 values of 2.5–71 nm against BT-474 cells.[29] Thus, the effect of growth inhibition of compound 2e was similar to a commercially available lapatinib.
Figure 4.
Effect of 4-trifluoromethyl cinnamoyl SNAc (1e) and 4-trifluoromethyl-cinnamoyl pyrone (2e) on apoptosis of BT-474 cells. Cells were treated with compounds 1e or 2e (each 5 µm) for 12 h with PDGF. After treatment, apoptosis was detected by: A) the live (green)/dead (red) assay (inset) and cell morphological changes, and B) p-Her2 (Tyr1221/1222), p-Akt (Ser473), and cleaved PARP signals
Figure 5.
A) Dose-dependent effect of 4-trifluoromethylcinnamoyl pyrone (2e) and AG-825 on growth inhibition of different breast cancer cell lines (MCF-7, BT-474, SK-BR-3, MDA-MB-453) and a nontransformed breast cell line, MCF-10A. B) Dose response curves of HER1 and HER2 kinase activity by 4-trifluoromethylcinnamoyl pyrone (2e) and AG-825. Enzyme inhibition was assessed by measuring the inhibition of biotinylated peptide substrate phosphorylation (DNEY*FY*V and DNDY*INA were used as substrates for HER2 and HER1, respectively)
To gain a preliminary mechanistic understanding of the effect of compound 2e, we evaluated the ability of the compound to inhibit HER1 and HER2 TK by using an ELISA assay (Figure 5b). Compound 2e was a potent inhibitor of both HER1 and HER2 TK, with IC50 values of 0.33 and 0.16 µm, respectively. This contrasts with AG-825, which had IC50 values of 23 and 0.59 µm for HER1 and HER2, respectively, thus indicating potent HER2 and only moderate HER1 TK inhibition. In addition, phosphorylation of kinase domain residue Tyr877 of HER2 was also inhibited by compound 2e, as well as AG-825 (Figure 4 in the Supporting Information). These results demonstrate that compound 2e is an effective dual inhibitor of the TK function of HER1 and HER2. This, in turn, enables growth inhibition against recalcitrant cell lines. It is important to note that while compound 2e is effective against MDA-MB-453 cells, this cell line is resistant to Herceptin (trastuzumab).
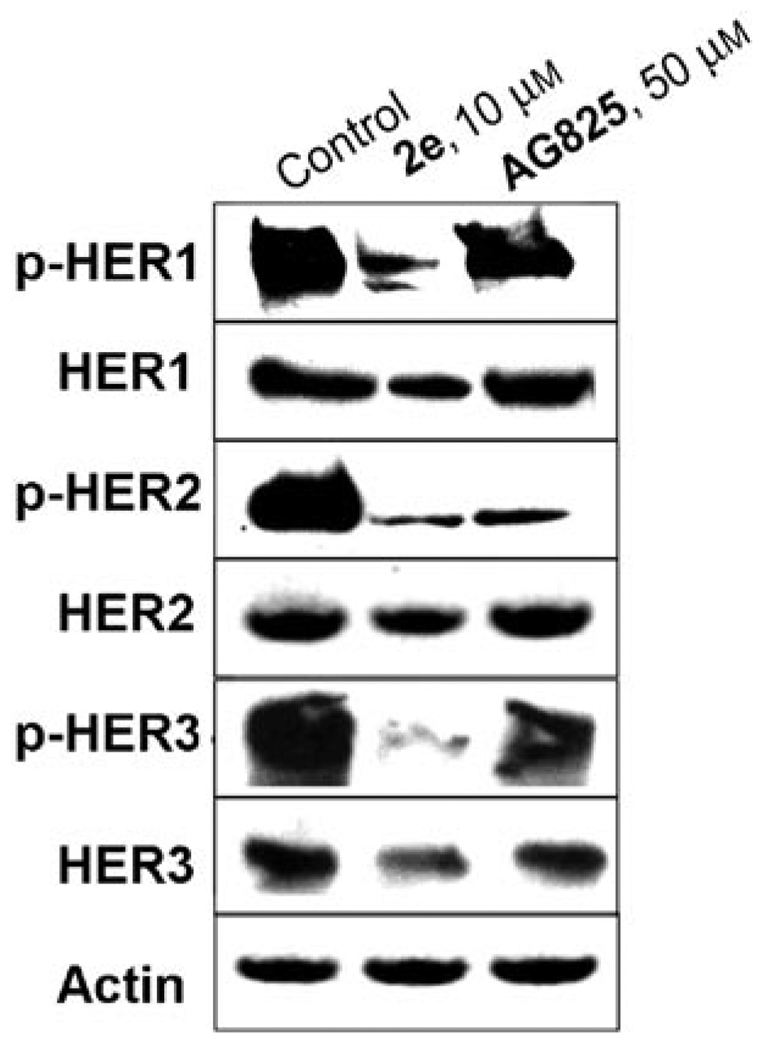
Because MDA-MB-453 cells were resistant against a HER2 specific inhibitor (AG-825, IC50 = 132 µm), these cells were chosen to examine the comparative effects of compound 2e and AG-825 on the phosphorylation of all three HER isoforms. Treatment of MDA-MB-453 cells with compound 2e strongly suppressed the phosphorylation of HER1, HER2, and HER3 (Figure 6). AG-825, however, did not suppress HER1 and HER3 phosphorylation (Figure 6); this is consistent with the aforementioned specificity of this compound against only the HER2 TK. The breadth of activity of compound 2e against all three HER isoforms might explain the activity against the Herceptin-resistant MDA-MB-453 cells. It is possible that compound 2e, by virtue of its broad specificity, can prevent both homodimerization (HER2/HER2) and heterodimerization (HER1/HER2 and HER2/HER3), resulting in growth inhibition of trastuzumab-resistant MDA-MB-453 cells although more experimental proof is needed.
Figure 6.
Inhibition of HER1, HER2, and HER3 in MDA-MB-453 cell lines by compound 2e and AG-825. Whole-cell lysates were subjected to SDS-PAGE analysis and probed with anti-phospho-HER1 (Tyr1148), anti-HER1, anti-phospho-HER2 (Tyr1221/1222), anti-HER2, anti-phospho-HER3 (Tyr1197), anti-HER3, and anti-actin, respectively.
Structural Features of Pyrones that Inhibit HER Tyrosine Kinase
The CF3 group (compound 2e) appears to be essential for high cell-based activity and selectivity, as replacing this group with CH3 resulted in a compound with an IC50 value of 55 µm—over 1800-fold less active than the CF3-containing analogue. The high activity of compound 2e was presumably due to potent inhibition of the HER2 receptor TK, and this was demonstrated by testing the compound against isolated HER2 kinase. The resulting IC50 value of 160 nm was substantially lower than that of compound 2g (9.4 µm, data not shown); this indicating that the CF3 group is critical in inhibiting TK activity. In addition to the CF3 group, the pyrone moiety is important. Comparison of compound 2e with its relevant SNAc starter substrate (compound 1e) revealed that the latter is approximately 30-fold less active than the former. As a result of the HER2 TK inhibition, downstream signaling is diminished. This was evident in diminished levels of p-HER2, p-Akt, and p-PI3K in the BT-474 cell line. Thus, deactivation of downstream signaling pathways occurs, presumably involving proapoptotic factors including p53, BAD, procaspase-9, and Forkhead (FKHR) transcription factor, as well as activation of transcription factors that involve cell survival, such as NF-kB, cyclic-AMP response element-binding protein, etc.[30]
Pyrones are commonly observed in polyketides generated through biosynthetic pathways of the type II[31] and type III PKSs,[19, 32] and have been found to have activity as HIV inhibitors,[33] acetylcholine esterase inhibitors (treatment of Alzhemier’s disease),[34] cholesterol esterase inhibitors (treatment of hypercholesterolemia),[35] and protein TK inhibitors (treatment of cancer).[36] Indeed, it is known that chalcone- or isoflavonoid-based pyrones appear to strongly interact with the adenine pocket of the ATP binding site of c-Src TK domains, presumably through both hydrophobic interactions and hydrogen bonding with hydrophobic residues (Leu275, Ala295, Leu395) and (Met343, Glu341, Thr340), respectively, resulting in TK competitive inhibition.[36] Incorporation of fluorobenzyl- or trifluoromethylbenzyl groups into existing TK inhibitors has been used to increase the selectivity of TK inhibition in addition to enhancing the potency of cellular activity, perhaps due to the enhanced hydrophobic interaction with the hydrophobic residues in the ATP-binding pocket of TK.[37] For example, the fluorobenzyl group of EGFR- and HER2-kinase inhibitors occupies a cleft formed by the hydrophobic residues (Met766, Leu777, Thr790, Thr854, and Phe856) in HER1 kinase domain.[37b] The fluorine moiety might also undergo hydrogen bonding to the Arg776 and Thr790 in the hydrophobic cleft, and is thus important for optimal inhibition of HER1; however, we cannot exculde the possibility that the CF3 moiety activates the pyrone ring as an electrophile. Further studies are underway wherein an X-ray structure of the HER kinase domain with compound 2e will be obtained, such that specific interactions of the compound with the ATP binding site can be elucidated.
Conclusion
In conclusion, we used an in vitro strategy to synthesize a library of polyketide analogs with alfalfa CHS[38] that employed twelve different aromatic-SNAc starter substrates with malonyl- or methylmalonyl-CoA as extender substrates. Testing of this library resulted in the identification of several unnatural polyketide-based polyphenols that suppressed the HER-PI3K-Akt signaling pathway, and had multiple HER TK inhibitory activity. The potent compounds were highly selective against breast-cancer cell lines, yet were essentially inactive against a nontransformed breast-cell line. Precursor-directed in vivo combinatorial biosynthesis is well known, yielding hybrid or “unnatural” natural products.[39] However, cell-based combinatorial synthesis platforms are subject to variabilities in the permeability of precursor substrates into the cells and potential toxicity of the precursors or their biosynthetic products.[40] Moreover, for complex products or product mixtures, purification is often process limiting. Thus, in vitro enzymatic synthesis might provide an efficient alternative to cell-based approaches, particularly in cases in which downstream tailoring reactions can be used to diversify further the product spectrum.
Experimental Section
Chemical Synthesis of N-acetylcysteamine Thioester (SNAc) Analogues
The two-step synthesis of the SNAc analogues (1a–1l, Fig. 1S in the Supporting Information) was carried out according to a modification of a previously reported method.[41] Briefly, N,N′-dicyclohexylcarbodiimide (1.2 equiv) was added to starting carboxylic acid (1 equiv) in ethyl acetate (25 mL) and the reaction mixture was stirred for 1 h followed by addition of N-hydroxysuccinimide (1.2 equiv) at RT. After 2 h, the substituted urea was filtered off and SNAc (1.2 equiv) was added to the reaction mixture and stirred for 4 h. The SNAc analogues were purified by using flash silica gel chromatography and identified by LC-ESI/MS and NMR. Compound characterization is in Supporting Information.
Expression and Purification of Chalcone Synthase (CHS)
Chalcone synthase (CHS) from alfalfa was produced by overexpression of the corresponding gene by using the pET-28 expression vector in E. coli. The protein was purified by using Ni-NTA column chromatography. Nickel-chelating ligand nitrilotriacetic acid (Ni-NTA) agarose beads were obtained from Invitrogen (Carlsbad, CA, USA) to give ~4 mg CHS/g bead.
In vitro Precursor-Directed CHS Enzyme Reaction
Tris-HCl buffer (100 µL, 50 mm, pH 7.5) was added to the reaction mixtures containing starter substrates (1 mm, 1a–1l), extender unit (3 mm, malonyl-CoA or methylmalonyl-CoA), and purified CHS (20 µL, ca. 2.0 mgmL−1) or Ni-NTA immobilized CHS (40 µg protein). The reaction mixtures were incubated at 37 °C for 2 h. The reactions were stopped by adding HCl (5 µL, 5% (v/v) 4n) and the products extracted into ethyl acetate (100 µL) and analyzed by LC-ESI/MS. To isolate polyketide products, we performed reactions (50 mL size). The resulting extracts were dried under nitrogen, redissolved in dichloromethane (DCM; 2 mL) containing traces of methanol, and the products were purified by using flash column chromatography with DCM/MeOH (from 20:1-3:1 (v/v)) as an eluent. Purified samples were dissolved in CD3OD (0.5 mL, Sigma, St. Louis, MO, USA) for NMR analysis (Bruker DPX500, GmbH, Germany). Compound characterization (2a-6k) is in Supporting Information.
Cells and Cell Culture
The human breast cancer cell lines used in this study were MCF-7, BT-474, SKBR-3, and MDA-MB-453. All cells were grown in DMEM (Invitrogen) supplemented with fetal bovine serum (10%; Hyclone, Waltham, MA, USA) and penicillin/streptomycin (1 × Pen/Strep). We also used a nontransformed MCF-10A cell line, which is phenotypically normal and nontumorigenic. MCF-10A cells were cultured in DMEM, insulin (10 µgmLa−1; Sigma, I1882), hEGF (20 ngmL−1; Trevigen, Gaithersburg, MD, USA), cholera toxin (100 ngmL−1; Sigma, C8052), hydrocortisone (500 ngmL−1, Sigma), Pen/Strep (1 ×; Cellgro, Herndon, VA, USA), and horse serum (5%; ATCC, Rockville, MD, USA).
Immunoblot Analysis
After exposing all of compounds with PDGF to breast cancer cells for 3 h, proteins from the cell lysates were separated by SDS-PAGE and transferred to a PVDF membrane. The membrane was blocked with skim milk (5%) in PBS-Tween 20 (0.1%, v/v) at 4°C overnight. The membrane was incubated with a primary antibody (1:500–1:1000; anti-phospho-Akt (Ser473), anti-Akt, anti-HER1, anti-phospho-HER1 (Tyr1148), antiphospho-HER2 (Tyr1221/1222), anti-phospho-HER2 (Tyr877), anti-HER2, anti-phospho-HER3 (Tyr1197), anti-HER3, anti-phospho-PI3K (Tyr508), anti-PI3K, anti-PARP, and anti-actin) at 4°C overnight. All antibodies were purchased from Cell Signaling Technology, Beverly, MA, USA. Horseradish peroxidase conjugated anti-rabbit or anti-mouse IgG was used as secondary antibody. Immunoreactive proteins were visualized by the chemiluminescence protocol (Super-Signal West, Pierce, Rockford, IL, USA).
Growth Inhibition Assays
Cells were grown in 96-well plates (1 × 104 cells/well) overnight, then treated with various concentrations of purified CHS products, and incubated for an additional 72 h. The effect of compounds on cell growth was examined by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay. Briefly, MTT solution (20 µL, 5 mgmL−1, Sigma) was added to each well and incubated for 4 h at 37°C. The supernatant was aspirated, and the MTT-formazan crystals formed by metabolically viable cells were dissolved in DMSO (200 µL). Finally, the absorbance was monitored by using a multi-well spectrophotometer (SpectraMax, Molecular Devices, Sunnyvale, CA, USA) at 595 nm.
HER Kinase Assay
Purified HER1 and HER2 TK and the corresponding biotinylated peptide substrates were purchased from Cell Signaling Technology and the kinase reaction was performed in duplicate. Various concentrations of trifluoromethylcinnamoyl pyrones (AG-825 and Compound 2e) were added to the HER kinase reaction mixture in 96-well polystyrene round-bottomed plates. The reaction mixture contained MgCl2 (5 mm), MnCl2 (5 mm), Na3VO4 (3 µm), DTT (1.25 mm), ATP (20 µm), peptide substrate (1.5 µm), and HER2 kinase (50 ng) in of HEPES buffer (50 mL, 60 mm, pH 7.5). The reaction was initiated by adding the peptide substrate and terminated after 30 min incubation at 23°C by adding EDTA (50 µL, 50 mm, pH 8.0). Each reaction mixture (25 µL) was diluted with deionized water (75 µL), and transferred into a 96-well streptavidin coated plate and incubated at 23°C for 60 min. After the plate was washed three times with PBS containing Tween 20 (0.1 %; PBST), primary antibody (100 µL; phospho-Tyrosine mAb, p-Tyr-100, 1:1000 in PBST with 1% BSA) was added and incubated at 23 °C for 60 min. After the plate was washed three times with PBST, of horseradish peroxidase-conjugated goat anti-mouse IgG (100 µL, 1:500 in PBST with 1% BSA) was added and the plate was reincubated at 23°C for 30 min. After the plate was washed five times with PBST, the TMB (3,3′,5,5″-tetramethylbenzidine) substrate was added and the plate was incubated at RT until color formed. The reaction was terminated by adding H2SO4 (50 µL, 1n) and the plate was read at 450 nm.
Live/Dead Cytotoxicity Assay
BT-474 cells were plated in slide chambers. After treatment with compounds 1e and 2e (5 µm,) for 12 h, ethidium homodimer-1 (10 µL, 2 mm) and calcein AM (5 µL, 4 mm) were added to PBS (2 mL), and 500 µL of this mixture was applied to each slide containing the cell monolayer. After incubation for 30 min at RT, the cells stained and visualized with an epifluorescence microscope (Micro Video Instruments Inc, Avon, MA, USA) equipped with an FITC-Texas red double filter.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (ES012619 and GM66712). M.I.K. acknowledges support from a Korea Research Foundation Grant funded by the Korean government (KRF-2006-352-D00047). We thank J.P. Noel for kindly supplying the chalcone synthase gene.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.200900433
References
- 1.Hynes NE, Lane HA. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.a) Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]; b) Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Nat. Rev. Mol. Cell. Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Delord JP, Allal C, Canal M, Mery E, Rochaix P, Hennebelle I, Pradines A, Chatelut E, Bugat R, Guichard S, Canal P. Ann. Oncol. 2005;16:1889–1897. doi: 10.1093/annonc/mdi405. [DOI] [PubMed] [Google Scholar]
- 5.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 6.a) Burris HA, 3rd, Hurwitz HI, Dees EC, Dowlati A, Blackwell KL, O'Neil B, Marcom PK, Ellis MJ, Overmoyer B, Jones SF, Harris JL, Smith DA, Koch KM, Stead A, Mangum S, Spector NL. J. Clin. Oncol. 2005;23:5305–5313. doi: 10.1200/JCO.2005.16.584. [DOI] [PubMed] [Google Scholar]; b) Nemunaitis J, Eiseman I, Cunningham C, Senzer N, Williams A, Lenehan PF, Olson SC, Bycott P, Schlicht M, Zentgraff R, Shin DM, Zinner RG. Clin. Cancer Res. 2005;11:3846–3853. doi: 10.1158/1078-0432.CCR-04-1950. [DOI] [PubMed] [Google Scholar]
- 7.a) Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aggarwal BB, Shishodia S. Biochem. Pharmacol. 2006;71:1397–1421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 10.a) Huynh H, Nguyen TT, Chan E, Tran E. Int. J. Oncol. 2003;23:821–829. [PubMed] [Google Scholar]; b) Shimizu M, Deguchi A, Lim JT, Moriwaki H, Kopelovich L, Weinstein IB. Clin. Cancer Res. 2005;11:2735–2746. doi: 10.1158/1078-0432.CCR-04-2014. [DOI] [PubMed] [Google Scholar]; c) Way TD, Kao MC, Lin JK. J. Biol. Chem. 2004;279:4479–4489. doi: 10.1074/jbc.M305529200. [DOI] [PubMed] [Google Scholar]
- 11.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Cell. Mol. Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee S, Li Y, Wang Z, Sarkar FH. Cancer Lett. 2008;269:226–242. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surh YJ. Nat Rev. Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Anticancer Res. 2004;24:2783–2840. [PubMed] [Google Scholar]
- 15.Arya P, Joseph R, Gan Z, Rakic B. Chem. Biol. 2005;12:163–180. doi: 10.1016/j.chembiol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Shang S, Iwadare H, Macks DE, Ambrosini LM, Tan DS. Org. Lett. 2007;9:1895–1898. doi: 10.1021/ol070405p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson B, Micklefield J. Nat. Chem. Biol. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 18.a) Kwon SJ, Lee MY, Ku B, Sherman DH, Dordick JS. ACS Chem. Biol. 2007;2:419–425. doi: 10.1021/cb700033s. [DOI] [PubMed] [Google Scholar]; b) Fernandes TG, Kwon SJ, Lee MY, Clark DS, Cabral JM, Dordick JS. Anal. Chem. 2008;80:6633–6639. doi: 10.1021/ac800848j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Boudet AM. Phytochemistry. 2007;68:2722–2735. doi: 10.1016/j.phytochem.2007.06.012. [DOI] [PubMed] [Google Scholar]; b) Huang YT, Hwang JJ, Lee PP, Ke FC, Huang JH, Huang CJ, Kandaswami C, Middleton E, Jr, Lee MT. Br. J. Pharmacol. 1999;128:999–1010. doi: 10.1038/sj.bjp.0702879. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Soobrattee MA, Bahorun T, Aruoma OI. Biofactors. 2006;27:19–35. doi: 10.1002/biof.5520270103. [DOI] [PubMed] [Google Scholar]
- 20.Austin MB, Noel JP. Nat. Prod. Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 21.Morita H, Takahashi Y, Noguchi H, Abe I. Biochem. Biophys. Res. Commun. 2000;279:190–195. doi: 10.1006/bbrc.2000.3920. [DOI] [PubMed] [Google Scholar]
- 22.Jez JM, Bowman ME, Noel JP. Proc. Natl. Acad. Sci. USA. 2002;99:5319–5324. doi: 10.1073/pnas.082590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe I, Takahashi Y, Lou W, Noguchi H. Org. Lett. 2003;5:1277–1280. doi: 10.1021/ol0300165. [DOI] [PubMed] [Google Scholar]
- 24.Eddy SF, Kane SE, Sonenshein GE. Cancer Res. 2007;67:9018–9023. doi: 10.1158/0008-5472.CAN-07-1691. [DOI] [PubMed] [Google Scholar]
- 25.Rameh LE, Cantley LC. J. Biol. Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 26.Kim MI, Kwon SJ, Dordick JS. Org. Lett. 2009;11:3806–3809. doi: 10.1021/ol901243e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murillo H, Schmidt LJ, Tindall DJ. Cancer Res. 2001;61:7408–7412. [PubMed] [Google Scholar]
- 28.a) Narayan M, Wilken JA, Harris LN, Baron AT, Kimbler KD, Maihle NJ. Cancer Res. 2009;69:2191–2194. doi: 10.1158/0008-5472.CAN-08-1056. [DOI] [PubMed] [Google Scholar]; b) Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Cancer Res. 2002;62:4132–4141. [PubMed] [Google Scholar]
- 29.a) Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, Halsey W, Sathe GM, Martin AM, Gilmer TM. Cancer Res. 2009;69:6871–6878. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]; b) Strecker TE, Shen Q, Zhang Y, Hill JL, Li Y, Wang C, Kim HT, Gilmer TM, Sexton KR, Hilsenbeck SG, Osborne CK, Brown PH. J. Natl. Cancer Inst. 2009;101:107–113. doi: 10.1093/jnci/djn436. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang D, Pal A, Bornmann WG, Yamasaki F, Esteva FJ, Hortobagyi GN, Bartholomeusz C, Ueno NT. Mol. Cancer Ther. 2008;7:1846–1850. doi: 10.1158/1535-7163.MCT-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Konecny GE, Pegram MD, Venkatesan N, Finn R, Yang G, Rahmeh M, Untch M, Rusnak DW, Spehar G, Mullin RJ, Keith BR, Gilmer TM, Berger M, Podratz KC, Slamon DJ. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 30.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Nat. Rev. Drug Discovery. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 31.Ridley CP, Khosla C. ACS Chem. Biol. 2007;2:104–108. doi: 10.1021/cb600382j. [DOI] [PubMed] [Google Scholar]
- 32.Austin MB, Izumikawa M, Bowman ME, Udwary DW, Ferrer JL, Moore BS, Noel JP. J. Biol. Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 33.Boyer FE, Vara Prasad JV, Domagala JM, Ellsworth EL, Gajda C, Hagen SE, Markoski LJ, Tait BD, Lunney EA, Palovsky A, Ferguson D, Graham N, Holler T, Hupe D, Nouhan C, Tummino PJ, Urumov A, Zeikus E, Zeikus G, Gracheck SJ, Sanders JM, VanderRoest S, Brodfuehrer J, Iyer K, Sinz M, Gulnik SV. J. Med. Chem. 2000;43:843–858. doi: 10.1021/jm990281p. [DOI] [PubMed] [Google Scholar]
- 34.Peng FC. J. Nat. Prod. 1995;58:857–862. doi: 10.1021/np50120a006. [DOI] [PubMed] [Google Scholar]
- 35.Deck LM, Baca ML, Salas SL, Hunsaker LA, Vander Jagt DL. J. Med. Chem. 1999;42:4250–4256. doi: 10.1021/jm990309x. [DOI] [PubMed] [Google Scholar]
- 36.Lin LG, Xie H, Li HL, Tong LJ, Tang CP, Ke CQ, Liu QF, Lin LP, Geng MY, Jiang H, Zhao WM, Ding J, Ye Y. J. Med. Chem. 2008;51:4419–4429. doi: 10.1021/jm701501x. [DOI] [PubMed] [Google Scholar]
- 37.a) Okram B, Nagle A, Adrian FJ, Lee C, Ren P, Wang X, Sim T, Xie Y, Xia G, Spraggon G, Warmuth M, Liu Y, Gray NS. Chem. Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]; b) Xu G, Searle LL, Hughes TV, Beck AK, Connolly PJ, Abad MC, Neeper MP, Struble GT, Springer BA, Emanuel SL, Gruninger RH, Pandey N, Adams M, Moreno-Mazza S, Fuentes-Pesquera AR, Middleton SA, Greenberger LM. Bioorg. Med. Chem. Lett. 2008;18:3495–3499. doi: 10.1016/j.bmcl.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Jez JM, Bowman ME, Noel JP. Biochemistry. 2001;40(49):14829–14838. doi: 10.1021/bi015621z. [DOI] [PubMed] [Google Scholar]
- 39.Walsh CT. ChemBioChem. 2002;3:124–134. [Google Scholar]
- 40.Weissman KJ, Bycroft M, Cutter AL, Hanefeld U, Frost EJ, Timoney MC, Harris R, Handa S, Roddis M, Staunton J, Leadlay PF. Chem. Biol. 1998;5(12):743–754. doi: 10.1016/s1074-5521(98)90666-4. [DOI] [PubMed] [Google Scholar]
- 41.Neises B, Steglich W. Angew. Chem. 1978;90:556–557. [Google Scholar]; Angew. Chem. Int. Ed. Engl. 1978;17:522–524. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.