Abstract
Like many positive-strand RNA viruses, brome mosaic virus (BMV) RNA replication occurs in membrane-invaginated vesicular compartments. BMV RNA replication compartments show parallels with membrane-enveloped, budding retrovirus virions, whose release depends on the cellular multivesicular body (MVB) sorting pathway. BMV RNA replication compartments are not released from their parent membranes, but might depend on MVB functions for membrane invagination. Prior results show that BMV RNA replication is severely inhibited by deletion of the crucial MVB gene DOA4 or BRO1. We report here that involvement of DOA4 and BRO1 in BMV RNA replication is not dependent on the MVB pathway's membrane-shaping functions but rather is due to their roles in recycling ubiquitin from MVB cargos. We show that deleting DOA4 or BRO1 inhibits the ubiquitination- and proteasome-dependent activation of homologous transcription factors Mga2p and Spt23p, which regulate many lipid metabolism genes, including the fatty acid desaturase gene OLE1, which is essential for BMV RNA replication. However, Mga2p processing and BMV RNA replication are restored by supplementing free ubiquitin, which is depleted in doa4Δ and bro1Δ cells. The results identify Mga2p and Spt23p processing and lipid regulation as sensitive targets of ubiquitin depletion and correctly predict multiple effects of modulating additional host genes RFU1, UBP6, and UFD3. Our results also show that BMV RNA replication depends on additional Mga2p-regulated genes likely involved in lipid metabolism beyond OLE1. Among other points, these findings show the potential for blocking viral RNA replication by modulating lipid synthesis at multiple levels.
INTRODUCTION
Positive-strand RNA viruses replicate their RNA genomes in close association with proliferated and rearranged host intracellular membranes (17, 47, 62). Accordingly, lipid synthesis and the lipid composition of intracellular membranes play important roles in RNA replication by many positive-strand RNA viruses (12, 13, 16, 23, 32, 38–40, 64, 66, 72–74). Revealing the critical interactions between membrane rearrangement, lipid synthesis/composition, and viral RNA replication will further understanding of viral replication and could provide foundations for novel broad-spectrum antiviral strategies.
Brome mosaic virus (BMV) has been used as a model system to study gene expression, RNA replication, virus-host interaction, and evolution of positive-strand RNA viruses (70). BMV has a tripartite RNA genome and a single subgenomic mRNA, RNA4. Genomic RNA1 and RNA2 encode the two BMV RNA replication proteins, 1a and 2apol, respectively. 2apol contains the viral RNA-dependent RNA polymerase domain. 1a, which contains RNA capping (3, 35) and NTPase/helicase domains (71), plays multiple, central roles in RNA replication complex assembly and function. For example, 1a directs itself, 2apol, and viral RNA replication templates to perinuclear endoplasmic reticulum (ER) membranes that become the sites of viral RNA synthesis both in BMV's natural plant hosts and in cells of the yeast Saccharomyces cerevisiae, which also support BMV RNA replication, subgenomic mRNA transcription, and selective RNA encapsidation (15, 18, 29, 37, 57, 58, 63). Moreover, at these perinuclear ER membranes, 1a induces formation of ∼60- to 75-nm spherular invaginations that serve as RNA replication compartments in yeast (63) and in plant hosts of BMV, such as Nicotiana benthamiana (X. Wang, A. Diaz, and P. Ahlquist, unpublished results). Many other positive-strand RNA viruses use similar vesicular membrane invaginations as RNA replication compartments (2, 17, 36).
BMV RNA replication is closely linked not only to formation of these vesicular RNA replication compartments (19, 42) but also to the lipid composition of the associated membranes (38–40). For example, even though normal numbers of replication compartments form, BMV RNA replication is inhibited by ≥20-fold when the activity of Δ9 fatty acid desaturase, which converts saturated to monounsaturated fatty acids (SFA and monoUFA, respectively), is reduced by mutations in yeast (39, 40) or by gene-specific RNA interference in N. benthamiana plants (X. Wang et al., unpublished data).
Previously, Kushner et al. (38) screened a yeast single-gene-deletion library and identified ∼100 genes whose deletion inhibited or enhanced BMV RNA replication by >3-fold. Among the implicated genes, we focused on DOA4 and BRO1, key components of the multivesicular body (MVB) pathway through which mono- or oligoubiquitinated cargo proteins, such as signaling receptors, are targeted to vacuoles/lysosomes for degradation (27, 33, 53). These cargoes are sorted into intralumenal vesicles that invaginate away from the cytoplasm into the endosomal lumen and then pinch off to create MVBs. This MVB pathway is hijacked by retroviruses and other enveloped viruses to support their topologically similar virion budding (10, 11, 48). For such viruses, disrupting viral recruitment or function of MVB pathway components arrests the release of budding virions at a late stage (10, 11, 48).
Before cargoes are sorted into intralumenal MVB vesicles and degraded, ubiquitin (Ub) is obligatorily removed from these cargoes and recycled by DOA4-encoded deubiquitinating enzyme Doa4p (5, 6, 21, 33, 43, 69). Doa4p is also involved in removing poly-Ub from proteins targeted for degradation by proteasomes (50). Doa4p is recruited to MVB sites primarily by Bro1p (4, 44, 49, 59), which also activates Doa4p's deubiquitinase activity (Fig. 1A) (59). Bro1p is the yeast ortholog of mammalian ALX1/AIP1, through which the MVB pathway is recruited by late domains of HIV and other retroviruses to mediate budding (14, 46, 48). As the assembly of BMV RNA replication complexes shares multiple similarities with budding retroviral virions (63), we investigated whether dependence of BMV RNA replication on DOA4 and BRO1 involved the MVB pathway. We report here that inhibition of BMV RNA replication by deleting DOA4 or BRO1 is independent of the MVB pathway's membrane-shaping functions, but rather is due to downregulated expression of Δ9 fatty acid desaturase and other genes on which BMV RNA replication depends. We further show that upon deletion of DOA4 or BRO1, a reduction in free Ub inhibits activation of specific transcription factors that induce expression of Δ9 fatty acid desaturase (Fig. 1A) and many other lipid metabolism genes. The results further demonstrate the crucial dependence of positive-strand RNA virus replication on a balanced spectrum of lipid synthesis and modification and the potential for blocking viral replication by modulating such synthesis at multiple levels.
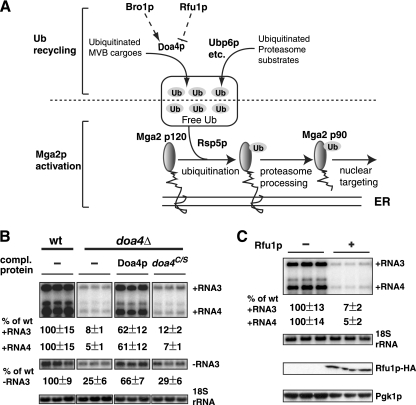
Fig. 1.
Doa4p and its deubiquitinating activity are required for BMV RNA replication. (A) Schematic illustrating the connection between free ubiquitin (Ub) levels and activation of transcription activator Mga2p (and Spt23p). Ub is removed, and thus recycled, from ubiquitinated MVB cargoes via Doa4p or from polyubiquitinated proteasomal substrates via Ubp6p and other deubiquitinating enzymes. While Bro1p recruits Doa4p to MVB and promotes Doa4p's enzymatic activity, Rfu1p inhibits Doa4p activity. Mga2p (Mga2 p120) is ubiquitinated by ubiquitin ligase Rsp5p. The C-terminal membrane-spanning domain is degraded by proteasomes, and the N-terminal activation domain (Mga2 p90) is released from perinuclear ER membranes and subsequently targeted to the nucleus. See the text for more details. (B) The BMV RNA replication defect is complemented (comp.) by wt Doa4p but not by the doa4C/S mutant in doa4Δ cells. wt DOA4 and the doa4C/S mutant were expressed from their endogenous promoter by using centrameric plasmids. (C) BMV RNA replication is severely affected in yeast cells overexpressing RFU1. RFU1 expression was driven by the GAL1 promoter in a high-copy-number plasmid. For both panels B and C, total RNA was extracted from yeast cells expressing BMV components. BMV positive- and negative-strand RNAs were detected by Northern blotting using probes specific to BMV RNAs. 18S rRNA bands were detected with an 18S rRNA probe. The measured BMV RNA signals were normalized to that of 18S rRNA. The blot showing negative-strand RNA accumulation was exposed longer than that for positive-strand RNA for comparison. Total proteins were extracted from equal numbers of yeast cells and analyzed by SDS-PAGE and immunoblot analysis with monoclonal anti-HA or anti-Pgk1p antibodies. All experiments were done multiple times in triplicate or duplicate (Fig. 8B). Representative blots are shown for each figure.
MATERIALS AND METHODS
Yeast strain and cell growth.
Yeast strain BY4743 and its various single-gene-deletion derivatives were used in all experiments. Cultures were grown at 30°C in defined synthetic medium containing 2% galactose as a carbon source. Leucine, uracil, histidine, or combinations thereof were omitted to maintain plasmid selection. Cells were grown in galactose medium for 2 passages (36 to 48 h) and harvested when the optical density at 600 nm (OD600) was between 0.4 and 1.0. To make medium supplemented with fatty acids, Tergitol NP-40 was added to a final concentration of 1% to solubilize the fatty acids. Equimolar amounts of palmitoleic acid (16:1) and oleic acid (18:1) were added to NP-40-containing medium to the specified concentrations (67).
Plasmids and plasmid construction.
Expression of BMV 1a and 2apol was driven by the galactose-inducible and glucose-repressible GAL1 promoter in most experiments. BMV 1a was expressed from plasmid pB1YT3 in all experiments that did not include expression of 2apol (71). To express both 1a and 2apol for assaying BMV RNA replication, plasmid pB12VG1 was used (38). CUP1 promoter-driven BMV RNA3 was launched from plasmid pB3VG128-H in most experiments in medium lacking copper (38). To assay 1a-mediated RNA3 stability, we used plasmid pB3MS82, which expresses BMV RNA3 from the GAL1 promoter (68).
Wild-type [wt] DOA4 was expressed from plasmid pDOA4-8 (50) or pCR30 (59), using its endogenous promoter. The doa4C/S and doa4AAFA mutants were expressed from pCR64 and pCR15-U, respectively (59). pCR15-U was made by subcloning the doa4AAFA mutant sequence from pCR15 (59) to pRS416, a low-copy-number centromeric plasmid. Ubiquitin was expressed from pYEP96-U, which expresses the ubiquitin gene from the CUP1 promoter (22). In all experiments, however, no additional copper was added to the medium. The pYEP96-U plasmid was made by subcloning the cassette containing the copper promoter, Ub open reading frame (ORF), and CYC1 terminator from pYEP96 (22) into pRS426, a high-copy-number 2μ plasmid. Myc-tagged Mga2p and Spt23p were expressed from 2μ plasmids YEpLac181-mycMGA2 and pRS426-mycSPT23HA, respectively (55). pRS426-mycSPT23HA was made by subcloning the mycSPT23HA fragment from pRS416-mycSPT23HA into pRS426. A construct overexpressing His6- and hemagglutinin (HA)-tagged RFU1 was purchased from Open Biosystem, and the RFU1 ORF was inserted into pBG1805, a 2μ plasmid under the control of the GAL1 promoter. The bro1-2 mutant, which has an early stop codon at position 820 amino acids (aa), was expressed from pSS41 and pGO369 for low and high expression levels, respectively (59). The bro1-C mutant (Bro1 aa 692 to 844) was expressed from pJEN6, a centromeric plasmid. Expression of both bro1-2 and bro1-C was driven by the endogenous promoter.
Cell fractionation assays.
Yeast spheroplasts were prepared from 5 OD600 units of cells and were lysed in lysis buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 10 mM dithiothreitol, and 1:200 dilution of yeast protease inhibitor mix [Sigma]). Half of the lysate was retained as the total extracted protein. The other half of the lysate was centrifuged at 4°C for 5 min at 20,000 × g. The supernatant was removed and retained, and the pellet was resuspended in lysis buffer. Equal volumes of the total (T), supernatant (S), and pellet (P) fractions were used for SDS-PAGE and Western blotting.
Western blotting.
Yeast cells were grown to an OD600 of 0.4 to 1.0, and 2 OD600 units of cells was harvested. Proteins were extracted as described previously (40), and equal volumes of extracted total proteins were used for electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane. Expression of target proteins was detected with the following antibodies and dilutions: rabbit anti-BMV 1a at 1:10,000, rabbit anti-Bro1p at 1:5,000 (a gift from Gregory Odorizzi, University of Colorado), mouse anti-BMV 2apol at 1:4,000, mouse anti-Pgk1p (A6457; Molecular Probes) at 1:10,000, mouse anti-Dpm1p (A6429; Molecular Probes) at 1:1,000, mouse anti-Myc (OP10; Calbiochem) at 1:2,000, mouse anti-Ub (P4D1; Santa Cruz Biotechnology) at 1:2,000, and mouse anti-HA (32-6700; Invitrogen) at 1:2,000, using horseradish peroxidase (HRP)-conjugated secondary antibodies and Supersignal West Femto substrate (Thermo Scientific).
RESULTS
Doa4p deubiquitinase activity is required for BMV RNA replication.
A previous genomewide yeast deletion mutant screen (38) revealed that BMV RNA replication depends on DOA4. In that screen, a BMV RNA3 derivative expressing Renilla luciferase (RLuc) was used as a viral RNA replication reporter. To eliminate the possibility that the BMV replication defect in cells with the complete DOA4 open reading frame deleted (doa4Δ) was specific to the Rluc reporter, we compared the accumulation of BMV RNA3 replication products in doa4Δ and wild-type (wt) cells expressing BMV 1a, 2apol, and template RNA3 (Fig. 1B). Viral RNA replication and subgenomic mRNA synthesis were assayed by strand-specific Northern blotting and normalized to 18S rRNA levels. In doa4Δ cells, positive-strand RNA3 accumulated to only 10 to 20% and RNA4 to 5 to 10% of wt replication levels (Fig. 1B), which agreed well with the previous Rluc-based measurements (38). Interestingly, negative-strand RNA3 in doa4Δ cells was detected at 20 to 30% of the wt level (Fig. 1B), indicating a more moderate reduction in this earlier replication step.
The BMV RNA replication defect in doa4Δ cells was largely complemented by expressing wt Doa4p (Fig. 1B). The nearly 70% complementation observed is close to the maximal possible restoration given the inevitable loss of the DOA4-expressing plasmid from some cells due to general inefficiencies in plasmid segregation during mitosis (24). To determine if Doa4p deubiquitinase activity was required for BMV RNA replication, we used a Doa4p doa4C/S mutant, which has a Cys781→Ser substitution that suppresses Doa4p's deubiquitinating activity (51) and its ability to support cargo protein sorting through the MVB pathway (59). To avoid possible nonphysiological effects from overexpression of doa4C/S (51), as from a high-copy-number plasmid, this mutant and wt DOA4 were independently expressed from the endogenous DOA4 promoter on low-copy-number centromeric plasmids. The doa4C/S mutant protein is stable in yeast (51), but in contrast to the strong complementation by wt DOA4, expression of doa4C/S only slightly increased the low levels of BMV RNA replication in doa4Δ cells (Fig. 1B). Thus, Doa4p's deubiquitinase activity is essential to support BMV RNA replication.
RFU1 (regulator of free ubiquitin chains 1) was recently identified as an inhibitor of Doa4p that helps maintain cellular Ub homeostasis (Fig. 1A) (34). Rfu1p interacts with Doa4p directly and inhibits Doa4p's enzymatic activity in vitro. Overexpression of RFU1 led to phenotypes similar to those of the doa4Δ mutant, including depleted free Ub levels and heat stress sensitivity (34; also see the next section). Therefore, we overexpressed HA-tagged RFU1 (Rfu1p-HA) (Fig. 1C) under the control of the strong GAL1 promoter in wt cells and checked its effect on BMV RNA replication. Overexpressing RFU1 inhibited BMV RNA replication by 13-fold (Fig. 1C), independently confirming that Doa4p's deubiquitinating activity is indeed required to support BMV RNA replication.
BMV RNA replication is restored in doa4Δ cells by added Ub.
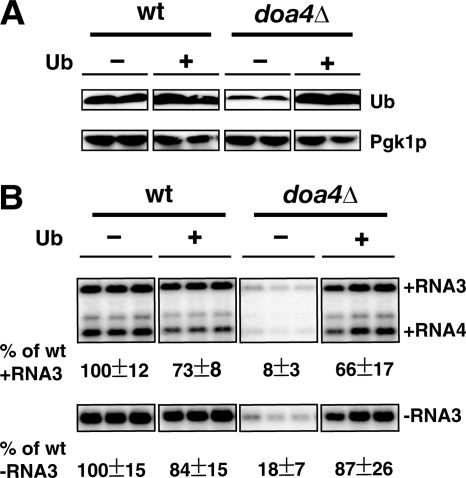
In addition to defects in MVB cargo protein sorting, deletion of DOA4 leads to depletion of free Ub, defective protein proteolysis in proteasomes and vacuoles/lysosomes, hypersensitivity to stress and amino acid analogs, and a strong sporulation defect (69). The Ub deficiency causes the remaining listed defects since supplementation with Ub largely restores the majority of affected functions (69). As expected, Western blotting confirmed an equivalent decrease in free Ub in the doa4Δ cells used here (Fig. 2A).
Fig. 2.
Supplemented ubiquitin complements BMV RNA replication defect in doa4Δ cells. (A) Reduced levels of free Ub are complemented by supplemented Ub. Total protein extraction and Western blotting were done as in Fig. 1 with monoclonal anti-Ub or anti-Pgk1p antibodies. (B) Supplemented Ub restores BMV RNA replication to close to wt levels. Ub was expressed from plasmid pYEP96-U under the control of the copper promoter and grown in medium lacking copper. RNA extraction and Northern blotting were done as in Fig. 1.
To determine whether this free Ub deficiency played a role in inhibiting BMV RNA replication in doa4Δ cells, Ub was expressed from plasmid pYEP96-U, which provides wt Ub levels (Fig. 2A) (69). In doa4Δ cells with supplemented Ub, all forms of BMV RNA, including positive- and negative-strand RNA3 and positive-strand RNA4, accumulated to levels similar to those in wt yeast cells harboring pYEP96-U (Fig. 2B). Similar to the maximum complementation achieved by expression of wt Doa4p (Fig. 1B), supplementation with Ub restored BMV RNA replication to ∼66% of that in wt cells, suggesting that a Ub-related pathway or pathways may be responsible for most or all inhibition of BMV RNA replication in doa4Δ cells.
BMV RNA replication deficiency in bro1Δ cells is linked to Doa4p activity and decreased free Ub.
Like DOA4, BRO1 was implicated in BMV RNA replication in a prior yeast deletion library screen (38). We found in repeated experiments that replication of wt BMV RNA3 was inhibited in bro1Δ cells by 2.5- to 4-fold (Fig. 3A), confirming the BRO1 dependence of BMV replication and showing that this effect was independent of the RLuc reporter used in the original primary screening.
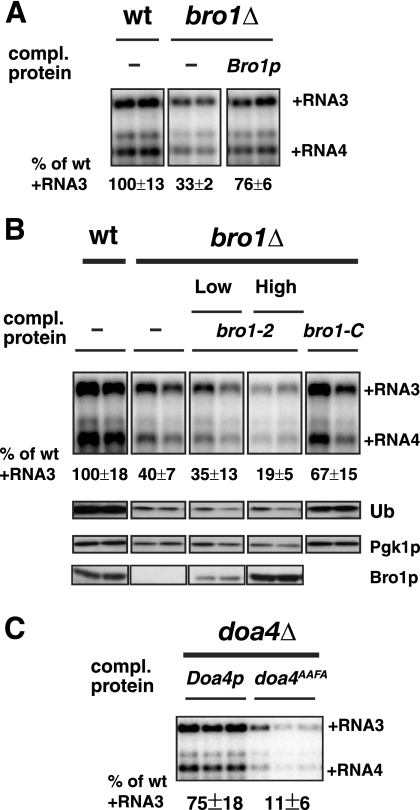
Fig. 3.
Inhibited BMV RNA replication in bro1Δ cells correlates with a lack of Doa4p's enzymatic activity and a decrease in free Ub levels. (A) Inhibited BMV RNA replication is restored by supplementation with wt Bro1p in bro1Δ cells. (B) BMV RNA replication is related to the levels of free Ub in bro1Δ cells. The bro1-2 mutant was expressed from either a low- or high-copy-number plasmid, and mutant bro1-C was expressed from a low-copy-number plasmid in bro1Δ cells. Note that the anti-Bro1p antiserum did not detect bro1-C. comp., complementing. (C) The functional interaction between the C termini of Doa4p and Bro1p is required for BMV RNA replication. The doa4AAFA mutant, which does not interact with Bro1p or support cargo sorting and deubiquitination, did not restore BMV RNA replication in doa4Δ cells. Accumulation of BMV RNAs and Ub, Bro1p, and Pgk1p levels were detected as in Fig. 1.
Bro1p has multiple functions in the MVB pathway, including activating Doa4p's deubiquitinase function and contributing to Doa4p recruitment to the MVB pathway (4, 59). Bro1p contributes to Doa4p recruitment by an interaction between their N termini, while Bro1p's crucial stimulation of Doa4p's deubiqutinase activity is mediated by interaction of the Bro1p C terminus with the Doa4p C-terminal catalytic domain (4, 44, 49, 59). Disruption of this C-terminal Bro1-Doa4p interaction drastically inhibits Doa4p's deubiquitinase and MVB cargo-sorting functions (49, 59).
To test the possible relationship between BRO1's effects on BMV RNA replication and DOA4, we performed complementation tests with the wt BRO1 allele and selected BRO1 mutants. The bro1-2 mutant has a Q820→Stop substitution that deletes the C-terminal 25 amino acids of Bro1p, blocks Doa4p deubiquitinase activation, and produces multiple phenotypes paralleling those of doa4Δ cells (59). As expected, expressing wt BRO1 in bro1Δ cells complemented the defect in BMV RNA replication (Fig. 3A). Unlike wt Bro1p, expression of bro1-2 from a low-copy-number plasmid had little or no significant effect on RNA3 replication, while expression of bro1-2 from a high-copy-number plasmid inhibited the already low BMV RNA replication a further 2-fold (Fig. 3B). The negative effects of overexpression of bro1-2, whose N terminus interacts with and recruits Doa4p without activation (59), might result from suppression of the low basal activity of Doa4p or from nonproductive sequestration of Doa4p from other roles on the proteasome and other sites (50).
To further assess the importance of Bro1p-mediated Doa4p deubiquitinase activation for BMV RNA replication, we also tested the converse approach of disrupting this activation by mutating DOA4 in the presence of wt BRO1. The DOA4 allele used was doa4AAFA, which substitutes AAFA for a C-proximal YPFL motif (aa 826 to 829) essential for the Doa4p-Bro1p interaction that stimulates Doa4p's enzymatic activity (59). The doa4AAFA mutant stably accumulates in cells, and like green fluorescent protein (GFP)-tagged wt Doa4p, GFP-tagged doa4AAFA properly localizes to MVBs (59). Unlike wt DOA4, however, expressing doa4AAFA in doa4Δ cells did not complement the defect in BMV RNA replication (Fig. 3C).
Next we expressed in bro1Δ cells a C-terminal fragment of Bro1p comprising aa 692 to 844, which lacks the N-terminal Bro1p sequence that facilitates Doa4p recruitment to MVB sites but retains all sequences needed to bind and activate Doa4p's C-terminal deubiquitinase domain (59). This C-terminal Bro1p fragment complemented BMV RNA replication nearly as efficiently as wt Bro1p (Fig. 3A and B).
Thus, the results with all three mutants were consistent with Bro1p activation of Doa4p deubiquitinase function being the major if not sole contribution of Bro1p to BMV RNA replication. This conclusion was further underscored by Western blots showing that, as in doa4Δ cells (Fig. 2B), free Ub levels in bro1Δ cells were greatly reduced relative to those in wt cells (Fig. 3B) (4). Moreover, bro1-2 expression, which did not restore BMV RNA replication, did not restore free Ub levels, whereas bro1-C expression stimulated BMV RNA replication and free Ub levels in parallel (Fig. 3B). Since multiple, independent findings implied that deletion of Bro1p affected BMV RNA replication indirectly, through the loss of its stimulating effect on Doa4p deubiquitinase, we next concentrated on how loss of Doa4p function inhibited BMV RNA replication.
Doa4p is not required for early steps of BMV RNA replication.
Loss of Doa4p or its activity increases the half-life of many cellular proteins (50, 51). Therefore, we checked whether deletion of DOA4 affects the accumulation of BMV replication proteins 1a and 2apol. Western blotting showed that 1a and 2apol levels were unaffected in doa4Δ cells (Fig. 4A), indicating that the BMV RNA replication defect in doa4Δ cells is not due to altered accumulation or balance of the viral replication proteins.
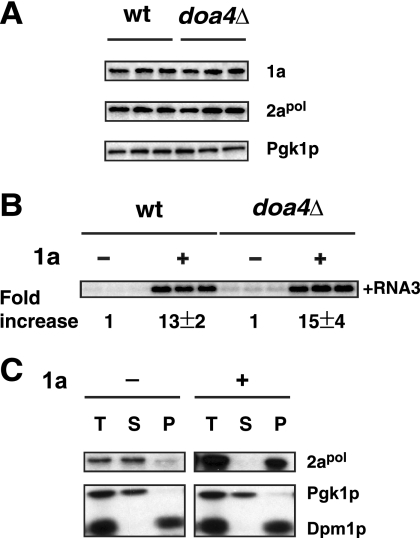
Fig. 4.
DOA4 is not required for early steps of BMV RNA replication. (A) BMV 1a and 2apol accumulate to equal levels in wt and doa4Δ cells. Accumulation of BMV 1a or 2apol protein was analyzed as in Fig. 1 with anti-1a or anti-2apol antibodies. (B) BMV RNA3 is stabilized by 1a in doa4Δ cells. BMV RNA3 only or RNA3 and 1a were expressed in wt and doa4Δ cells. Accumulation of RNA3 was detected by using a BMV-specific probe as in Fig. 1. (C) BMV 2apol is recruited to the membrane fraction by 1a in doa4Δ cells. Yeast cells expressing 2apol only or coexpressing 2apol and 1a were spheroplasted and lysed to yield a total protein fraction (T). The lysate was centrifuged to obtain membrane-depleted supernatant (S) and membrane-enriched pellet (P) fractions. Equal volumes of total, supernatant, and pellet fractions were subjected to SDS-PAGE, and Pgk1p, Dpm1p, and BMV 2apol were detected by using antibodies against each protein. Note that Pgk1p is a soluble cytoplasmic protein, while Dpm1p is an ER membrane protein.
BMV 1a recruits viral RNA replication templates into a membrane-associated, RNase-resistant state with a likely physical location inside the viral spherules (63). This is reflected by a dramatic 8- to 20-fold increase in viral RNA template accumulation depending on the amount of 1a protein expressed (28, 63). BMV genomic RNA3 accumulation in the presence of 1a increased 15-fold in doa4Δ cells, very similar to the 13-fold increase in wt cells (Fig. 4B), indicating that recruitment of viral RNA replication templates was not affected by deletion of DOA4.
BMV 1a also stimulates 2apol accumulation and recruits 2apol to ER membranes through an interaction between the 1a C terminus and 2apol N terminus (15, 31). We examined the distribution of 2apol in doa4Δ cells without or with 1a by cell fractionation and Western blotting (Fig. 4C). In doa4Δ cells without 1a, 2apol was mainly detected in the membrane-depleted supernatant, similar to soluble cytoplasmic protein Pgk1p and opposite from ER marker Dpm1p, which accumulates primarily in the membrane-enriched pellet fraction (Fig. 4C). As in wt cells, 2apol accumulation increased several-fold and shifted to the membrane-enriched pellet upon 1a expression in doa4Δ cells, indicating that recruitment of 2apol to membranes by 1a was not affected in doa4Δ cells (Fig. 4C).
Collectively, the above data indicate that in doa4Δ cells BMV RNA replication was inhibited within the short interval after 1a-mediated recruitment of RNA templates and 2apol to the ER membrane, but before negative-strand RNA synthesis.
Activation of transcription factor Mga2p is inhibited in doa4Δ cells.
Pathway and literature analysis revealed that the regulated expression of Δ9 fatty acid desaturase, which is required for BMV RNA replication in yeast (39, 40) and plants (X. Wang et al., unpublished data), was a candidate pathway connecting DOA4 function and Ub depletion to BMV RNA replication (Fig. 1A). Fatty acid desaturase and other lipid synthesis genes are regulated by conserved processes that retain key features from yeast to Drosophila to humans, including the proteolytic release and activation of membrane-anchored, cytosolic transcription factors that then translocate into the nucleus to activate transcription of lipid metabolism genes (1, 20, 56). Specifically, in yeast, Δ9 fatty acid desaturase gene OLE1 and other lipid synthesis genes are regulated by the homologous, functionally related transcription activators Mga2p and Spt23p (75). Mga2p and Spt23p precursor forms (p120s) are sequestered in the ER membrane through C-terminal membrane-spanning domains (Fig. 1A). Upon changes in lipid composition or environmental stresses, Mga2p and Spt23p are mono- or oligoubiquitinated by ubiquitin ligase Rsp5p, the C-terminal membrane anchors are recognized and degraded by the proteasome, and their N-terminal transcription activation domains (p90s) are released and translocated into the nucleus to induce transcription of OLE1 and other genes linked to lipid metabolism (Fig. 1A) (7, 8, 25, 26, 54–56, 65). Loss of Doa4p function might inhibit Mga2p and Spt23p activation and thus OLE1 expression because consequent reduction of free Ub levels may inhibit Mga2p and Spt23p ubiquitination. Alternatively, or in addition, since a subset of Doa4p is associated with proteasomes (50), deletion of DOA4 might alter proteasome function and lead to inefficient Mga2p and Spt23p processing. Moreover, as shown above, loss of DOA4 function mirrors the effects of reduced Ole1p activity in inhibiting BMV RNA replication at a point between RNA replication complex assembly and negative-strand RNA synthesis (40).
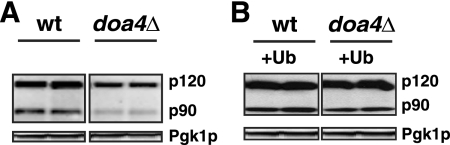
To compare levels of accumulation of the transcriptionally inactive precursor p120 and processed, active p90 forms of Mga2p and Spt23p in wt and doa4Δ cells, we expressed N-terminally Myc-tagged Mga2p and Spt23p, allowing both p120 and p90 to be detected using an anti-Myc antibody (26, 55). For standard growth conditions and the yeast BY4743 strain used here, Mga2p produced a much stronger Western blot signal and so was used for further analysis (Fig. 5). While the levels of accumulated BMV 1a, 2apol, and host protein Pgk1p (Fig. 4A and 5) were similar in both wt and doa4Δ cells, accumulation of both the p120 and p90 forms of Mga2p decreased in doa4Δ cells. However, the decrease in accumulation of the transcriptionally active p90 form was more dramatic (Fig. 5A), consistent with a decrease in Mga2p processing. The reason for the reduction in p120 levels in doa4Δ cells is currently unclear, but this reduction was minor (22 to 28%) compared to the 80 to 90% inhibition of BMV RNA replication. To determine if limited free Ub caused the reduced activation of Mga2p in doa4Δ cells, Ub was supplemented from plasmid pYEP96-U to wt levels (Fig. 2A). This increased accumulation of Mga2p p90 close to wt levels (Fig. 5B), showing that the reduced in free Ub in doa4Δ cells is the major cause of inhibited Mga2p activation.
Fig. 5.
Activation of transcription factor Mga2p is inhibited in doa4Δ cells. N-terminally Myc-tagged Mga2p was expressed in wt and doa4Δ cells alone in panel A or along with Ub and BMV 1a in panel B. Accumulation of Mga2p p120 and p90 was detected with an anti-Myc monoclonal antibody by Western blotting.
Inhibition of BMV RNA replication in doa4Δ cells is linked to inhibition of OLE1 expression.
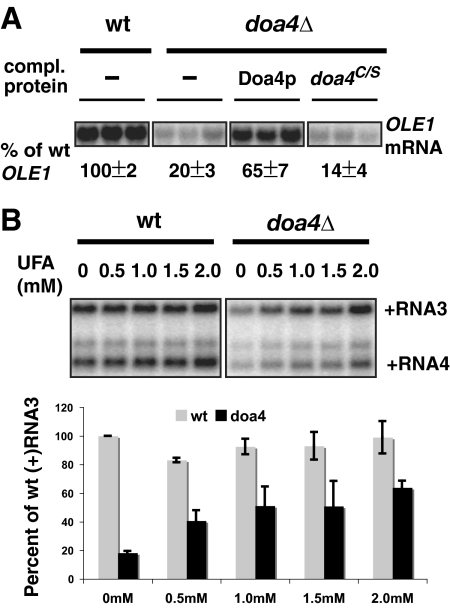
Since Mga2p and Spt23p regulate OLE1 transcription, we analyzed OLE1 mRNA levels. In doa4Δ cells, OLE1 mRNA accumulation was typically reduced 3-fold or more below wt levels (Fig. 6A). We similarly observed a 3-fold reduction of OLE1 transcript levels in cells overexpressing RFU1 (data not shown). This is consistent with earlier work showing that overexpression of RFU1 inhibited Doa4p activity and decreased Ub levels (34). Plasmid-based expression of wt DOA4 in doa4Δ cells restored accumulation of OLE1 mRNA (Fig. 6A) in parallel to BMV RNA replication (Fig. 1B). Moreover, accumulation of OLE1 mRNA was not increased in doa4Δ cells expressing the enzymatically inactive doa4C/S mutant (Fig. 6A), demonstrating a clear correlation between Doa4p enzymatic activity, OLE1 mRNA accumulation, and BMV RNA replication.
Fig. 6.
Inhibited BMV RNA replication in doa4Δ cells is linked to inhibition of OLE1 expression. (A) Decreased accumulation of OLE1 mRNA correlates with defective BMV RNA replication in doa4Δ cells. OLE1 mRNA was detected by Northern blotting with an OLE1-specific probe. The RNA signals were normalized to that of 18S rRNA. (B) BMV RNA replication was measured after addition of increasing concentrations of UFA (an equimolar mixture of palmitoleic and oleic acids) to the yeast growth medium. Accumulation of BMV RNA was detected as in Fig. 1. Values represent the means of four independent repeats.
To test if decreased BMV RNA replication was directly related to the downregulation of OLE1 activity in doa4Δ cells, we provided increasing amounts of the unsaturated fatty acid (UFA) products of OLE1 (an equimolar mixture of palmitoleic acid and oleic acid) in the growth medium. Adding up to 2 mM UFA did not affect the growth of wt and doa4Δ strains (data not shown). Supplemented UFA did not enhance BMV RNA replication in wt cells, but restored BMV RNA replication in a dose-dependent manner in doa4Δ cells (Fig. 6B). Accumulation of positive-strand viral RNA3 increased more than 3-fold at the highest UFA concentration, to about 65% of wt RNA replication levels. Levels of RNA4, which was more strongly inhibited than RNA3 in doa4Δ cells (Fig. 1B), saw a correspondingly greater 4-fold increase upon UFA addition, from ∼10% to 40% compared to wt levels (Fig. 6B). However, we never observed full complementation of BMV RNA replication with addition of UFA, as can be achieved in the ole1w and ole1Δ mutants (40). This implied that decreased OLE1 expression only partially accounts for the inhibition of BMV RNA replication in doa4Δ cells, as is addressed in the next section.
Other Mga2p-regulated genes are involved in BMV RNA replication.
Besides OLE1, whose expression has been extensively studied, Mga2p and Spt23p collectively regulate at least 30 genes involved in lipid metabolism, including those synthesizing fatty acids (FAS1 and ACC1), sterols (ERG1, -3, -5, and -26), and sphingolipids (SUR1 and -4) (7). However, Mga2p and Spt23p regulate overlapping but not exactly matching sets of genes. While OLE1 expression is induced by either Mga2p or Spt23p, other lipid metabolism genes, such as SUR4 and ERG5, are preferentially induced by Mga2p (7). To test if other Spt23p- and/or Mga2p-regulated genes besides OLE1 might be partially responsible for inhibiting BMV RNA replication in doa4Δ cells, we tested BMV RNA replication in mga2Δ and spt23Δ cells. In both cell types, OLE1 transcripts accumulated to wt levels (Fig. 7), which was expected since either Mga2p or Spt23p is sufficient to activate OLE1 expression (75).
Fig. 7.
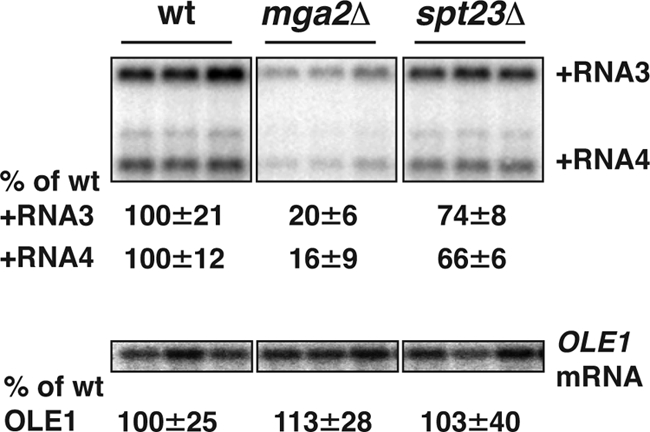
BMV RNA replication is inhibited in mga2Δ mutant cells. BMV components were expressed in wt, mga2Δ, and spt23Δ cells. Accumulation of BMV RNAs and OLE1 mRNA was assessed as in Fig. 1 with BMV RNA- or OLE1-specific probes.
Despite unaltered levels of OLE1 mRNA, deletion of MGA2 inhibited BMV RNA replication by more than 5-fold, compared to a mild decrease (25%) upon deletion of SPT23 (Fig. 7). This implies that one or more other genes specifically or preferentially regulated by Mga2p play important roles in BMV RNA replication.
DOA4-independent depletion of free Ub inhibits Mga2p activation and BMV RNA replication.
We tested if deleting free Ub would affect Mga2p and Spt23p activation, the consequent OLE1 expression, and BMV RNA replication as a general phenomenon not specific to deletion or inhibition of DOA4. Among 60 host gene deletion mutants previously found to inhibit BMV RNA replication (38), the ufd3Δ and ubp6Δ mutants have depleted free Ub levels (30, 41, 61). Ubp6p is a deubiquitinating enzyme and an accessory component of the proteasome (Fig. 1A) (5, 41). Ufd3p is involved in protein turnover via an interaction with CDC48, a critical component of a major pathway for delivering substrate proteins to proteasomes (30, 41, 61). Ufd3p also regulates Spt23p stability (61). Deletion of UFD3 and UBP6 may affect Spt23p and Mga2p activation in at least two ways. First, as in doa4Δ cells, decreased free Ub levels in ufd3Δ and ubp6Δ cells may inhibit ubiquitination and subsequent activation of Mga2p and Spt23p. Alternatively, or in addition, deletion of UBP6 may affect proteasome activity and thus Mga2p and Spt23p processing (41), while deletion of UFD3 may affect the stability of the processed, active Spt23p and Mga2p p90 forms (61).
Consistent with the results of Kushner et al. (38), BMV RNA replication was inhibited in ufd3Δ and ubp6Δ mutants to 5-fold and 10- to 20-fold, respectively (Fig. 8A). Also consistent with previous reports (30, 41, 61), free Ub levels were much reduced in both ufd3Δ and ubp6Δ mutants (Fig. 8B). While Mga2p p120 accumulated to near wt levels in ufd3Δ and ubp6Δ mutants, Mga2p p90 was much reduced (Fig. 8B), confirming that Mga2p processing was inhibited.
Fig. 8.
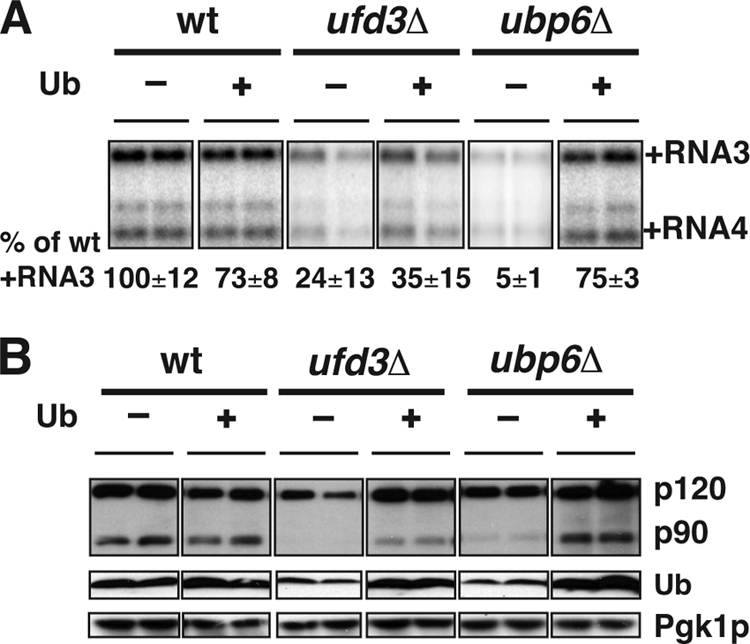
Inhibited Mga2p activation and BMV RNA replication can be restored or partially restored in ubp6Δ and ufd3Δ cells by supplementation with Ub. (A) BMV RNA replication and (B) Mga2p activation were assessed in wt, ufd3Δ, and ubp6Δ cells with or without the addition of Ub. BMV 1a was expressed in all cells in panel B. Accumulation of BMV RNAs and activation of Mga2p were analyzed as in Fig. 1 and 5, respectively.
To test if reduced Mga2p p90 accumulation and BMV RNA replication could be restored in these mutants by supplementation with Ub, pYEP96-U was used to express Ub to wt levels in both mutants (Fig. 8B). In ubp6Δ cells, providing Ub increased Mga2p p90 to near wt levels. On the other hand, supplementation with Ub only slightly increased Mga2p p90 accumulation in ufd3Δ cells, suggesting that deletion of UFD3 may affect both Mga2p activation and stability. Consistent with Mga2 p90 accumulation, BMV RNA replication was restored to wt levels in ubp6Δ cells but only increased from 20% to 35% in ufd3Δ cells (Fig. 8A).
DISCUSSION
The RNA replication complexes of BMV and many other positive-strand RNA viruses (17, 47, 62, 63), budding enveloped virions (10, 11, 48), and MVB intralumenal vesicles (27, 33, 53) are all topologically similar in being formed by invaginating membranes away from the cytoplasm. Just as budding of many enveloped virions depends on MVB components, prior genomewide screening showed that efficient BMV RNA replication requires MVB genes (38), and recent results show that MVB components also are transiently recruited by another positive-strand RNA virus, tomato bushy stunt virus, to peroxisome membranes, where they promote RNA replication (9).
Here we investigated how two MVB genes, the deubiquitinase gene DOA4 and the multifunctional gene BRO1, contribute to BMV RNA replication. The mammalian homologs of DOA4 and BRO1 are the genes coding for Ubpy and Alix (59, 60). Notably, Alix has emerged as a key player in the MVB-dependent budding of multiple retroviruses and some other viruses, although the precise nature of its functions in such viral budding remain under study (14, 46, 48). Nevertheless, many features of BRO1/ALIX and DOA4/Ubpy function appear conserved from yeast to mammals. Among these, in parallel to Bro1p-mediated recruitment of Doa4p for MVB cargo deubiquitination (59), recruitment of Bro1p homolog Alix to HIV-1 Gag promotes Gag deubiquitination (45).
Despite the topological similarities of membrane-bound BMV RNA replication compartments and MVB compartments, we found that the roles of DOA4 and BRO1 in BMV RNA replication were independent of the protein- and membrane-sorting functions of the MVB pathway. Rather, as detailed below, defects in Doa4p or Bro1p or their interactions disrupted BMV RNA replication by depleting free Ub levels and thereby inhibiting induction of OLE1 fatty acid desaturase and other lipid synthesis genes required by BMV RNA replication (Fig. 2, 5, 6, and 7). Possible implications of these results for virus control are also considered.
DOA4-, BRO1-, and Ub-dependent activation of lipid synthesis genes.
Multiple results showed that the BMV RNA replication defects in doa4Δ and bro1Δ cells were linked to free Ub levels and the activation of lipid synthesis/modification genes, but not to the membrane-shaping functions of the MVB pathway. BMV RNA replication required Doa4p with a catalytically active deubiquitinase (Fig. 1B), a function that is dispensable for invaginating intralumenal vesicles to form MVBs (59) but essential to maintain normal free Ub levels (69). In keeping with this, across varied circumstances in doa4Δ cells there was a tight correlation between BMV RNA replication and the levels of free Ub (Fig. 2), the Ub-dependent activation of lipid synthesis regulator Mga2p (Fig. 5B), and the accumulation of OLE1 fatty acid desaturase mRNA (Fig. 6A). The mechanistic linkage between these observations was confirmed by showing that the BMV RNA replication defect in doa4Δ cells was complemented by supplementation of free Ub levels (Fig. 2), which restored activation of Mga2p (Fig. 5B), or by feeding the UFA products of Ole1p fatty acid desaturase (Fig. 6B), thus bypassing Ub levels, Mga2p activation, and OLE1 expression.
BRO1, which was also required for BMV RNA replication, provides multiple functions to the MVB pathway. For example, unlike DOA4, deletion of BRO1 blocks the formation of normal MVB membrane compartments, yielding instead aberrant “class E” endosomes that are flattened cisternae without intralumenal vesicles (27, 33, 53). However, using mutations that selectively ablate specific functions, we found that BRO1 contributes to BMV RNA replication indirectly by interacting with Doa4p to stimulate its deubiquitinase activity and the accumulation of free Ub (Fig. 3B). Thus, in bro1Δ cells, BMV RNA replication was restored by expressing a Bro1p C-terminal fragment, bro1-C, that neither supports formation of functional MVB membrane compartments nor facilitates Doa4p recruitment to MVB sites, but interacts with and activates the Doa4p catalytic domain (59) and stimulates free Ub levels (Fig. 3B). Conversely, BMV RNA replication in bro1Δ cells was not restored by expressing the bro1-2 C-terminal-truncation mutant (Fig. 3B), which supports intralumenal vesicle and MVB compartment formation but does not stimulate Doa4p's deubiquitinase activity (59). In keeping with this, the doa4AAFA mutant, which disrupts the Doa4p side of the activating interaction with wt Bro1p, similarly blocked BMV RNA replication in doa4Δ cells (Fig. 3C). Thus, Doa4p's deubiquitinating activity, not Bro1p's possible role in inducing membrane curvature, is required for supporting BMV RNA replication.
Thus, all of the above results indicated that DOA4 and BRO1 primarily affected BMV RNA replication through their effects on free Ub levels and the Ub-dependent activation of lipid synthesis regulators Mga2p and Spt23p. While this underlying linkage between Ub levels, lipid synthesis, and BMV RNA synthesis was clear, some variations were noted between the levels of free Ub and associated effects on BMV RNA replication in the doa4Δ and bro1Δ mutants (Fig. 2A and 3B). One likely explanation for some variations in both Ub levels and inhibition of BMV RNA replication would be variations in culture growth conditions, which alter free Ub levels in the doa4Δ mutant. Specifically, Ub levels in the doa4Δ strain reach ∼1/3 of those in wt cells during log-phase growth, but fall to only ∼1/10 of those in wt cells in the stationary phase (69).
We further tested and validated the linkage of free Ub levels, lipid synthesis activation, and BMV RNA replication by using mutations in other host genes that modulate free Ub levels. As predicted, overexpressing RFU1, which inhibits Doa4p's enzymatic activity and depletes free Ub (34), markedly reduced OLE1 transcript levels (results not shown) and BMV RNA replication in wt cells (Fig. 1C). Moreover, DOA4-independent inhibition of BMV RNA replication in ubp6Δ and ufd3Δ cells also strongly correlated with decreased free Ub levels and deficient Mga2p activation (Fig. 8).
In both plant cells and yeast, subgenomic RNA4 accumulation is much more sensitive than genomic RNA3 accumulation to slight variations in inoculation, growth conditions, host genotype, etc. In keeping with this, DOA4 deletion inhibited RNA4 accumulation more significantly than RNA3 accumulation, as shown in Fig. 1B and elsewhere. Thus, while both RNA3 synthesis and RNA4 synthesis are inhibited by DOA4 deletion and restored by supplying Ub or unsaturated fatty acids, they do have different sensitivities to these and other effects.
Mga2p-regulated genes beyond OLE1 contribute to BMV RNA replication.
OLE1 mutations inhibit BMV RNA replication ≥20-fold, but are completely complemented by supplying the UFA products of OLE1 in the medium (40). In contrast, the BMV RNA replication defect in doa4Δ cells was only partially complemented by supplementing cells with UFA (Fig. 6B), implying that OLE1 was not the only BMV-required gene affected. Consistent with this, Mga2p and Spt23p control many genes involved in fatty acid, sphingolipid, and sterol metabolism (7), and BMV RNA replication depends on many lipid synthesis modification and transport genes beyond OLE1 (38). Furthermore, although both accumulated wt levels of OLE1 mRNA, BMV RNA replication was inhibited 5-fold in mga2Δ cells but by only ∼25% in spt23Δ cells.
Accordingly, genes other than OLE1 that are preferentially controlled by Mga2p might account for the residual inhibition of BMV RNA replication that is not complemented by UFA feeding (Fig. 7). Mga2p was predominantly expressed over Spt23p in the BY4743 strain used here (Fig. 5B), which may partly account for the greater defect in mga2Δ cells. Additionally, the promoters of multiple lipid metabolism genes are more strongly bound by Mga2p than Spt23p, including genes involved in synthesizing sterols important for the replication of other positive-strand RNA viruses, including hepatitis C virus (HCV) (73) and tomato bushy stunt virus (64).
Relevance to bromovirus replication in plants.
The results presented here obtained with yeast reveal new aspects of host function required for BMV RNA replication and interconnections between key pathways required by BMV, including unsaturated fatty acid synthesis and Ub-dependent proteasomal processing. As noted above, these pathways show broad conservation across eukaryotes, and other recent results from our laboratory show that BMV RNA replication in plants also depends critically on both of these pathways. We find, e.g., that inhibiting of proteasome function suppresses BMV RNA replication in barley protoplasts (B. Gancarz and P. Ahlquist, unpublished data), confirming that proteasome-dependent pathways are essential for BMV replication in cells of a natural plant host. Moreover, in a particularly direct connection to the present results, we find that knocking down fatty acid desaturase activity dramatically inhibits BMV RNA replication in N. benthamiana (X. Wang, A. Diaz, and P. Ahlquist, unpublished data). Thus, as was originally found (39, 40) and further extended here using yeast, membrane lipid composition and unsaturated fatty acids in particular are also crucial for BMV RNA replication in a plant host. Together, these results and recent studies of the involvement of sterol synthesis in tombusvirus replication (64) show that multiple membrane-associated pathways linked to virus replication in yeast are also essential in natural hosts.
Relationship to other positive-strand RNA viruses.
Membrane lipid synthesis and composition are critical for RNA replication by many if not all positive-strand RNA viruses (13, 32, 38, 64, 66, 72–74). Inhibition of one or more aspects of fatty acid synthesis, e.g., inhibits replication of many viruses, including poliovirus (23), Semliki Forest virus (52), cowpea mosaic virus (12), BMV (39, 40), HCV (32), and Drosophila C virus (16). Similarly, cholesterol metabolic pathways are required by many viruses, such as HCV, for entry, RNA replication, and egress (73). Accordingly, the results presented here with a variety of host genes highlight broader potentials to control viral RNA replication and other aspects of infection by manipulating lipid synthesis and its regulation in novel ways. Advancement of understanding of the lipid dependencies of specific virus replication steps and the cellular regulatory circuits that control different aspects of lipid metabolism and their interaction with other pathways should enhance the potential for virus control while minimizing host toxicity. The wide importance of these pathways suggests the possibility of approaches effective against multiple viruses, and the broad conservation of many features of lipid regulation from yeast to humans (1, 20, 56) should facilitate generalizing such strategies across a range of practically important hosts.
ACKNOWLEDGMENTS
We thank Wai-Ming Lee for providing the OLE1 plasmid, Ling Liu, Jordan Maresh, and Nick Zaban for general assistance and all members of the Ahlquist lab for discussions and suggestions. We thank Daniel Finley of Harvard University, Mark Hochstrasser of Yale University, Stefan Jentsch of Max-Planck Institute of Biochemistry, Germany, and Greg Odorizzi of University of Colorado for providing related plasmids and antiserum.
This work was supported by NIH grant GM35072. A.D. was partially supported by NIH training grant T32 AI078985. P.A. is an investigator of Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Aguilar P. S., de Mendoza D. 2006. Control of fatty acid desaturation: a mechanism conserved from bacteria to humans. Mol. Microbiol. 62:1507–1514 [DOI] [PubMed] [Google Scholar]
- 2. Ahlquist P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahola T., Ahlquist P. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amerik A., Sindhi N., Hochstrasser M. 2006. A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J. Cell Biol. 175:825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amerik A. Y., Li S. J., Hochstrasser M. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981–992 [DOI] [PubMed] [Google Scholar]
- 6. Amerik A. Y., Nowak J., Swaminathan S., Hochstrasser M. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11:3365–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Auld K. L., Brown C. R., Casolari J. M., Komili S., Silver P. A. 2006. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell 21:861–871 [DOI] [PubMed] [Google Scholar]
- 8. Auld K. L., Silver P. A. 2006. Transcriptional regulation by the proteasome as a mechanism for cellular protein homeostasis. Cell Cycle 5:1503–1505 [DOI] [PubMed] [Google Scholar]
- 9. Barajas D., Jiang Y., Nagy P. D. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bieniasz P. D. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calistri A., Salata C., Parolin C., Palu G. 2009. Role of multivesicular bodies and their components in the egress of enveloped RNA viruses. Rev. Med. Virol. 19:31–45 [DOI] [PubMed] [Google Scholar]
- 12. Carette J. E., Stuiver M., Van Lent J., Wellink J., Van Kammen A. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castorena K. M., Stapleford K. A., Miller D. J. 2010. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics 11:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chatellard-Causse C., et al. 2002. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J. Biol. Chem. 277:29108–29115 [DOI] [PubMed] [Google Scholar]
- 15. Chen J., Ahlquist P. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cherry S., et al. 2006. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. den Boon J., Diaz A., Ahlquist 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. den Boon J. A., Chen J., Ahlquist P. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diaz A., Wang X., Ahlquist P. 2010. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. U. S. A. 107:16291–16296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B. 2002. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296:879–883 [DOI] [PubMed] [Google Scholar]
- 21. Dupre S., Haguenauer-Tsapis R. 2001. Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol. Cell. Biol. 21:4482–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finley D., et al. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14:5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guinea R., Carrasco L. 1990. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 9:2011–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hegemann J. H., et al. 1999. A fast method to diagnose chromosome and plasmid loss in Saccharomyces cerevisiae strains. Yeast 15:1009–1019 [DOI] [PubMed] [Google Scholar]
- 25. Hitchcock A. L., et al. 2001. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12:3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoppe T., et al. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102:577–586 [DOI] [PubMed] [Google Scholar]
- 27. Hurley J. H., Emr S. D. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35:277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janda M., Ahlquist P. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. U. S. A. 95:2227–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janda M., Ahlquist P. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961–970 [DOI] [PubMed] [Google Scholar]
- 30. Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456 [DOI] [PubMed] [Google Scholar]
- 31. Kao C. C., Ahlquist P. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66:7293–7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kapadia S. B., Chisari F. V. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katzmann D. J., Odorizzi G., Emr S. D. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905 [DOI] [PubMed] [Google Scholar]
- 34. Kimura Y., et al. 2009. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137:549–559 [DOI] [PubMed] [Google Scholar]
- 35. Kong F., Sivakumaran K., Kao C. 1999. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200–210 [DOI] [PubMed] [Google Scholar]
- 36. Kopek B. G., Perkins G., Miller D. J., Ellisman M. H., Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krol M. A., et al. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. U. S. A. 96:13650–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kushner D. B., et al. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764–15769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee W. M., Ahlquist P. 2003. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 77:12819–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee W. M., Ishikawa M., Ahlquist P. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leggett D. S., et al. 2002. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10:495–507 [DOI] [PubMed] [Google Scholar]
- 42. Liu L., et al. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5:e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Losko S., Kopp F., Kranz A., Kolling R. 2001. Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell 12:1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luhtala N., Odorizzi G. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin-Serrano J., Perez-Caballero D., Bieniasz P. D. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuo H., et al. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303:531–534 [DOI] [PubMed] [Google Scholar]
- 47. Miller S., Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morita E., Sundquist W. I. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425 [DOI] [PubMed] [Google Scholar]
- 49. Nikko E., Andre B. 2007. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot. Cell 6:1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Papa F. R., Amerik A. Y., Hochstrasser M. 1999. Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol. Biol. Cell 10:741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Papa F. R., Hochstrasser M. 1993. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366:313–319 [DOI] [PubMed] [Google Scholar]
- 52. Perez L., Guinea R., Carrasco L. 1991. Synthesis of Semliki Forest virus RNA requires continuous lipid synthesis. Virology 183:74–82 [DOI] [PubMed] [Google Scholar]
- 53. Piper R. C., Katzmann D. J. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Piwko W., Jentsch S. 2006. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat. Struct. Mol. Biol. 13:691–697 [DOI] [PubMed] [Google Scholar]
- 55. Rape M., et al. 2001. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107:667–677 [DOI] [PubMed] [Google Scholar]
- 56. Rape M., Jentsch S. 2004. Productive RUPture: activation of transcription factors by proteasomal processing. Biochim. Biophys. Acta 1695:209–213 [DOI] [PubMed] [Google Scholar]
- 57. Restrepo-Hartwig M., Ahlquist P. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Restrepo-Hartwig M. A., Ahlquist P. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Richter C., West M., Odorizzi G. 2007. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 26:2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Row P. E., Prior I. A., McCullough J., Clague M. J., Urbe S. 2006. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281:12618–12624 [DOI] [PubMed] [Google Scholar]
- 61. Rumpf S., Jentsch S. 2006. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 21:261–269 [DOI] [PubMed] [Google Scholar]
- 62. Salonen A., Ahola T., Kaariainen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schwartz M., et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 64. Sharma M., Sasvari Z., Nagy P. D. 2010. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J. Virol. 84:2270–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shcherbik N., Zoladek T., Nickels J. T., Haines D. S. 2003. Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr. Biol. 13:1227–1233 [DOI] [PubMed] [Google Scholar]
- 66. Stapleford K. A., Rapaport D., Miller D. J. 2009. Mitochondrion-enriched anionic phospholipids facilitate Flock House virus RNA polymerase membrane association. J. Virol. 83:4498–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stukey J. E., McDonough V. M., Martin C. E. 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264:16537–16544 [PubMed] [Google Scholar]
- 68. Sullivan M. L., Ahlquist P. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Swaminathan S., Amerik A. Y., Hochstrasser M. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10:2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang X., Ahlquist P. 2008. Brome mosaic virus (Bromoviridae), p. 381–386 In Mahy B. W. J., van Regenmortel M. H. V. (ed.), Encyclopedia of virology, 3rd ed. Academic Press, New York, NY [Google Scholar]
- 71. Wang X., et al. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu S. X., Ahlquist P., Kaesberg P. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. U. S. A. 89:11136–11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ye J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ye J., et al. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. U. S. A. 100:15865–15870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang S., Skalsky Y., Garfinkel D. J. 1999. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151:473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]