Abstract
The 288-nucleotide (nt) 3′ untranslated region (UTR) in the genome of the bovine coronavirus (BCoV) and 339-nt 3′ UTR in the severe acute respiratory syndrome (SARS) coronavirus (SCoV) can each replace the 301-nt 3′ UTR in the mouse hepatitis coronavirus (MHV) for virus replication, thus demonstrating common 3′ cis-replication signals. Here, we show that replacing the 209-nt MHV 5′ UTR with the ∼63%-sequence-identical 210-nt BCoV 5′ UTR by reverse genetics does not yield viable virus, suggesting 5′ end signals are more stringent or possibly are not strictly 5′ UTR confined. To identify potential smaller, 5′-common signals, each of three stem-loop (SL) signaling domains and one inter-stem-loop domain from the BCoV 5′ UTR was tested by replacing its counterpart in the MHV genome. The SLI/II domain (nucleotides 1 to 84) and SLIII domain (nucleotides 85 to 141) each immediately enabled near-wild-type (wt) MHV-like progeny, thus behaving similarly to comparable 5′-proximal regions of the SCoV 5′ UTR as shown by others. The inter-stem-loop domain (nt 142 to 173 between SLs III and IV) enabled small plaques only after genetic adaptation. The SLIV domain (nt 174 to 210) required a 16-nt extension into BCoV open reading frame 1 (ORF1) for apparent stabilization of a longer BCoV SLIV (nt 174 to 226) and optimal virus replication. Surprisingly, pleiomorphic SLIV structures, including a terminal loop deletion, were found among debilitated progeny from intra-SLIV chimeras. The results show the inter-stem-loop domain to be a potential novel species-specific cis-replication element and that cis-acting SLIV in the viral genome extends into ORF1 in a manner that stabilizes its lower stem and is thus not 5′ UTR confined.
INTRODUCTION
cis-acting structures in positive-strand [(+)-strand] RNA virus genomes that function as signals for genome translation, transcription, and replication are potential sites for antiviral drug design. The fact that replication signals from evolutionarily divergent coronaviruses can be exchanged, for example, from the 3′ untranslated region (UTR) of the (group 2a) bovine coronavirus (BCoV) to the (group 2a) mouse hepatitis coronavirus (MHV) (21–22) or from the (group 2b) severe acute respiratory syndrome (SARS) coronavirus (SCoV) to MHV (15), would suggest that designed therapeutic agents against common signals might be broadly effective. Several studies have identified cis-replication structures in the 3′ UTR of MHV, heretofore recognized as the group 2 type species, and in the 3′ UTR of BCoV that are likely involved in the initial steps of genome translation and initiation of negative-strand [(−)-strand] synthesis. These elements, identified in either a helper virus-dependent defective interfering (DI) RNA of MHV or BCoV or by reverse genetics in the full-length MHV genome, are the following: (i) the 3′ poly(A) tail (26, 34), (ii) the 3′-terminal 55 nucleotides (nt) of the 3′ UTR (14, 23, 26, 42), and (iii) an upstream bulged stem-loop (SL) and associated hairpin pseudoknot (13, 21–22, 36). Curiously, an ∼140-nt hypervariable region in the 3′ UTR that comprises part of a 3′-proximal bulged stem-loop and harbors a coronavirus-universal octamer sequence (GGAAGAGC) is not required for MHV replication in cell culture but plays a role in MHV pathogenesis (14).
cis-replication structures have also been identified in the 5′-proximal region of group 2 coronaviruses, but few studies have characterized their common signaling features. In BCoV, the very 5′ end of the genome exhibits a puzzling hypervariability during virus replication in that 5′ extensions of up to 9 nt and varying sequences within the first 14 nt are found, but the full significance of this in virus replication is unknown (19). The higher-order RNA structures within the BCoV 5′ UTR were initially predicted by the Tinoco algorithm (35) and more recently by the mfold algorithm of Zuker (31, 41). They have been characterized as SLs I to IV (Fig. 1B) and were shown to be consistent with enzyme structure probing analyses (5–6, 32–33). More recent characterizations based on comparisons with MHV have shown SLI to be comprised of two smaller stem-loops, named SL1 and SL2 (Fig. 1B) (24–25, 27–28). In the context of the DI RNA in BCoV-infected cells, it was not feasible to determine whether SLI or SLII is a bona fide cis-replication element, since SLI resides within the 65-nt leader and SLII resides largely within the leader, and transfected DI RNAs containing an experimentally mutated leader rapidly acquired the leader sequence of the helper virus genome in a process known as “leader switching” (5–6, 29). SLII, furthermore, harbors the leader-associated transcription regulatory core sequence (UCUAAAC) responsible for the RNA-dependent RNA polymerase (RdRp) template switch that causes leader switching (Fig. 1B) (37, 39). It was shown, however, that the 5′-terminal 13 nt which comprise part of SL1 are required for DI RNA replication, making the formerly defined SLI a probable cis-replication element (5). By contrast, short forms of SLIII and SLIV (indicated by the shaded areas in Fig. 1B) were shown by mutation analyses to be required for DI RNA replication and were therefore characterized as cis-replication signals (32–33). BCoV SLIII and its predicted homolog in other coronaviruses have associated with them the start codon for a short upstream open reading frame (uORF) also found in the 5′ UTRs of virtually all coronaviruses (32). The coronavirus short uORF potentially encodes typically 8 to 11 amino acids (aa), but its function has not been established. Recent mfold analyses have predicted BCoV SLIII to be 46 nt in length rather than 20 nt as previously predicted (Fig. 1B) (32), and still other algorithms have predicted it to be 53 nt in length (28). Between SLIII and a predicted SLIV in BCoV is a 44-nt region for which no function has yet been ascribed and within which the 3′-proximal 32-nt portion appears relatively unstructured by mfold prediction (Fig. 1B and data not shown). SLIV in BCoV and its predicted homolog in other coronaviruses (33) have associated with them the start codon for open reading frame 1 (ORF1), a ∼20-kb ORF encoding 16 nonstructural proteins (nsp's) that make up the replicase/transcriptase complex (16). Downstream of SLIV and within the partial ORF1 in BCoV DI RNA (a region that is also within the nonstructural protein 1 [nsp1] cistron) (1, 4) are SLV and SLVI, which have been shown by stem disruption and restoration studies to be cis-replication structures for DI RNA (3, 17).
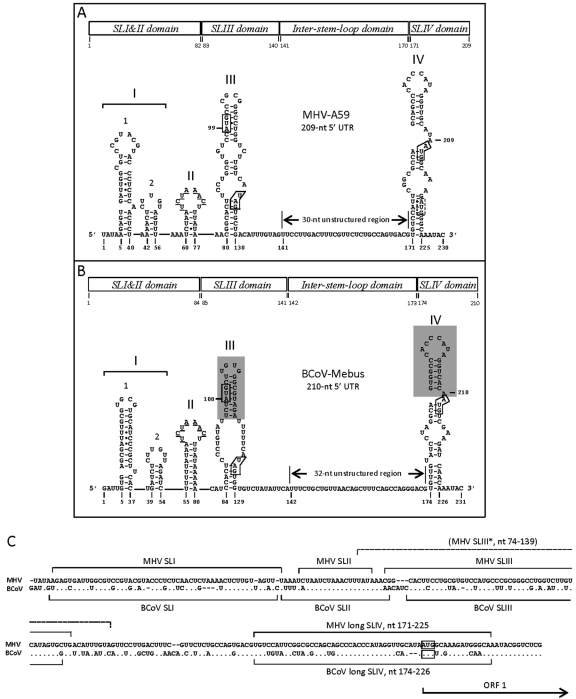
Fig. 1.
RNA structures in the 5′ untranslated region of MHV and BCoV genomes. Higher-order structures are shown for MHV-A59 (A) and BCoV-Mebus (B). For both viruses, the structures of stem-loops I (comprised of two smaller stem-loops, named 1 and 2), II, III, and IV, an ∼30-nt inter-stem-loop domain between stem-loops III and IV, and a short piece of ORF1 are shown. The solid circles in stems 1, II, and III of MHV and stem 1 of BCoV represent non-Watson-Crick base pairings as identified by Liu et al. (28) and Li et al. (25). The bases making up the UCUAAAC transcription regulatory core sequence are underlined. The previously-described “short” forms of stem-loops III and IV for BCoV as identified by Raman et al. (32–33) are highlighted in gray. The AUG start and UAG stop codons for the SLIII-associated intra-5′ UTR ORF and the SLIV-associated AUG start codon for the nsp1 ORF are boxed. A second in-frame AUG codon within the MHV ORF1 is boxed with a dashed line. The domains used for testing cis-replication function are defined at the top of each figure. (C) Sequence alignments of the 5′-proximal ∼237 nt of MHV-A59 and BCoV-Mebus. Sequences are aligned with gaps to maximize nucleotide identities, and differences are noted. The nucleotide positions of stem-loops as drawn in panels A and B are indicated. The asterisk indicates a 66-nt SLIII in MHV-A59, predicted and named SL4 by Kang et al. (24), is identified with a dashed bracket. The AUG start codons for ORF 1 are boxed.
In MHV, a series of elegant physical-chemical studies has been carried out in conjunction with consensus covariation modeling and reverse genetics to characterize cis-replication stem-loops in the 5′-terminal 140 nt of the 209-nt 5′ UTR (Fig. 1A) (24–25, 27–28). With solution nuclear magnetic resonance (NMR) spectroscopy, the 36-nt SL1 (nucleotides 5 to 40) was shown to be functionally and structurally bipartite (25). That is, for virus replication, the upper part of SL1 must be stable with Watson and Crick base pairing whereas the lower part of the stem must maintain a certain level of lability. A mutation that stabilized the lower stem of SL1 led to progeny carrying destabilizing compensatory mutations in the lower stem and also to mutations at one of two sites in the extreme 3′ end of the 3′ UTR, suggesting a possible physical interaction between the 5′ and 3′ UTRs (25). The 15-nt SL2 (nt 42 to 56) was shown to carry a pentaloop (C47-U48-U49-G50-U51) on a 5-base-pair stem, wherein the loop contains a 5′ YNMG(N)n tetraloop motif, where n is 0 or 1, a highly conserved motif in SL2 structures of group 1 and group 2 coronaviruses (27–28). Both SL1 and SL2 are predicted regulators of (−)-strand synthesis (25, 28). The MHV homolog of BCoV SLII, an 18-nt structure as predicted by mfold (nt 60 to 77), was predicted by Kang et al. (24) to be a single-stranded or weakly folded structure. The MHV homolog of BCoV SLIII, a 51-nt structure as predicted by mfold (nt 80 to 130), was predicted by Kang et al. (24) using alternative algorithms to be a 66-nt-long stem-loop (nt 74 to 139, named SL4) (Fig. 1C).
Overall, the predictions for the 5′ UTR higher-order structures in BCoV and MHV show similarities, suggesting they might be functionally conserved among group 2a coronaviruses. In the study by Kang et al. (24), SCoV stem-loops 1, 2, and 4 (SL4 is equivalent to SLIII in the current report) were able to substitute for their MHV counterparts in the MHV background when studied by reverse genetics. The SCoV SLIV, which in mfold predictions by us (33) and others (7) appeared to be more group 1-like, failed to support virus replication in the MHV background (24).
Here we postulated that since the group 2a MHV and BCoV genomes are structurally similar, their 5′ UTRs would be interchangeable, as were their 3′ UTRs (21–22). An initial set of experiments, however, demonstrated that a precise replacement of the 209-nt MHV 5′ UTR with the 210-nt BCoV 5′ UTR yielded no viable progeny. We therefore made replacements of regionally characterized cis-acting BCoV 5′ UTR structures in an effort to identify common 5′-proximal cis-replication signals that would function for MHV genome replication. From these and extended analyses, which included determining the sequence of a 5′-proximal 1,072 nt (nt 22 to 1093) and 3′-proximal 500 nt [excluding the poly(A) tail] in progeny genomes, we learned the following: (i) that the combined SLI-SLII region and the SLIII region can directly functionally replace homologous regions in the MHV background, (ii) that the 32-nt relatively unstructured region between SLs III and IV supports production of virus with slow-growing features after two blind cell passages and genetic adaptation, and (iii) that only after a 16-nt extension of the BCoV SLIV into BCoV ORF1 to form a predicted stabilizing “long” SLIV did BCoV SLIV function in the MHV background to form an apparently fully fit phenotype without concurrent adaptive mutations. This led to the hypothesis that stability in the lower stem contributes to fitness. Additionally, a study of six intra-SLIV BCoV/MHV chimeras yielded results that, with one exception, supported this hypothesis. Furthermore, a study of two chimeras with mutations covering extensive regions of SLIV revealed, surprisingly, that variant SLIV structures in progeny genomes, including internal deletions, can be tolerated for virus replication. These results together establish that the inter-stem-loop domain between SLIII and SLIV is a potential novel species-specific cis-replication element and that the most fit BCoV or MHV cis-replication SLIV in the MHV background spans the junction of the 5′ untranslated region and ORF1 in a manner that stabilizes the lower stem.
MATERIALS AND METHODS
Cells.
Delayed brain tumor (DBT) cells (18), mouse L2 cells (12), and baby hamster kidney cells expressing the MHV receptor (BHK-MHVR) (8–9) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% defined fetal calf serum (FCS) (HyClone) and 20 μg/ml gentamicin (Invitrogen). Cells were maintained at 37°C with 5% CO2 for all experiments. BHK-MHVR cells were maintained in selection medium containing 0.8 mg/ml Geneticin (G418 sulfate; Invitrogen) (40).
Viruses and GenBank accession numbers.
GenBank accession numbers for virus sequences analyzed were as follows: NC_012936 for Parker rat CoV, NC_001846 for MHV-A59, NC_006852 for MHV-JHM, AF201929 for MHV-2, NC_006577 for human coronavirus HKU1 (HCoV-HKU1), NC_010327 for equine coronavirus (ECoV), U00735 for BCoV-Mebus, NC_003045 for BCoV-ENT, NC_005147 for HCoV-OC43, FJ415324 for HECoV-4408, NC_007732 for PHEV, and NC_004718 for SCoV-Tor2.
RNA structure prediction and sequence alignments.
The mfold program of Zuker (http://www.bioinfo.rpi.edu/zukerm/) (31, 41) was used for RNA structure predictions. 5′ UTR sequence alignments were made using Vector NTI Suite 8 software (Invitrogen).
Plasmid construction for mutations in fragment A of the infectious clone MHV-A59-1000.
Fragments A through G for the infectious clone of MHV-A59-1000 (icMHV) (40) were a kind gift from Ralph Baric, University of North Carolina. cDNA plasmid A (containing fragment A), which contains an upstream T7 promoter and nucleotides 1 to 4882 of the MHV genome and was prepared in plasmid pCR-XL-TopoA (Invitrogen), was used to replace 5′-proximal regions of the MHV genome with the homologous region of the BCoV-Mebus genome. A plasmid A equivalent for the BCoV-Mebus genome, nucleotides 1 to 4750, also containing an upstream T7 promoter, was prepared and used as the source for the BCoV sequences. Site-directed mutagenesis was carried out by overlap PCR with the appropriate primers containing the mutated sequences as described below. All PCRs used AccuPrime Taq polymerase (high fidelity) (Invitrogen) under conditions recommended by the manufacturer. Briefly, the first PCR was done with the primer T7startBCoV (5′GTCGGCCTCTTAATACGACTCACTATAGGATTGTGAGCGATTTGC3′) or T7startMHV (5′GTCGGCCTCTTAATACGACTCACTATAGTATAAGAGTGATTGGCGTCCG3′) and the designated reverse (+) primer, and the second PCR was done with primer MHV-Sca I (5′CGCCTTGCAGGAGTACTTACCCTTC3′) and the designated forward (−) primer. The (+) and (−) in the primer names here and throughout designate the polarity of the RNA to which the primer binds. PCR fragments were chromatographically purified (Qiaex II; Qiagen) and used in the third PCR with the primers T7startBCoV (or T7startMHV) and MHV-Sca I. The resulting PCR fragments were TA cloned into pCR-XL-Topo, followed by plasmid preparation and sequence confirmation. With purified cloned DNA, a restriction fragment exchange was made in order to minimize unwanted mutations in the wild-type (wt) icMHV plasmid DNA. Briefly, both wt icMHV plasmid A DNA and site-directed mutagenized Topo plasmid DNA were digested with the restriction enzymes SfiI and Sca I (New England BioLabs), whose sites are in the vector at nucleotide 1616 upstream from the T7 promoter and at nucleotide 910 in the MHV genome, respectively. The ∼2.5-kb SfiI-ScaI fragment from Topo clones was ligated with the ∼6.5-kb SfiI-ScaI-linearized fragment from the wt icMHV plasmid A DNA. The BCoV/MHV chimeric plasmid A was then used to create a recombinant virus with fragments B through G in the MHV-A59-1000 reverse genetics system.
Assembly of full-length MHV-A59-1000 infectious recombinant virus RNA.
wt and chimeric MHV viruses were produced as described by Yount et al. (40) and as modified by Denison et al. (11). Briefly, plasmids containing the seven cDNA cassettes (A [wt or mutagenized], B, C, D, E, F, and G) of the MHV-A59-1000 genome were digested with the appropriate restriction enzymes (New England BioLabs), followed by gel electrophoresis purification of the fragments (QIAEX II; Qiagen). Gel-purified DNA fragments were in vitro ligated with T4 DNA ligase (New England BioLabs) in a total reaction volume of 210 μl overnight at 4°C. Ligated cDNA was chloroform extracted, isopropanol precipitated, and size was confirmed by electrophoresis in a 0.6% nondenaturing agarose gel. Infectious MHV genomic RNA was generated in vitro with the mMessage mMachine T7 transcription kit (Ambion) from the ligated cDNA in a total reaction volume of 50 μl supplemented with 7.5 μl of 30 mM GTP. In vitro transcription was done at 40.5°C for 30 min, 37°C for 60 min, 40.5°C for 30 min, 37°C for 30 min, and 40.5°C for 30 min. In parallel, RNA transcripts encoding the MHV mRNA for the nucleocapsid protein (N) were generated from a cloned N cDNA plasmid containing an upstream T7 promoter using the mMessage mMachine in a total reaction volume of 25 μl at 37°C for 3 h. Both icMHV genomic RNA and N mRNA were treated with 5 μl Turbo DNase (Ambion) at 37°C for 30 min and mixed for electroporation. To prepare for transfection by electroporation, freshly confluent BHK-MHVR cells were separated by trypsinization, washed twice with phosphate-buffered saline (PBS), and resuspended in PBS at a concentration of ∼1 × 107 cells/ml. The RNA mixture was added to 650 μl of the resuspended BHK-MHVR cells in a 4-mm-gap cuvette (Phenix) and pulsed three times at 850 V with 25 μF in a Gene Pulser electroporator (Bio-Rad). Electroporated cells were then seeded onto a layer of ∼1 × 106 DBT cells in a 75-cm2 flask and incubated at 37°C. Virus viability was determined by syncytium development (cytopathic effect [CPE]) in cells in 72 h or less. Cultures that did not develop syncytia within 72 h of electroporation were blind passaged at 48-h intervals at least three times at 37°C in a further attempt to recover infectious virus. Unless otherwise noted, three independent experiments with this protocol were carried out before a mutated MHV genome was claimed to be nonviable.
Characterization of mutant (BCoV/MHV chimera) progeny by genome sequence analysis.
Routinely, supernatant fluids from cells that first showed CPE (either the transfected cells or cells after blind passaging) was named virus passage zero (VP0). When 80 to 100% of the VP0 virus-infected cells showed CPE, intracellular RNA was TRIzol (Invitrogen)-extracted and the viral genome was sequenced by reverse transcription-PCR (RT-PCR) for the 5′-proximal nucleotides 22 to 1093 and the 3′-terminal 500 nt [excluding the poly(A) tail]. VP0 virus was then used for virus titration, to determine plaque morphology, and as starting material for growth kinetics on DBT cells. If the sequenced regions of the genome showed evidence of mixed progeny genotypes, selected plaque-purified virus was grown and sequenced.
For analysis of the 5′ nucleotides 22 to 1093 of progeny virus genomes, extracted cellular RNA was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) using primer MHV-1094(+) (5′CGATCAACGTGCCAAGCCACAAGG3′), which binds MHV genomic nucleotides 1094 to 1117, and cDNA was PCR amplified with the primers MHV-leader(−) (5′TATAAGAGTGATTGGCGTCCG3′), which binds nucleotides 1 to 21 of the MHV antileader, or BCoV-leader(−) (5′GATTGTGAGCGATTTGCGTGCG3′), which binds nucleotides 1 to 22 of the BCoV antileader, and MHV-1094(+). PCR products were gel purified (Qiaex II; Qiagen) prior to automated sequencing with primers MHV(261-284)(−) (5′CCATGGATGCTTCCGAACGCATCG3′) and MHV(605-623)(+) (5′GTTACACAGGCAGACGCGC3′).
For analysis of the 3′ 500 nt of progeny virus genomes, the same procedure was used except that total RNA was reverse transcribed using the primer DI3(+) (5′CGGGATCCGTCGACACGCGTTTTTTTTTTTTTTTTTTTT3′), which binds the poly(A) tail, and the cDNA was PCR amplified with the primers MHV(30811-30830)(−) (5′GGATGGTGGTGCAGATGTGG3′), which binds the negative strand of virus nucleotides 30811 to 30830, and primer DI3(+). Gel-purified PCR products were sequenced with the primer MHV(30811-30830)(−).
Characterization of mutant (BCoV/MHV chimera) progeny by virus titration, plaque morphology, growth kinetics, and Northern analysis.
Virus titration and plaque characterizations were carried out on L2 cells. Briefly, freshly confluent L2 cells in 6-well flasks were inoculated with virus serially diluted in DMEM for 2 h at 37°C, washed once with PBS, and overlaid with DMEM supplemented with 1% low-melting-point agarose and 2% FCS. After incubation at 37°C for ∼60 h, the overlay was removed and cells were fixed with 10% formaldehyde at room temperature for 30 min, stained with 1% crystal violet solution for 10 min, washed with water, and air dried. Plaque sizes were identified as large (wt) if ≥2.5 mm, medium if 1.5 to 2.5 mm, or small if ≤1.5 mm in diameter. Representative plaque images were taken with a Nikon digital camera and prepared using Adobe Photoshop CS software.
To determine growth kinetics, freshly confluent DBT cells in 25-cm2 flasks (∼4 × 106 cells) were inoculated with wt or chimeric MHV VP0 virus in DMEM at a multiplicity of infection (MOI) of 0.01 for 2 h at 37°C. Cells were washed twice with DMEM, and 5 ml of DMEM containing 2% FCS was added; incubation was continued at 37°C. One-hundred-microliter samples of supernatant fluid were collected at the indicated times postinoculation, and viral titers were determined by plaque assays on L2 cells.
Northern analyses were done as previously described (20). Briefly, freshly confluent DBT cells in 25-cm2 flasks (∼4 × 106 cells) were infected with wt or chimeric viruses at an MOI of 0.01 PFU/cell. At 20 h postinfection (hpi), intracellular RNA was TRIzol extracted and one-tenth of the total RNA from one 25-cm2 flask (∼60 μg RNA total per 25-cm2 flask) was resolved by electrophoresis in a 1.0% agarose-formaldehyde gel at 150 V for 4 h. RNA was transferred to a HyBond N+ nylon membrane (Amersham Biosciences) by vacuum blotting for 3 h followed by UV cross-linking. After prehybridization of the membrane with NorthernMax prehybridization/hybridization buffer (Ambion) at 55°C for 4 h, the blot was probed at 55°C overnight with 20 pmol (∼4 × 105 cpm/pmol) of γ-32P-5′end-labeled 3′ UTR-specific oligonucleotide MHV(31094–31122)(+) (5′CAGCAAGACATCCATTCTGATAGAGAGTG3′), which binds MHV genomic nucleotides 31094 to 31122. Probed blots were exposed to Kodak XAR-5 film at −80°C for imaging, and images were prepared using Adobe Photoshop CS software.
RESULTS
A precise 210-nt BCoV 5′ UTR failed to functionally replace the 209-nt MHV 5′ UTR in the MHV genome.
Since the 288-nt 3′ UTR of BCoV can functionally replace the 301-nt 3′ UTR of MHV in the MHV genome (21–22) and the BCoV DI RNA can replicate in MHV-infected cells, indicating that BCoV cis-replication signals can be recognized by the MHV RNA replication machinery (38), we reasoned that a precise substitution of the BCoV 5′ UTR in the MHV genome might function for genome replication as well. To test this, a 5′ UTR replacement was made using the MHV reverse genetics system developed by Ralph Baric and colleagues (40). Replacement of the 209-nt MHV 5′ UTR with the 210-nt BCoV 5′ UTR as represented in chimera B1–210/M (Fig. 2A and B) yielded no viable virus as determined by syncytium formation during three blind cell passages in three separate transfection trials. We therefore chose to separately analyze shorter domains of the 5′ UTR to determine which components, if any, were compatible based on the identity of cis-replication elements described for BCoV DI RNA (5–6, 32–33).
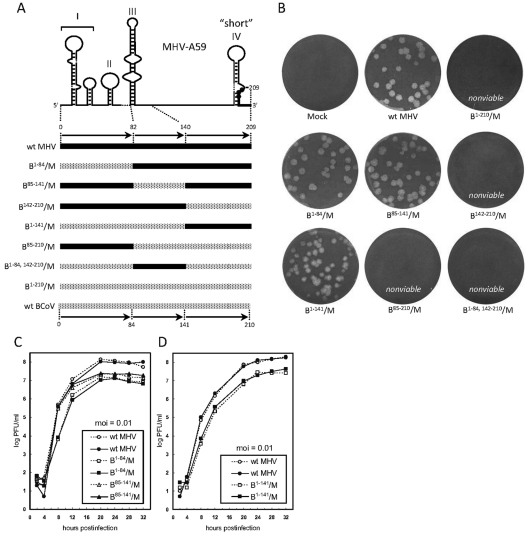
Fig. 2.
The BCoV SLI/II domain and SLIII domain, individually and together, support virus replication in the MHV background. (A) Scheme showing RNA domains in the BCoV 5′ UTR tested by reverse genetics for support of replication in the MHV genomic background. Stem-loop structures coinciding with domains in MHV (solid line) and BCoV (stippled line) are depicted. Filled circles on SLIV identify the AUG start codon for the nsp1 ORF. Note that the short form of stem-loop IV is depicted. (B) Plaque phenotypes for wt MHV and corresponding chimeras. Nonviable chimeras formed no visible syncytia during the first through third blind cell passages, and transfected cells appeared indistinguishable from mock-infected cells. (C) Growth kinetics of wt MHV, chimera B1–84/M, and chimera B85–141/M. (D) Growth kinetics of wt MHV and chimera B1–141/M. For growth kinetics, the results of two tests for each construct are shown.
The BCoV 5′ UTR SLI/II domain (nt 1 to 84) and SLIII domain (nt 85 to 141) each immediately functionally substituted for its MHV counterpart, but the 32-nt inter-stem-loop domain (nt 142 to 173) in combination with the 37-nt 5′ UTR-confined SLIV domain (nt 174 to 210) did not.
To test the hypothesis that individual parts of the BCoV 5′ UTR can functionally replace homologous regions in the MHV genome, the BCoV 5′ UTR was initially divided into three segments, the SLI/II domain (nucleotides 1 to 84), the SLIII domain (nucleotides 85 to 141), and the 32-nt inter-stem-loop domain between stem-loops III and IV (nucleotides 142 to 173) joined with the 37-nt 5′ UTR-confined SLIV domain (nucleotides 174 to 210) to form a 69-nt 142 to 210 segment as shown in Fig. 2A, and each was tested separately in a BCoV/MHV chimera. The choice to use MHV nucleotide 140 as the downstream border for MHV SLIII (Fig. 1C) was based on a prediction of MHV stem-loop III size (66 nt) by Kang et al. (24) that utilized two predictive algorithms in addition to mfold. (Note that in the report of Kang et al. [24], SLIII is named SL4.) In our tests, syncytia were detected within 24 h postelectroporation (hpe) from wt genomic RNA and within 24 to 48 hpe from cells electroporated with chimeric genomic RNAs containing the BCoV SLI/II domain (B1–84/M) or the BCoV SLIII domain (B85–141/M). In each case, wt-like large plaques appeared from VP0 virus (Fig. 2B). RT-PCR sequence analyses of progeny genomes from cells producing VP1 and VP10 for both chimeras revealed no additional mutations within genomic 5′ nucleotides 22 to 1093 and the 3′ 500 nt (data not shown), although they might have been present within the first 21 nt or within the remaining unsequenced part of the genome. The rationale for examining the 5′ nucleotides 22 to 1093 and 3′ 500 nt in progeny here and throughout this study was to identify potential reversions to MHV-like sequences within the transplanted domains and to identify potential compensatory mutations in cis-replication elements elsewhere within the 5′ UTR and adjacent nsp1 cistron (3, 17). Reports of long-range interactions between MHV genomic 5′ and 3′ ends (25) made it important to seek altered sequences within the 3′ UTR as well. For progeny of both B1–84/M and B85–141/M, growth kinetics using an MOI of 0.01 PFU/cell were nearly identical, with maximal titers of ∼1.0 × 107 PFU/ml, ∼10-fold less than for the wt, peaking at 20 hpi (Fig. 2C). With an MOI of 5 PFU/cell, results were nearly the same except that titers peaked at 12 hpi (not shown). A chimera with joined SLI/II and SLIII domains, B1–141/M (Fig. 2A), behaved similarly to B1–84/M and B85–141/M in that syncytia were observed within 48 hpe, large plaques were produced (Fig. 2B), and growth kinetics with an MOI of 0.01 PFU/cell showed a final titer of ∼1.0 × 107 PFU/ml (Fig. 2D). These results demonstrated that while the BCoV SLI/II and SLIII domains both differ in nucleotide sequence from their MHV counterparts by ∼36% (including spaces for alignment) (Fig. 1C), both supported replication of the chimera to yield slightly less fit virus than the MHV wt without reversions or compensatory nucleotide changes within the 5′-terminal 22 to 1,093 nt or 3′-terminal 500 nt. How nucleotides 1 to 21 within progeny from B1–84/M or B1–141/M might have changed was not determined at this time.
The B142–210/M chimera harboring the BCoV inter-stem-loop and SLIV domains, on the other hand, failed to develop CPE by 72 hpe, and no virus was recovered during three blind cell passages in three separate transfection trials (Fig. 2A and B). Similarly, chimeras harboring the BCoV SLIII, inter-stem-loop, and SLIV domains, B85–210/M, the BCoV SLI/II, inter-stem-loop, and SLIV domains, B1–84, 142–210/M, or all domains excepting the inter-stem-loop domain, B1–141, 174–210/M, yielded no viable progeny (Fig. 2A and B and 3A and B). These results together identified one or more regions within the BCoV inter-stem-loop domain or the BCoV SLIV domain or both that are incompatible with the remainder of the MHV genome.
The BCoV inter-stem-loop domain (nucleotides 142 to 173) and the BCoV long SLIV (nucleotides 174 to 226) together (B142–226/M) enabled viable but debilitated chimeric progeny.
Since mfold predicts a long SLIV for both MHV and BCoV which is formed in part by a 16-nt extension into its cognate ORF1 (Fig. 1A and B), we postulated that the 53-nt intra-SLIV chimera alone (wherein the first 37 nt are from BCoV and the second 16 nt are from MHV [Fig. 4B, left panel]) might be incompatible with the remaining genome. We therefore tested the 53-nt-long BCoV SLIV, in which the SLIV domain was extended 16 nt into the BCoV ORF1 in two chimeric constructs. In the first, the entire BCoV 1-to-226-nt region was tested in the chimera B1–226/M, but no viable progeny were generated (Fig. 3A and B). In the second, the long BCoV SLIV was joined with the BCoV 32-nt inter-stem-loop domain to form chimera B142–226/M, from which viral syncytia appeared after one blind cell passage and plaques produced from VP0 virus were heterogeneous in size (ranging from small to large) (Fig. 3B) and yielded a titer of 1.5 × 104 PFU/ml (not shown). Additionally, potential compensatory mutations were found both outside (C30A and A363G) and within (C158G, A163U, and G169U) the BCoV 32-nt inter-stem-loop region (described in the last section of Results). Growth of the chimera, therefore, although debilitated, indicated that the long BCoV SLIV does provide some survival benefit to the chimera since by comparison the B142–210/M chimera was nonviable (Fig. 2B). Because the entire 142-to-226-nt region was still not fully compatible with the MHV background as judged from the fitness of progeny for growth in cell culture, we separately tested the BCoV 32-nt inter-stem-loop domain and the BCoV long SLIV alone in the chimeras B142–173/M and B174–226/M, respectively. The results with the BCoV long SLIV in the MHV background are described first.
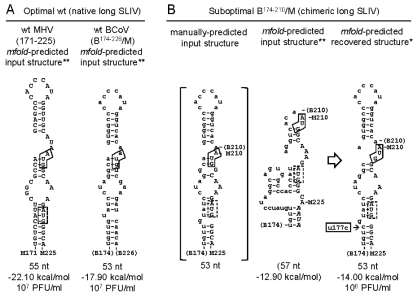
Fig. 4.
Optimal and suboptimal forms of SLIV in the MHV background. (A) Optimal forms of the native “long” SLIV are the wt MHV SLIV (left) and the BCoV SLIV (right) as mfold predicted in the context of nucleotides 1 to 400 of the MHV genome and as a standalone stem-loop. The negative free energy of the stem-loops and the titer of the wt or chimeric virus is shown. (B) Suboptimal form of the chimeric BCoV/MHV SLIV fused precisely at the junction between the BCoV 5′ UTR and the MHV ORF1. An inspection-based postulated structure (left) is compared with the mfold-predicted structure in the context of nucleotides 1 to 400 and as a standalone structure (center) and with the mfold-predicted stand-alone structure from progeny (right). Note that both the terminal loop and the stem are greatly rearranged in the mfold-predicted SLIV structure in the input chimera. Also note that virus from VP0-infected cells has a mutation in the base of the stem (U177C, boxed) which stabilizes the SL according to mfold prediction (right). The ORF1 start codon is boxed with a solid line, and a second, in-frame, intra-SLIV AUG codon in the MHV ORF1 is boxed with a dashed line. Structures identified by a double asterisk (**) are predicted by mfold as the most stable form both in the context of nucleotides 1 to 400 and as a standalone structure, and that identified by a single asterisk (*) is predicted to be the most stable form in a standalone structure. Nucleotides of MHV origin are denoted by uppercase letters, and those of BCoV origin are denoted by lowercase letters. Numbers identifying nucleotide positions in the BCoV genome are enclosed in parentheses.
Fig. 3.
BCoV “long” (i.e., 16-nt-extended) SLIV immediately supports MHV wt-like virus replication in the MHV background, but the BCoV 32-nt inter-stem-loop domain requires blind passage adaptation before progeny appear. (A) Scheme showing chimera constructs. Note that the long form of SLIV is depicted. (B) Plaque phenotypes of wt MHV and the identified chimeras. (C) Growth kinetics of wt MHV and B174–226/M chimera (left panel) and Northern analysis showing RNA patterns (right panel). (D) Growth kinetics of wt MHV and chimera B1–141, 174–226/M (left panel) and Northern analysis showing RNA patterns (right panel). For growth kinetics, the standard error of three measurements is shown. For Northern analyses, an MHV 3′-UTR-specific probe was used.
The BCoV long SLIV (in which SLIV is extended 16 nt into BCoV ORF1) alone in the MHV background (B174–226/M) is immediately optimal for virus replication, whereas the intra-SLIV BCoV/MHV chimera (B174–210/M) is suboptimal and fosters a putative compensatory nucleotide change in the lower stem.
When the chimera with the BCoV long SLIV alone, B174–226/M (Fig. 3A), was tested, syncytia formed within 24 hpe and plaques made from VP0 virus were wt MHV-like (Fig. 3B). Furthermore, the sequence of progeny genomes demonstrated no other mutations within the 5′ nucleotides 22 to 1093 or 3′ 500 nt (data not shown), growth kinetics were indistinguishable from those of wt MHV (Fig. 3C, left panel), and Northern blot analysis of RNA from infected cells displayed wt-like genomic and subgenomic mRNA patterns (Fig. 3C, right panel). These results therefore demonstrate that BCoV long SLIV alone is fully functional in the context of the MHV background despite a 19-nt (35%) sequence difference between the BCoV and MHV long SLIV structures (Fig. 1C) and a differing 5′-terminal nsp1 amino acid sequence of MSKINK for BCoV versus MAKMGK for MHV. It also indicates that the 5′-terminal 16 nt of the nsp1 cistron are a functioning part of the native cis-acting SLIV and suggests that the BCoV native long SLIV in the nonviable B1–226/M chimera (Fig. 3B) or in the debilitated B142–226/M chimera (Fig. 3B) is not the incompatible component.
To determine whether the chimeric BCoV/MHV long SLIV alone is compatible with the remaining MHV genome, the chimera B174–210/M was tested. Interestingly, in this chimera a wt-like long SLIV structure (Fig. 4A) is not mfold predicted as expected (Fig. 4B, left panel) either in the context of nucleotides 1 to 400 of the B174–210/M chimera or as a standalone structure. Rather, in both a distorted structure is predicted (Fig. 4B, center panel). Nevertheless, from B174–210/M, syncytia appeared within 24 h and VP0 virus generated large plaques and a titer of 106 PFU/ml (data not shown). Interestingly, in progeny genomes, mutations were present in the base of the long stem (U177C) (Fig. 4B, right panel) and within SL1 (A36C), indicating genetic adaptation had occurred early. mfold analyses showed that the U177C mutation alone within SLIV increased the predicted stability of the overall stem-loop from a ΔG of −12.90 to −14.00 kcal/mol with a return to an mfold-predicted wt-like long SLIV structure (Fig. 4B, right panel). The role of the A36C mutation was not evaluated further, although it would be expected to destabilize the lower stem of SL1. Together these results suggest that the native SLIV structure in both MHV and BCoV probably extends 16 nt into its cognate nsp1 cistron in a manner that stabilizes its lower stem and contributes to virus fitness.
Predicted stability in the lower stem of SLIV correlates with fitness for virus growth in cell culture.
To examine the role of stability in the lower stem for fitness, two sets of three constructs each with mutations in the lower stem of wt MHV SLIV were examined. In the first, the MHV sequence in both sides of the lower fourth of SLIV (B174–182, 219–226/M), the lower left fourth only (B174–182/M), or the lower right fourth only (B219–226/M) was converted to BCoV sequence (Fig. 5A, B, and C, respectively). The bases differing from those in wt MHV SLIV are encircled, and the mfold-predicted standalone structures are shown. Following transfection, syncytia were observed within 48 hpe for each. However, VP0 virus from only the chimera with BCoV sequence in both sides of the lower fourth (B174–182, 219–226/M, where SLIV ΔG = −20.10 kcal/mol) and the lower left fourth only (B174–182/M, where SLIV ΔG = −17.00 kcal/mol) made primarily large plaques and produced titers of 7.2 × 106 and 1.0 × 107 PFU/ml, respectively (Fig. 5A and B). In contrast, the chimera with BCoV sequence on the lower right fourth (B219–226/M, where SLIV ΔG = −14.00 kcal/mol) made medium plaques and produced a titer of only 5.0 × 104 PFU/ml (Fig. 5C). Note that the mfold-predicted lower stems in the first two chimeras resemble that in wt BCoV long SLIV (Fig. 4A, right panel) in that there is a single nucleotide bulge (although a G and not a U in the second instance) and a total of eight base pairings. The lower stem in the third chimera, by contrast, has a 7-nt inner loop and a total of five base pairings. The lower titer and the smaller plaques from the third SLIV chimera therefore correlate with a predicted less-stable lower stem and are consistent with the results for the chimera studied in Fig. 4, in which stability in the lower stem appeared to contribute to virus fitness for growth in cell culture.
Fig. 5.
Predicted stability in the lower stem of SLIV correlates with fitness for virus growth in cell culture. (A) Both sides of the lower fourth of SLIV are BCoV sequence. (B) The left side of the lower fourth of SLIV is BCoV sequence. (C) The right side of the lower fourth of SLIV is BCoV sequence. (D) Both sides of the lower half of SLIV are BCoV sequence. (E) The left side of the lower half of SLIV is BCoV sequence. (F) The right side of the lower half of SLIV is BCoV sequence. Structures identified by an asterisk (*) are predicted by mfold to be the most stable standalone form. The structure in panel E, left part, is drawn to conform to those in panels A through D and F. The AUG boxed with a solid line identifies the ORF1 start codon, and a second, in-frame, intra-SLIV AUG codon is boxed with a dashed line. Nucleotides from MHV are shown in uppercase letters, and those from BCoV are shown in lowercase letters. Numbers identifying nucleotide positions in the BCoV genome are enclosed in parentheses. Encircled nucleotides are those in the BCoV sequence that differ from their MHV counterparts.
To determine if stability in the extended lower stem is required for fitness as well, a second set of three chimeras was studied. For this, the lower half of both sides of SLIV (B174–187, 214–226/M, where SLIV ΔG = −17.40 kcal/mol), the lower left half (B174–187/M, where SLIV ΔG = −16.40 kcal/mol), and the lower right half (B214–226/M, where SLIV ΔG = −15.80 kcal/mol) were made with BCoV sequence and tested, and in each, syncytia appeared within 48 hpe (Fig. 5D, E, and F, respectively). However, only progeny of the first two chimeras, B174–187, 214–226/M and B174–187/M, made primarily large plaques and produced titers of 2.0 × 106 and 2.5 × 106 PFU/ml, respectively (Fig. 5D and E), whereas progeny from the third chimera (B214–226/M) made primarily small and medium plaques and produced a titer of only 6.5 × 103 PFU/ml (Fig. 5F). A surprise from this set of chimeras was that whereas the predicted stabilities of the first two were higher than that of the third, only the mfold-predicted structure of the first was comparable to those in Fig. 5A and B with a predicted stable lower stem. The mfold-predicted structure of the second chimera, B174–187/M, was distorted (Fig. 5E, right panel). That is, in this structure there is no clearly defined lower stem. Still, the results for the first chimera (with a predicted stable lower stem) and the third chimera (with a predicted unstable lower stem) conformed to the notion that a stable lower stem contributes to progeny fitness (Fig. 5D versus F). In no progeny from the six chimeras tested were there mutations found within the 5′-terminal 22 to 1,093 nt or 3′-terminal 500 nt. Therefore, with the exception of B174–187/M, these results show a positive correlation between predicted stability in the lower stem of SLIV and fitness of virus for growth in cell culture.
Surprising SLIV pleiomorphic structures, including internal deletions, are tolerated in survivors of chimeras with mutations throughout the long SLIV.
To examine the influence on fitness of structure in the upper stem-loop along with those in the lower stem, two sets of changes throughout the full length of SLIV were tested in separate chimeras. In the first, the entire left side of SLIV was made with BCoV sequence to form the chimera B174–199/M (Fig. 6A), which is identical to construct B174–187/M (Fig. 5B) except that nucleotides 191 and 192 are U and G, respectively, causing base pairings of U-G and G-U rather than C-G and A-U in the upper stem (Fig. 6A). Thus, the modified upper stem has three consecutive G-U base pairings, which likely weakens the entire upper helix. Interestingly, the mfold-predicted structure of this SLIV, although it has an mfold-predicted ΔG value of −15.10 kcal/mol, is distorted (Fig. 6A, second panel), with a pattern very similar to that for chimera B174–210/M (Fig. 4B, center panel). With chimera B174–199/M, syncytia were found after one blind cell passage in one of the four transfection trials, and medium plaques were generated by VP0 virus (data not shown). Progeny genomes showed mixed sequences in the 5′ nucleotide-22-to-1093 region (data not shown); thus, plaque-purified virus populations were sequenced that revealed two distinct deletion genotypes, 1 and 2, despite nearly identical plaque appearances (Fig. 6A). From plaque 1, 18 nt (nucleotides 176 to 193) had been deleted from the left side of the stem, and from plaque 2, 5 nt (nucleotides 177 to 181) had been deleted from the left side (Fig. 6A). Viral titers for these reached 2.2 × 107 and 5.0 × 106 PFU/ml, respectively, and in each, no additional mutations were found within the genomic 5′ nucleotides 22 to 1093 and 3′ 500 nt.
Fig. 6.
Recovered variant SLIV structures obtained from extensively mutated input chimeric SLIV. (A) Results for chimera B174–199/M, in which the upstream half of SLIV is made with BCoV sequence. The SLIV sequences of two separate plaque-purified isolates from VP0 virus show an 18-nt upstream deletion in one (plaque 1) and a 5-nt upstream deletion in the other (plaque 2). (B) Results for chimera B200–226/M, in which the downstream half of SLIV is made with BCoV sequence. The SLIV sequence of a plaque isolate from VP0 virus shows a 20-nt deleted upstream region that includes the terminal loop. Structures identified by a double asterisk (**) are predicted by mfold to be the most stable form both in the context of nucleotides 1 to 400 and as a standalone structure, and those identified by a single asterisk (*) are predicted to be the most stable form in a standalone structure. In both panels A and B, nucleotides from MHV are shown in uppercase letters and those from BCoV are shown in lowercase letters. Encircled nucleotides are those in the BCoV sequence that differ from their MHV counterparts.
In the second chimera, the entire right side of the long SLIV was made with BCoV sequence to form B200–226/M, in which the SLIV ΔG is −8.60 kcal/mol (Fig. 6B, left panel) and the viral titer was 2.5 × 107 PFU/ml. From this chimera, syncytia were observed within 48 hpe in one of four transfection trials and plaques from VP0 virus were medium in size. Sequencing revealed a 20-nt deletion within SLIV that included the entire 8-nt terminal loop (Fig. 6B, right panel). No additional mutations within the genomic 5′ nucleotides 22 to 1093 and 3′ 500 nt were found. Furthermore, for each of the three deletion genotypes described for Fig. 6, a chimera virus was made that carried only the partially deleted SLIV in the MHV background and progeny matching parental phenotypes and SLIV genotypes were recovered within 48 hpe, suggesting that compensatory mutations may not have occurred elsewhere in the MHV genome (data not shown). At this time, no further characterizations of the extensively deleted SLIV progeny have been made. These results together indicate that although possession of extensive intra-SLIV deletions may yield a less-fit virus, they are not necessarily lethal for MHV replication and there is a certain tolerance for SLIV pleiomorphism.
The BCoV 32-nt inter-stem-loop domain between SLIII and SLIV (nt 142 to 173) behaves as a novel species-specific cis-replication element.
Given that chimeras containing the BCoV SLI/II and SLIII domains together (B1–141/M) and the BCoV long SLIV domain alone (B174–226/M) were immediately functional for virus replication in the MHV background but those containing the BCoV inter-stem-loop domain (B1–226/M or B142–226/M) were not (Fig. 2 and 3), we hypothesized that the inter-stem-loop domain may represent a separate virus-specific cis-replication element. The BCoV inter-stem-loop domain was therefore tested for replication function in three separate constructs. In the first two, incremental 3′-ward extensions of the BCoV sequence in B1–141/M were made to form B1–158/M (a 17-nt extension) and B1–173/M (a full 32-nt extension) (Fig. 3A). In results consistent with our hypothesis, the chimera B1–158/M enabled progeny with heterogeneous (small, medium, and large) plaque sizes after two blind cell passages (Fig. 3B) and sequences from two isolated plaques revealed three 5′ UTR-mapping mutations, two of which were C145U and G147U within the inter-stem-loop domain (Fig. 7B). For B1–173/M, no viable virus was obtained from two transfection trials (Fig. 3B). In the third chimera, the BCoV 32-nt inter-stem-loop domain alone was placed in the MHV background to form B142–173/M (Fig. 3A), and from this, syncytia appeared after two blind cell passages (Fig. 3B) and virus from five isolated plaques had up to four mutations each in the 5′ UTR, at least one of which in each was C145U, C158G, A163U, or G169U within the inter-stem-loop domain (Fig. 7B). Interestingly, three of the five inter-stem-loop-located changes in progeny from B1–158/M and B142–173/M made the transplanted BCoV domain more MHV-like (see underlined bases in Fig. 7B). To test the viral specificity of the inter-stem-loop domain by another approach, growth of a chimera in which the MHV 30-nt inter-stem-loop domain was placed within the long SLIV-containing BCoV 5′ UTR in the MHV background, to form B1–141, 174–226/M (Fig. 3A), was analyzed. This chimera yielded syncytia within 48 hpe and plaques of medium size (Fig. 3B), progeny with a maximal titer of 1 × 106 PFU/ml (Fig. 3D, left panel) and a full complement of subgenomic RNAs (sgmRNAs) (Fig. 3D, right panel), but no mutations were found within the 5′-proximal nucleotides 22 to 1093 or 3′ 500 nt (data not shown). These results together show that the transplanted 32-nt BCoV inter-stem-loop domain becomes compatible within the MHV background concurrent with the acquisition of potential intradomain compensatory mutations, whereas the MHV 30-nt inter-stem-loop domain immediately enables progeny without mutations within the 5′-proximal nucleotides 22 to 1093 or 3′ 500 nt. For still-undetermined reasons, however, the progeny are less fit than wt MHV (note the lower peak titer of 106 PFU/ml, versus 108 PFU/ml in Fig. 3D, left panel, and the less-abundant sgmRNAs in Fig. 3D, right panel). Thus, the inter-stem-loop domain behaves as a cis-replication element in a virus-specific manner. Interestingly, an alignment of the inter-stem-loop region of 11 group 2a coronaviruses (Fig. 7A) shows the viruses to be classifiable as MHV-like, BCoV-like, or intermediate, suggesting the inter-stem-loop domain may prove to be a cis-replication element specific for BCoV-like or MHV-like (or other) group 2a coronaviruses.
Fig. 7.
Alignment of 5′ UTR inter-stem-loop domains. (A) Alignment of the 5′ UTR inter-stem-loop domains among 11 group 2a coronaviruses. Identical nucleotides are shaded. Abbreviations: Rat, Parker rat coronavirus; A59, mouse hepatitis virus-A59 strain; JHM, mouse hepatitis virus JHM strain; 2, mouse hepatitis virus 2 strain; HKU1, human coronavirus HKU1; ECoV, equine coronavirus; Meb., bovine coronavirus Mebus strain; ENT, bovine coronavirus Enteric strain; OC43, human coronavirus OC43; 4408, human enteric coronavirus-4408; PHEV, porcine hemagglutinating encephalomyelitis virus. (B) Alignment of the 5′ UTR inter-stem-loop domains of MHV-A59 and BCoV-Mebus with arrows identifying the genetic adaptations found within this domain in progeny from chimeras B1–158/M and B142–173/M. Underlined nucleotides identify potential compensatory mutations that make the transplanted BCoV domain more MHV-like.
DISCUSSION
In this study we asked whether cis-acting higher-order RNA structures in the BCoV 5′ UTR, previously identified as cis-acting replication elements in a BCoV defective interfering RNA, would function in the MHV genome to support virus replication, thereby defining common 5′ cis-replication signals for these two group 2a coronaviruses. The study was initiated since a precise replacement of the 209-nt MHV 5′ UTR with the 210-nt BCoV 5′ UTR did not function in the MHV genome as had been expected since (i) the two viruses are phylogenetically relatively closely related (10), (ii) the BCoV DI RNA has been shown to replicate in the presence of MHV helper virus, indicating compatible replication signals (38), and (iii) the 3′ UTR of BCoV, as well as the 3′ UTR in the more distant SCoV (15), is functional in the MHV genome (21–22). By replacement of individual homologous regions in the MHV 5′ UTR with cis-acting domains in the BCoV 5′ UTR, it was learned that whereas the SLI/II and SLIII domains worked well despite nucleotide identity differences of 36% in both, a relatively unstructured 32-nt inter-stem-loop domain between SLs III and IV with a difference in nucleotide identity of 41% yielded a viable virus only after two blind cell passages and genetic adaptation, and the SLIV domain with a nucleotide identity difference of 32% was immediately functional without adaptation after it had been extended into the BCoV nonstructural protein 1 coding region by 16 nt to form an intact BCoV “long” SLIV.
In a study by Kang et al. (24) that similarly tested 5′ UTR structures from SCoV in the MHV genome, it was learned that regions equivalent to SL1, SL2, and SLIII (as depicted in Fig. 1A, above) enabled replication of the chimera, whereas the region containing the SCoV transcription regulatory core sequence (ACGAAC) (mapping between SL2 and SLIII) did not, nor did structures downstream of nucleotide 140, namely, the region equivalent to the inter-stem-loop and SLIV domains as depicted in Fig. 1A. Thus, since the results of our experiments with BCoV are similar to those of Kang et al. (24) for SCoV with regard to cis-acting SL1, SL2, and SLIII, it would appear that these signaling structures, along with those in the 3′ UTR, might be common among coronavirus subgroups 2a and 2b and therefore potential sites for broad-spectrum antiviral targeting. The other regions of the 5′ UTR so far appear less attractive as common targets for reasons described below.
The first major conclusion from the current study is that the relatively unstructured BCoV 32-nt inter-stem-loop domain potentially behaves as a coronavirus-specific cis-replication element. This is supported by the finding that the chimera with the MHV inter-stem-loop domain (B1–141, 174–226/M in Fig. 3B and D) but not the chimera with the BCoV inter-stem-loop domain (B142–173/M in Fig. 3B) replicated immediately after transfection, albeit with less than wt-like features (note the medium-size plaques and the 100-fold-lower virus titer at 28 hpi). What the 32-nt inter-stem-loop domain does for virus replication is not known at this time, but the fact that it appears tailored to different species within subgroup 2a viruses (Fig. 7A) would suggest that it has evolved with other viral elements. Perhaps this explains why MHV supported the replication of BCoV DI RNA less well than did the other BCoV-like coronaviruses, such as ECoV, HCoV-OC43, and HECoV-4408 (38). In a follow-up study, (B.-J. Guan, Y.-P. Su, H.-Y. Wu, and D. A. Brian, to be reported elsewhere), we are examining other elements that may interact with the inter-stem-loop domain.
The second major conclusion from the current study is that for the first time it has been demonstrated in the context of the coronavirus genome that SLIV is a cis-acting replication element and furthermore that it is comprised of sequences from both the 5′ UTR and the most 5′ region of ORF1. Evidence for the latter lies in the fact that the 16 nt from BCoV ORF1 functions with the rest of BCoV SLIV to provide optimal MHV replication in the MHV background (Fig. 3C), whereas the 16 nt from MHV ORF1 with the rest of BCoV SLIV, although viable after transfection, is recovered with an apparent intra-SLIV stabilizing mutation that arises soon after transfection (Fig. 4B, right panel). With one exception, an examination of six SLIV constructs with mutations in the lower stem showed a positive correlation between a predicted higher stability in the lower stem and virus fitness. B174-187/M, which exhibits a near-wt-like phenotype (Fig. 5E), curiously has an mfold-predicted distorted SLIV structure that appears much like that for B174-199/M, from which partially debilitated progeny with partial SLIV deletions are obtained after blind cell passaging (Fig. 6A). B174-199/M, however, has three tandem G-U base pairs in the upper stem that may contribute to further unpredicted distortions in overall SLIV structure and function. The predicted distorted structures for both B174-187/M and B174-199/M are similar to that for B174-210/M shown in Fig. 4B, center panel. Although we are unable at this time to explain the exceptionally fit phenotype of B174-187/M, we conclude that in general the fitness of a chimeric SLIV virus correlates directly with the stability of SLIV in its lower stem.
The idea that the 5′-terminal part of ORF1 (encoding the N-terminal region of nsp1) should provide an important cis-replication function has been hinted at in several earlier studies: (i) a 5′-terminal portion of the nsp1 cistron was consistently found as a part of group 1 and group 2 coronavirus DI RNAs (group III coronaviruses lack an nsp1 cistron) (1, 30), (ii) the 5′-terminal portion of the nsp1 cistron, and its translation, was found to be necessary for BCoV DI RNA replication (4), (iii) SLs V and VI in the BCoV DI RNA, as higher-order RNA structures, have been shown to behave as cis-replication elements (3, 17), and (iv) in MHV, mutations in the 5′ terminal region of the nsp1 cistron have been shown to be detrimental to MHV replication by an unknown mechanism (2).
The occurrence of the nsp1 start codon in the downstream side of SLIV leaves open the possibility that SLIV functions as a regulatory structure for initiation of ORF1 translation or for initiation of genome synthesis from the (−)-strand antigenome. If so, it is surprising that so many partial deletion variants of the long SLIV as found here (Fig. 6) enabled virus replication with apparently few disabilities. Knowledge of how the pleiomorphic SLIV structures are able to carry out a replication function may shed light on the mechanistic features of wt SLIV. Studies with the BCoV SLIV to date have shown the binding of six unidentified cellular proteins ranging from 25 to 58 kDa in molecular mass but no viral proteins (33). Identifying these and noting how protein binding patterns change with the variant SLIV structures may also give clues to SLIV function.
It is noteworthy that the packaging of the MHV genome in the absence of a terminal loop on SLIV (as shown in Fig. 6B) is consistent with a proposal by Chen and Olsthoorn (7) which states that SLIV (equivalent to SL5 in their study) may function as a packaging signal when it contains one or more copies of a conserved UUYCGU motif. Multiple copies of this motif are found in certain group 1 coronaviruses (porcine transmissible gastroenteritis coronavirus, for example) and group 2 coronaviruses (SCoV, for example). SLIV of BCoV and that of MHV do not contain this motif.
With a focus on stem-loop IV, the current study indicates that the structure in the lower stem of SLIV acts in concert with the whole of SLIV to provide a mechanism for the optimal replication of group 2a coronaviruses. Therefore, a detailed understanding of the function of this cis-replication element should be pursued in the interest of finding sites for designing inhibitors of coronaviruses closely related to MHV and BCoV, which include the human pathogens HCoV-OC43, HECoV-4408, and HCoV-HKU1.
ACKNOWLEDGMENTS
We thank Agnieszka Dziduszko, Kortney Gustin, Yu-Pin Su, Yi-Hsin Fan, and Kim Nixon for many helpful discussions.
This work was supported by Public Health Service grant AI014367 from the National Institute of Allergies and Infectious Diseases and by funds from the University of Tennessee, College of Veterinary Medicine, Center of Excellence Program for Livestock Diseases and Human Health.
Footnotes
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Brian D. A., Baric R. S. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brockway S. M., Denison M. R. 2005. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 340:209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown C. G., Nixon K. S., Senanayake S. D., Brian D. A. 2007. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 81:7716–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang R. Y., Brian D. A. 1996. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 70:2201–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang R. Y., Hofmann M. A., Sethna P. B., Brian D. A. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 68:8223–8231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang R. Y., Krishnan R., Brian D. A. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 70:2720–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen S. C., Olsthoorn R. C. 2010. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology 401:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen W., Baric R. S. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen W., Madden V. J., Bagnell C. R., Jr., Baric R. S. 1997. Host-derived intracellular immunization against mouse hepatitis virus infection. Virology 228:318–332 [DOI] [PubMed] [Google Scholar]
- 10. Chouljenko V. N., Lin X. Q., Storz J., Kousoulas K. G., Gorbalenya A. E. 2001. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 82:2927–2933 [DOI] [PubMed] [Google Scholar]
- 11. Denison M. R., et al. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaertner D. J., Winograd D. F., Compton S. R., Paturzo F. X., Smith A. L. 1993. Development and optimization of plaque assays for rat coronaviruses. J. Virol. Methods 43:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goebel S. J., Hsue B., Dombrowski T. F., Masters P. S. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goebel S. J., Miller T. B., Bennett C. J., Bernard K. A., Masters P. S. 2007. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 81:1274–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goebel S. J., Taylor J., Masters P. S. 2004. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 78:7846–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gorbalenya A. E., Enjuanes L., Ziebuhr J., Snijder E. J. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gustin K. M., Guan B. J., Dziduszko A., Brian D. A. 2009. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 83:6087–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirano N., Fujiwara K., Matumoto M. 1976. Mouse hepatitis virus (MHV-2). Plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219–225 [PubMed] [Google Scholar]
- 19. Hofmann M. A., Senanayake S. D., Brian D. A. 1993. A translation-attenuating intraleader open reading frame is selected on coronavirus mRNAs during persistent infection. Proc. Natl. Acad. Sci. U. S. A. 90:11733–11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann M. A., Sethna P. B., Brian D. A. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 64:4108–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsue B., Hartshorne T., Masters P. S. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsue B., Masters P. S. 1997. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 71:7567–7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson R. F., et al. 2005. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570–14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang H., Feng M., Schroeder M. E., Giedroc D. P., Leibowitz J. L. 2006. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 80:10600–10614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li L., et al. 2008. Structural lability in stem-loop 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J. Mol. Biol. 377:790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin Y. J., Liao C. L., Lai M. M. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu P., et al. 2009. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J. Virol. 83:12084–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu P., et al. 2007. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA 13:763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Makino S., Lai M. M. 1989. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J. Virol. 63:5285–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Masters P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathews D. H., Sabina J., Zuker M., Turner D. H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911–940 [DOI] [PubMed] [Google Scholar]
- 32. Raman S., Bouma P., Williams G. D., Brian D. A. 2003. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 77:6720–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raman S., Brian D. A. 2005. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 79:12434–12446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spagnolo J. F., Hogue B. G. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tinoco I., Jr., et al. 1973. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 246:40–41 [DOI] [PubMed] [Google Scholar]
- 36. Williams G. D., Chang R. Y., Brian D. A. 1999. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 73:8349–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu H. Y., Brian D. A. 2007. 5′-proximal hot spot for an inducible positive-to-negative-strand template switch by coronavirus RNA-dependent RNA polymerase. J. Virol. 81:3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu H. Y., et al. 2003. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology 315:174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu H. Y., Ozdarendeli A., Brian D. A. 2006. Bovine coronavirus 5′-proximal genomic acceptor hotspot for discontinuous transcription is 65 nucleotides wide. J. Virol. 80:2183–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yount B., Denison M. R., Weiss S. R., Baric R. S. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zust R., Miller T. B., Goebel S. J., Thiel V., Masters P. S. 2008. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 82:1214–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]