Abstract
Model membrane and cellular detergent extraction studies show (n-3) PUFA predominately incorporate into nonrafts; thus, we hypothesized (n-3) PUFA could disrupt nonraft organization. The first objective of this study was to determine whether (n-3) PUFA disrupted nonrafts of EL4 cells, an extension of our previous work in which we discovered an (n-3) PUFA diminished raft clustering. EPA or DHA treatment of EL4 cells increased plasma membrane accumulation of the nonraft probe 1,1′-dilinoleyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate by ~50–70% relative to a BSA control. Förster resonance energy transfer imaging showed EPA and DHA also disrupted EL4 nanometer scale nonraft organization by increasing the distance between nonraft molecules by ~25% compared with BSA. However, changes in nonrafts were due to an increase in cell size; under conditions where EPA or DHA did not increase cell size, nonraft organization was unaffected. We next translated findings on EL4 cells by testing if (n-3) PUFA administered to mice disrupted nonrafts and rafts. Imaging of B cells isolated from mice fed low- or high-fat (HF) (n-3) PUFA diets showed no change in nonraft organization compared with a control diet (CD). However, confocal microscopy revealed the HF (n-3) PUFA diet disrupted lipid raft clustering and size by ~40% relative to CD. Taken together, our data from 2 different model systems suggest (n-3) PUFA have limited effects on nonrafts. The ex vivo data, which confirm previous studies with EL4 cells, provide evidence that (n-3) PUFA consumed through the diet disrupt B cell lipid raft clustering.
Introduction
EPA and DHA, the bioactive (n-3) PUFA of fish oil, are increasingly available and consumed by the general public as over-the-counter supplements (1, 2). Clinically, EPA and DHA have applications for the prevention and/or treatment of some metabolic diseases (3–6); in addition, they have potential utility for treating the symptoms associated with inflammatory and autoimmune disorders (7–9). However, one major limitation of further developing these fatty acids for clinical use is an incomplete understanding of their targets and molecular mechanisms.
An emerging mechanism of the action of (n-3) PUFA, due to their unique molecular structure, is modification of plasma membrane lipid rafts (10), which are sphingolipid-cholesterol enriched domains that compartmentalize signaling proteins (11). We recently discovered an (n-3) PUFA disrupted lipid raft clustering of EL4 cells (12). The data raised a new question, i.e. could (n-3) PUFA also disrupt the organization of nonraft domains. These membrane domains are broadly defined as those regions that are not enriched in sphingolipids and cholesterol that also compartmentalize specific proteins (e.g. major histocompatibility complex (MHC) class I, Toll-like Receptor 4, etc.) (13). There were 2 reasons to hypothesize (n-3) PUFA would disrupt nonraft organization. First, experiments using model membranes demonstrated DHA acyl chains, due to their structural incompatibility with cholesterol, primarily incorporated into nonrafts to enhance nonraft formation (14–16). Second, biochemical detergent extraction studies showed a large fraction (up to 70%) of EPA and DHA localized into nonrafts (12, 17–19). Thus, these studies suggest that a major role of (n-3) PUFA acyl chains is to modify nonraft domain organization.
The first objective of this study was to extend our previous work by determining if EPA and DHA treatment disrupted nonraft organization of EL4 cells. The second objective was to translate the findings on EL4 cells by testing the impact of dietary (n-3) PUFA on both nonraft and lipid raft organization in an animal model. To address our objectives, we relied on quantitative imaging methods of confocal and Förster resonance energy transfer (FRET)4 microscopy. Application of these methods to the study of (n-3) PUFA and membrane domains advances the field by overcoming the use of cold detergent extraction as a primary method of studying how (n-3) PUFA modify membrane domains. Although detergent resistance has great predictive value, the detergent can induce artifacts (20–22). Furthermore, the biochemical detergent method does not report on the effects of (n-3) PUFA on the appropriate length scales on which membrane domains form (11). Therefore, we used more appropriate imaging methods to address the effects of (n-3) PUFA on membrane domain organization.
Materials and Methods
Cells.
EL4 cells were maintained in RPMI 1640–1× (Mediatech) with 10% heat-inactivated defined FBS (Hyclone), 2 mmol/L l-glutamine (Mediatech), and 1% penicillin/streptomycin (Mediatech) at 37°C in a 5% CO2 incubator. The lipid composition of the FBS was as previously reported (12).
Fatty acid treatment.
A total of 9–10 × 105 EL4 cells was treated for 15.5 h with 25 μmol/L FFA (Nu-Check Prep) complexed to fatty acid-free BSA (Roche Biochemicals) in serum-free medium as previously described (12). For select experiments, EL4 cells were treated in the presence of 10% FBS. The rationale for selecting the fatty acid concentration and time of treatment was to be consistent with our previous study on EL4 cells and membrane domains (12). Oleic (OA) and arachidonic (AA) acids were tested to rule out general effects of fatty acid treatment and to ensure specificity of EPA and DHA.
Mice, diets, and B cell isolation.
Male C57BL/6 mice (Charles River), aged 4–6 wk (~18 g), were fed the following diets (Harlan-Teklad) for 3 wk: a purified control diet (CD) (5% fat wt:wt), an (n-6) PUFA diet (20% fat wt:wt), a low-fat (LF) (n-3) PUFA diet (5% fat wt:wt), or a high fat (HF) (n-3) PUFA diet (20% fat wt:wt) (23) (Table 1). For the CD and LF (n-3) PUFA diets, ~13% of the total energy was from fat. For the HF (n-6) and (n-3) PUFA diets, ~41% of the total energy was from fat. For the LF (n-3) PUFA diet, 3.3% of the total energy was from α-linolenic acid, 1% from EPA, and 0.6% from DHA (23). For the HF (n-3) PUFA diet, 10.5% of the total energy was from α-linolenic acid, 3% from EPA, and 2% from DHA (23). The diets were analyzed for their fatty acid composition (Supplemental Table 1). For B cell isolation, mice were killed using CO2 inhalation after 3 wk of feeding. Naïve B220+ B cells (>90% purity) from splenocytes were purified with negative selection (Miltenyi Biotec) as previously described (23). All experiments with mice fulfilled guidelines established by the East Carolina University for euthanasia and humane treatment.
TABLE 1.
Composition of experimental diets1
| Ingredients | CD | HF (n-6) PUFA | LF (n-3) PUFA | HF (n-3) PUFA |
| g/kg | ||||
| Casein | 185.0 | 220.0 | 185.0 | 220.0 |
| l-Cystine | 2.5 | 3.0 | 2.5 | 3.0 |
| Corn starch | 370.0 | 173.9 | 370.0 | 173.9 |
| Maltodextrin | 140.0 | 140.0 | 140.0 | 140.0 |
| Sucrose | 150.0 | 150.0 | 150.0 | 150.0 |
| Cellulose (fiber) | 50.0 | 50.0 | 50.0 | 50.0 |
| Flaxseed oil | 0.0 | 0.0 | 23.1 | 92.5 |
| Fish oil (Menhaden) | 0.0 | 0.0 | 23.1 | 92.5 |
| Soybean oil | 50.0 | 30.0 | 3.75 | 15.0 |
| Safflower oil | 0.0 | 125.0 | 0.0 | 0.0 |
| Hydrogenated coconut oil | 0.0 | 45.0 | 0.0 | 0.0 |
| Mineral mix, AIN-93M2 | 35.0 | 42.0 | 35.0 | 42.0 |
| Vitamin mix, AIN-932 | 15.0 | 18.0 | 15.0 | 18.0 |
| Choline bitartate | 2.5 | 3.0 | 2.5 | 3.0 |
| TBHQ | 0.02 | 0.06 | 0.02 | 0.06 |
C57BL/6 mice were fed for 3 wk CD, HF (n-6) PUFA, LF, and HF (n-3) PUFA diets.
Composition of mineral and vitamin formulas are as reported in (24).
Fatty acid analysis.
For analysis of diets, ∼0.01–0.05 g of the diet pellets was homogenized using a Dounce homogenizer prior to extraction. Total lipids were extracted and analyzed relative to standards (Restek) from the differing cell types or diets with GC using our previously published protocol (12, 23). Areas of identified peaks were summed and each peak area is expressed as the percentage of total peak area for a given treatment (12, 23).
Column chromatography.
To assess incorporation of EPA and DHA into neutral lipid, FFA, and polar lipid fractions, 2 × 106 EL4 cells were treated with fatty acids spiked with 185 kBq [14C]EPA or [14C]DHA (American Radiolabeled Chemicals). Extracted lipids were separated into different lipid classes using aminopropyl beads (Sigma) loaded in a Pasteur pipette with a fiberglass plug and elution with HPLC-grade organic solvents (Fisher Scientific) as previously described (23, 25).
Staining and imaging with 1,1#x2032-dilinoleyl-3,3,3#x2032,3#x2032-tetramethylindocarbocyanine perchlorate and 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid.
A total of 2 × 106 cells was washed twice with cold PBS, stained with 0.66 mg/L of the nonraft probe 1,1′-dilinoleyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (FAST-DiI; Invitrogen) or 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid (C16-Bodipy; Invitrogen) for 5 min on ice followed by 2 washes with cold PBS (26, 27). Cells were fixed in 4% paraformaldehyde (Fisher Scientific) for 1 h, washed twice, placed in Vitrotubes (Fiber Optic Center), and mounted onto slides with nail polish. All imaging studies, described below, relied on a Zeiss LSM510 confocal microscope using a 100× oil objective. Pinhole, detector gain, and laser settings were kept constant between samples for a given experiment. For some experiments, images were acquired as z-stacks.
Sample preparation for FRET microscopy.
MHC class I molecules were used as the probe to measure nanometer scale molecular proximity, because these molecules are localized to nonrafts and are established to serve as excellent reporters for measuring changes in membrane organization (28–30). Cells were stained with varying concentrations of Cy3 (donor) and Cy5 (acceptor) separately labeled M1/42.3.9.8 anti-MHC class I antibodies (BioXCell). Fluorophores were conjugated to the antibody using a standard fluorophore conjugation kit (GE Healthcare). Specificity of the antibody was confirmed by using isotype controls (BioXCell) (12). Total antibody levels were held constant at 0.02 mg/sample using unlabeled antibody as donor:acceptor ratios were varied from 1:1, 1:2, and 1:3 in 1 × 10−4 L PBS (31, 32). Cells were stained for 15 min on ice, washed twice with PBS, fixed, washed, and mounted to slides as described above. A positive control M1/42.3.9.8 antibody was generated by simultaneously conjugating Cy3 and Cy5 fluorophores, yielding a dual labeled antibody to ensure high FRET. Negative control samples were labeled with donor fluorophore only to check for donor channel bleaching during acceptor fluorophore bleaching and acceptor fluorophore only to optimize bleaching time.
FRET microscopy.
FRET was measured in terms of the efficiency of energy transfer (E) from donor (Cy3) to acceptor (Cy5) fluorophores separated by distance r, given by the following equation: E = 1/{1 + (r/Ro)6}, where Ro is the Förster radius of the donor and acceptor fluorophore pairs and is 54 Å for Cy3/Cy5 (33, 34). The filter sets were 560- to 615-nm band pass for Cy3, and 650-nm long pass for Cy5 in 2 separate tracks. Excitation relied on 543- and 633-nm lasers, respectively, for Cy3 and Cy5. Eight-bit 512 × 512 pixel images were acquired. Acceptor photobleaching was carried out by iterative scanning of the 633-nm laser for ~2 min. Gain settings were optimized to ensure images were not saturated after photobleaching.
Lipid raft staining.
B cells were labeled with cholera toxin subunit B-FITC (Invitrogen) for cross-linking lipid rafts and imaged as previously described (12).
Image analysis.
All images were analyzed with NIH ImageJ (35). Approximately 20–33 cells per treatment per experiment were analyzed as previously described (12). For FAST-DiI and C16-Bodipy studies, differential interference contrast images were routinely used to confirm plasma membrane compared with intracellular staining. For image intensity analysis, images were background subtracted and manually thresholded for regions of interest either inside the cell or on the plasma membrane. The approach was confirmed by selecting several regions of interest manually drawn on the cell surface and inside the cell. Cells were also scored as high intensity if they were in the top one-third of the distribution of fluorescence intensities.
FRET images were analyzed with FRETcalc v3.0 plug-in for NIH ImageJ (36). Some cells could not be properly registered and were not included in the analysis. Images were registered by manual translation or rotation. All images were background subtracted and smoothed using a 3 × 3 filter. The threshold values for the donor and acceptor images were determined manually. This was required to calculate FRET efficiency of the entire cell by the FRETcalc plug-in (36).
Images of lipid rafts were background subtracted and raft size was determined in terms of Feret diameter using NIH ImageJ as previously described (12).
Cell growth and apoptosis measurements.
Cell growth was determined by counting cells in duplicate or triplicate using a hemacytometer. Dead cells were excluded with Trypan blue (HyClone, Fisher Scientific) staining. Measurements were routinely confirmed by a second person using blinded samples. We verified that this approach gave the same results as a cell proliferation testing kit (GenScript). The advantage of counting cells over the kit was the absolute number of cells could be determined rather than relative changes. Cell survival was measured in terms of Annexin V-Cy5/Sytox Blue (BD Pharmingen) staining with a BD LSRII flow cytometer as previously described (23).
Statistical analysis.
Reported values are means ± SEM from several independent experiments. For animal studies, independent experiments were conducted using 1 mouse from each diet group. All statistical analyses were conducted using Excel and GraphPad Prism (GraphPad Software). Parametric statistics were used, because the data were normally distributed. Unequal variances were tested for prior to ANOVA using a Levene’s test. For FRET studies, efficiency values varied between experiments due to variation in the fluorophore to antibody ratios; thus, significance was established against the control using repeated-measures 1-way ANOVA followed by a Dunnett’s t test. For cell growth and apoptosis measurements as a function of time, 2-way ANOVA analysis was used followed by a Bonferroni t test. The 2-way ANOVA used treatment and time as factors and there was no interaction effect. For all other studies, significance was established against the control using a 1-way ANOVA followed by a Dunnett’s t test. P-values < 0.05 were considered significant.
Results
EPA and DHA treatment disrupted nonraft organization of EL4 cells.
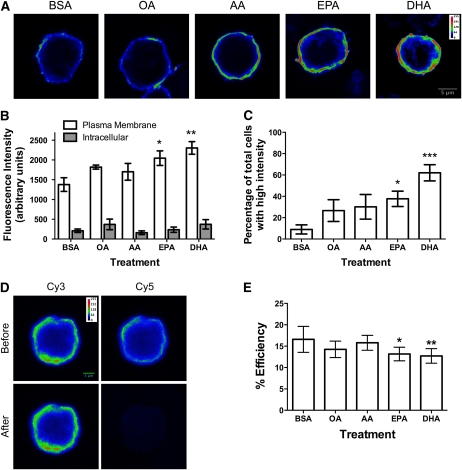
EPA and DHA treatment of EL4 cells increased accumulation of the nonraft probe FAST-DiI in the plasma membrane by ~50–70% relative to the BSA control (Fig. 1A,B). The OA and AA treatments did not significantly increase FAST-DiI uptake compared with BSA (Fig. 1B). The intensity of FAST-DiI inside of the cell was not changed by treatment with any of the fatty acids (Fig. 1B). The increase in FAST-DiI plasma membrane binding with EPA and DHA was also confirmed by scoring the cells for intensity. EPA and DHA treatment resulted in the largest percentage of cells with a high intensity of plasma membrane staining compared with BSA (Fig. 1C).
FIGURE 1.
EPA and DHA treatment disrupted nonraft organization of EL4 cells. (A) Fluorescence images of EL4 cells treated with BSA, OA, AA, EPA, or DHA and stained with FAST-DiI. (B) FAST-DiI intracellular and plasma membrane image intensity. (C) Percentage of cells with high FAST-DiI intensity as a function of treatment. (D) Sample images of BSA-treated cells from acceptor photobleaching FRET. (E) FRET efficiency values for treated EL4 cells. Images are on a rainbow palette to discriminate differences in relative fluorescence intensity. Red and blue values indicate high and low intensity, respectively. Data are means ± SEM, n = 3–4. Asterisks indicate different from BSA: *P < 0.05, **P < 0.01, ***P < 0.001.
A major concern was that the EPA and DHA treatment exerted a general effect on fluorescent probe uptake. Therefore, we tested the effects of fatty acid treatment on the uptake of a nonspecific probe, C16-Bodipy (37). C16-Bodipy did not merely report on the same subcellular organization as FAST-DiI. Colocalization analysis of z-stacks of EL4 cells costained with FAST-DiI and C16-Bodipy showed the percent colocalization between the 2 probes was ~32%, as measured by Mander’s coefficients (Supplemental Fig. 1A) (12). Relative to BSA, EPA and DHA treatment did not promote uptake of C16-Bodipy into the plasma membrane or inside the cell (Supplemental Fig. 1B). OA and AA treatments also had no effect.
We also determined if EPA and DHA treatment modified nonraft organization on a nanometer scale using FRET microscopy. Positive control experiments showed the acceptor photobleaching approach with Cy3 and Cy5 antibodies increased FRET (Supplemental Fig. 2A). The distribution of MHC class I molecules on the surface of EL4 cells was then determined as random, clustered, or a mixture of random and clustered by increasing the donor:acceptor fluorophore ratios. FRET efficiency did not increase with increasing donor:acceptor ratios (Supplemental Fig. 2B), which is the signature pattern of a random distribution of molecules (31, 32). This allowed us to combine FRET acceptor:donor ratios in the subsequent analyses.
Analysis of FRET efficiency values from FRET images (Fig. 1D) in every single experiment showed EPA and DHA treatment consistently decreased FRET by 25–30% relative to the BSA control (Fig. 1E). OA and AA treatments did not lower FRET relative to BSA. The most plausible explanation for an increase in the distance between neighboring MHC I molecules with (n-3) PUFA was an increase in cell size, which was investigated next.
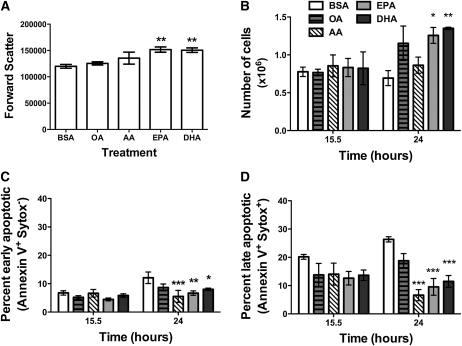
EPA and DHA treatment increased cell size and growth.
Cell size was measured in terms of forward scatter with flow cytometry (Fig. 2A). EPA and DHA treatment increased cell size by ~26% relative to BSA after 15.5 h of treatment. OA and AA treatment had no effect. A similar trend was confirmed with microscopy measurements (data not shown). EPA and DHA, in addition to AA, treatment also increased side scatter relative to BSA (data not shown).
FIGURE 2.
EPA and DHA treatment increased EL4 cell size and growth and prevented cell death. (A) Median forward scatter values for EL4 cells treated with BSA, OA, AA, EPA, or DHA. (B) Cell growth, (C) early apoptosis, and (D) late apoptosis as a function of time for the different treatment groups. Data are means ± SEM, n = 4. Asterisks indicate different from BSA: *P < 0.05, **P < 0.01, ***P < 0.001.
The subsequent experiment tested if the increase in cell size related to a change in cell growth (Fig. 2B). At 15.5 h of treatment, cell number between the BSA control and the differing fatty acids did not differ. After 24 h, EPA and DHA treatment increased the number of viable cells. In contrast, OA and AA treatment did not increase cell growth compared with BSA. We then tested if the increased ability to grow was driven by an ability to prevent cell death. At 15.5 h of treatment, the cells were equally viable when measured for early and late apoptosis (Fig. 2C,D) (12). After 24 h of treatment, all 3 PUFA (AA, EPA, and DHA) prevented early and late apoptosis relative to the BSA control (Fig. 2C,D). OA treatment had no effect relative to BSA.
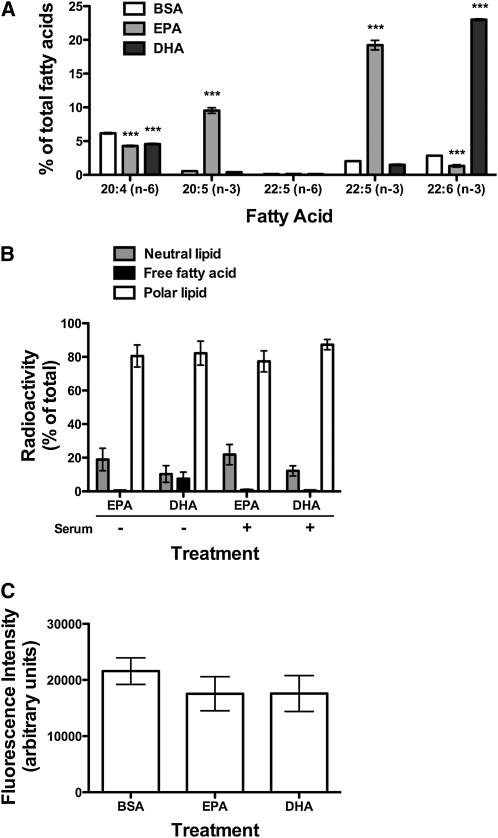
EPA and DHA treatment did not modify nonraft organization when cell size was unchanged.
We determined if preventing the expansion of cell size prevented changes in nonraft organization. Treatment conditions were first optimized to prevent an increase in cell size but still allowed for efficient uptake of the fatty acids. Cells were treated in the presence of 10% FBS, which significantly elevated EPA and DHA levels (Fig. 3A). Uptake of EPA and DHA into polar lipids did not change relative to treatment in serum free conditions, as measured with radiolabeled fatty acids (Fig. 3B). Forward scatter values did not differ between EPA and DHA treatment compared with the BSA control (data not shown). Image analysis of cells stained with FAST-DiI (Fig. 3C) showed no change in fluorescence intensity with EPA- or DHA-treated cells relative to BSA. Similarly, subsequent FRET imaging showed treatment of EL4 cells with (n-3) PUFA did not lower FRET relative to BSA. FRET efficiency values were ~10% for BSA-, EPA-, and DHA-treated cells.
FIGURE 3.
EPA and DHA treatment had no effect on nonraft organization when cell size was unchanged. (A) Total levels of 20:4, 20:5, 22:5, and 22:6 upon treatment of EL4 cells with BSA, EPA, or DHA in serum-containing medium. (B) Incorporation of radiolabeled EPA and DHA into neutral lipids, FFA, and polar lipid fractions in serum-free and serum-containing medium. (C) FAST-DiI intensity of EL4 cells treated with BSA, EPA, or DHA in serum-containing medium. Data are means ± SEM, n = 3. Asterisks indicate different from BSA: ***P < 0.001.
In vivo administration of (n-3) PUFA did not modify nonraft organization but disrupted lipid raft clustering.
Body weights of C57BL/6 mice fed the HF (n-6), LF (n-3), and HF (n-3) PUFA diets did not differ from the CD group after 3 wk (~22 ± 0.3 g). Energy intakes also did not differ between these groups and the CD group (11.4 ± 0.1 kcal/d). GC measurements confirmed uptake of fatty acids from the diet into B cells (Table 2). Cell size did not differ between the B cells from the CD group and those from the other groups (data not shown).
TABLE 2.
Fatty acid analysis of B cells isolated from mice fed diets differing in fat level and composition1,2
| Fatty acid | CD | HF (n-6) PUFA | LF (n-3) PUFA | HF (n-3) PUFA |
| % of total fatty acids | ||||
| 14:0 | 0.9 ± 0.2 | 0.7 ± 0.1 | 0.6 ± 0.2 | 0.8 ± 0.1 |
| 16:0 | 24.9 ± 1.7 | 23.1 ± 2.0 | 25.8 ± 0.8 | 24.6 ± 0.5 |
| 16:1 | 0.8 ± 0.2 | 0.5 ± 0.1 | 1.0 ± 0.2 | 1.1 ± 0.1 |
| 18:0 | 16.5 ± 1.2 | 18.5 ± 2.3 | 18.5 ± 4.0 | 19.4 ± 2.8 |
| 18:1 trans | 0.7 ± 0.2 | 0.9 ± 0.6 | 0.9 ± 0.4 | 0.5 ± 0.2 |
| 18:1 cis | 14.2 ± 2.4 | 9.7 ± 0.6 | 13.2 ± 0.6 | 12.4 ± 0.6 |
| 18:1 (n-7) | 1.9 ± 0.5 | 0.8 ± 0.2 | 1.7 ± 0.1 | 1.7 ± 0.1 |
| 18:2 (n-6) | 17.6 ± 1.6 | 16.5 ± 1.3 | 17.1 ± 2.0 | 18.5 ± 1.0 |
| 18:3 (n-6) | 0.5 ± 0.3 | 1.7 ± 0.7 | 3.6 ± 0.8** | 1.6 ± 0.6 |
| 18:3 (n-3) | 1.6 ± 0.2 | 1.1 ± 0.1 | 1.7 ± 0.2 | 2.3 ± 0.3 |
| 20:1 | 1.1 ± 0.4 | 1.4 ± 0.4 | 0.4 ± 0.1 | 0.7 ± 0.2 |
| 20:2 (n-6) | 1.4 ± 0.7 | 1.6 ± 0.7 | 0.5 ± 0.3 | 0.7 ± 0.5 |
| 20:3 (n-6) | 3.5 ± 1.6 | 0.5 ± 0.2 | 0.9 ± 0.3 | 1.0 ± 0.3 |
| 20:3 (n-3) | 1.1 ± 0.5 | 1.4 ± 0.2 | 0.7 ± 0.2 | 0.8 ± 0.3 |
| 20:4 (n-6) | 5.1 ± 1.6 | 3.5 ± 2.0 | 3.2 ± 0.4 | 2.4 ± 0.7 |
| 20:5 (n-3) | 0.2 ± 0.0 | 0.4 ± 0.1 | 2.0 ± 0.2* | 3.8 ± 0.6*** |
| 22:2 (n-6) | 1.2 ± 0.4 | 0.4 ± 0.2 | 0.1 ± 0.0* | 0.4 ± 0.1 |
| 22:4 (n-6) | 1.2 ± 0.2 | 1.1 ± 0.4 | 0.5 ± 0.0* | 0.5 ± 0.1* |
| 22:5 (n-6) | 0.3 ± 0.1 | 2.3 ± 1.7 | 0.3 ± 0.1 | 0.4 ± 0.1 |
| 22:5 (n-3) | 1.0 ± 0.4 | 1.2 ± 0.1 | 2.7 ± 0.7 | 2.4 ± 0.4 |
| 22:6 (n-3) | 1.7 ± 0.4 | 1.5 ± 0.3 | 3.0 ± 0.4 | 3.5 ± 0.6* |
| 24:1 | 0.9 ± 0.2 | 0.2 ± 0.1* | 0.4 ± 0.0 | 0.4 ± 0.1 |
| ∑SFA | 44.6 ± 2.3 | 46.2 ± 5.2 | 45.6 ± 4.5 | 45.8 ± 2.4 |
| ∑MUFA | 19.8 ± 3.2 | 13.6 ± 1.3 | 17.8 ± 0.6 | 16.9 ± 0.5 |
| ∑(n-6) PUFA | 30.6 ± 2.5 | 26.3 ± 3.9 | 26.5 ± 3.1 | 25.6 ± 2.0 |
| ∑(n-3) PUFA | 5.0 ± 0.7 | 4.3 ± 0.7 | 10.1 ± 0.9** | 11.7 ± 0.8*** |
| ∑PUFA | 35.6 ± 3.0 | 30.7 ± 4.6 | 36.6 ± 4.0 | 37.3 ± 2.0 |
| (n-6):(n-3) PUFA | 6.6 ± 1.0 | 6.1 ± 0.2 | 2.6 ± 0.1** | 2.2 ± 0.2*** |
Values are mean ± SEM, = 4–5. Asterisks indicate different from CD, *P < 0.05, **P < 0.01, ***P < 0.001.
Fatty acids with values < 0.5 are not shown for clarity.
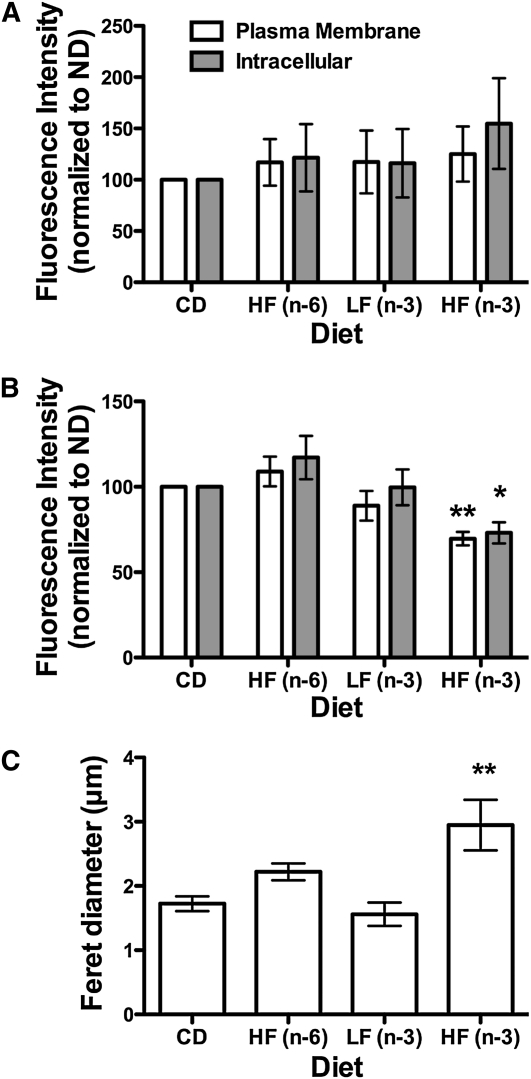
Relative to CD, the HF (n-6), LF (n-3), and HF (n-3) PUFA diets did not increase FAST-DiI uptake into B cells (Fig. 4A). Unexpectedly, the HF (n-3) PUFA diet decreased plasma membrane and intracellular uptake of C16-Bodipy by ~33% (Fig. 4B). The LF (n-3) and HF (n-6) PUFA diets did not affect C16-Bodipy binding. Sample images of FAST-DiI and C16-Bodipy staining are presented in Supplemental Figure 3. To verify the conclusion that (n-3) PUFA did not disrupt nonrafts, MHC I FRET experiments were conducted with the HF (n-3) PUFA diet, because it lowered C16-Bodipy uptake. FRET imaging showed nearly identical FRET efficiency values (~12%) for B cells isolated from the CD and HF (n-3) PUFA diet-fed mice.
FIGURE 4.
(n-3) PUFA diets did not modify nonraft organization, but the HF (n-3) PUFA diet disrupted lipid raft clustering. (A) FAST-DiI intracellular and plasma membrane image intensity of B cells isolated from mice fed CD, HF (n-6), LF (n-3), and HF (n-3) PUFA diets. (B) C16-Bodipy intracellular and plasma membrane image intensity. Fluorescent values in A and B are normalized to CD, because gain settings were not the same between differing sets of experiments. (C) Feret diameter of lipid rafts. Data are means ± SEM, n = 3–5. Asterisks indicate different from CD: *P < 0.05, ** P < 0.01.
Finally, we investigated if the HF (n-3) PUFA diet disrupted B cell lipid raft organization. Microscopy images showed the HF (n-3) PUFA diet decreased the clustering of lipid rafts (Supplemental Fig. 4) and increased the Feret diameter of the domains by ~40% (Fig. 4C) compared with CD. In contrast, the HF (n-6) and LF (n-3) PUFA diets, relative to CD, did not affect lipid raft organization.
Discussion
In this study, we addressed if (n-3) PUFA disrupted nonraft organization of EL4 cells followed by translational studies that determined the impact of (n-3) PUFA on nonraft and lipid raft organization ex vivo. The approach relied on quantitative imaging methods to overcome limitations of using detergent extraction (20, 21). As discussed below, the data point to an emerging model in which lipid rafts appears more sensitive to disruption than nonrafts in response to (n-3) PUFA intervention in EL4 and primary B cells.
Disruption of nonraft organization in vitro.
In serum free conditions, EPA and DHA treatment modified nonraft organization of EL4 cells. There were several possibilities by which EPA or DHA treatment increased FAST-DiI in the plasma membrane. One possibility was uptake of FAST-DiI with (n-3) PUFA represented EPA- and/or DHA-rich domains (13, 14). Alternatively, increased FAST-DiI accumulation with EPA or DHA represented a dissolving of rafts. Thus, the area of nonrafts could have increased upon treatment, but these nonrafts were not organizationally distinct domains. To address these 2 possibilities, specific fluorescent probes will have to be designed, synthesized, and extensively tested in the future.
Some other interpretations of the results with FAST-DiI staining were ruled out. One possibility was BSA, OA, and AA treatment made FAST-DiI staining of the membrane less efficient than EPA or DHA. This was highly unlikely, because we observed similar uptake of C16-Bodipy for most of the treatments. A second possibility was EPA and DHA decreased the microviscosity of the membrane, which allowed for more uptake of the probe (38). Although this was possible, one would then have expected increased internalization of the probe with EPA or DHA treatment compared with BSA; furthermore, increased uptake of C16-Bodipy would have occurred.
The FRET data with EL4 cells revealed EPA and DHA could modify membrane organization on a nanometer scale. To the best of our knowledge, this was the first study to use FRET imaging to study nanometer scale organization of (n-3) PUFA in a cellular system. The data highlight the use of this methodology to determine effects of (n-3) PUFA on the appropriate size scale. The FRET studies showed EPA and DHA increased the distance between randomly distributed nonraft MHC class I molecules.
The increase in cell size with EPA or DHA treatment did not maintain the relative density of MHC class I surface molecules compared with the BSA control. We previously reported MHC I surface levels on EL4 cells increased by ~30–35% with EPA or DHA treatment (12). Here, we found cell size increased by ~26–30% with EPA or DHA. Given that forward scatter roughly reports on the diameter of the cell and if we assume the cell is a sphere, a ~30% increase in diameter of the cell with EPA and DHA treatment should increase surface area by ~60%. Thus, MHC I surface expression should have increased >30–35% with EPA or DHA treatment to maintain the same density of molecules as the BSA-treated cells. Therefore, as cell size increased, MHC I molecules were pushed further apart.
Nonraft organization was not disrupted by (n-3) PUFA in vitro or ex vivo when cell size was unaffected.
Mechanistically, we speculate an increase in cell size with EPA and DHA treatment under serum free conditions caused a redistribution in the lateral organization of lipids and proteins. The increase in cell size appeared to be related to the ability of EPA and DHA to promote cell growth and prevent cell death. The growth data were consistent with previous studies that showed EPA and DHA enhanced cell growth in some cell types (39, 40). Although the numbers of EL4 cells were equivalent across the treatment groups at 15.5 h (time point at which studies were conducted), by 24 h, there were differences in cell number. Therefore, the data suggest cells treated with EPA or DHA at 15.5 h grew in size at a faster rate than the BSA-treated cells and were likely poised to divide. It has been reported with primary cells that cell size, measured with forward scatter, increased as cells started to divide, because a homogenous population of cells can have varying thresholds for entry into the cell cycle (41).
The ex vivo data also showed (n-3) PUFA administered to mice at 2 different doses had no effect on nonraft organization. The ex vivo data were consistent with the in vitro studies in which EPA and DHA did not affect cell size but inconsistent with the in vitro studies in which cell size increased. Several possibilities, which may not be mutually exclusive, could explain the discrepancy. First, there was the obvious difference in cell size, as discussed above. Second, (n-3) PUFA in vitro were administered as single fatty acids, whereas the diet provided a complex mixture of differing fatty acids. Third, B cells from the mice incorporated much lower levels of (n-3) PUFA compared with EL4 cells. However, in vivo levels were more physiologically relevant than those administered in vitro. Indeed, 2 independent imaging experiments showed treatment of EL4 cells in serum free conditions with a lower dose (10 μmol/L EPA or DHA) did not effect FAST-DiI uptake relative to BSA (data not shown). Overall, the in vitro studies using serum free conditions suggest the changes in nonraft organization had limited physiological relevance. Of course, our studies do not rule out the possibility that nonraft organization could be disrupted with (n-3) PUFA in other cell types.
Lipid raft clustering was sensitive to (n-3) PUFA.
Unexpectedly, the HF (n-3) PUFA diet lowered uptake of C16-Bodipy in B cells. It was unlikely the lowered uptake was due to a modification in nonraft organization, because we measured no change in FAST-DiI intensity or FRET. One possibility could be that as (n-3) PUFA increased raft size, it made the membrane more ordered, which prevented uptake of C16-Bodipy. Overall, it was unclear why this probe was not efficiently recruited into the cell, which we aim to investigate in the future. Nevertheless, the HF (n-3) PUFA diet disrupted lipid raft organization, which was highly consistent with our previous data with EL4 cells in which DHA diminished raft clustering (12). The data were also consistent with several studies using in vitro or fat-1 transgenic mouse model systems that showed (n-3) PUFA disrupted lipid raft molecular organization (18, 42–46). Specifically, the ex vivo data from this study agreed with an electron microscopy study that demonstrated treatment of HeLa cells with DHA had a more robust effect on cholesterol-dependent raft domains compared with cholesterol-independent nonraft domains of the inner plasma membrane leaflet (47).
The discovery that the HF (n-3) PUFA diet made rafts appear larger was the first visual evidence of (n-3) PUFA dietary intervention having an effect on lipid rafts in an animal model. Note that although we interpret the rafts to be larger in size, it is possible that they could represent rafts that are much smaller in size but only seem larger due to increased levels of GM1. Future studies will have to address whether the disruption in raft organization represents the formation of highly ordered raft domains or many small raft domains. We also propose lipid raft clustering was diminished, because it was reported that cholera toxin binds more efficiently to GM1 raft molecules when they are declustered (48). Surprisingly, the LF (n-3) PUFA diet did not exert an effect on raft organization. Thus, the levels of (n-3) PUFA in this diet may not have been high enough to modify lipid rafts. Future studies will have to determine at what dose (n-3) PUFA serve to disrupt rafts.
Implications.
Although numerous studies have established that (n-3) PUFA modify the global membrane parameter of microviscosity (38), the specific effects of (n-3) PUFA on membrane domain organization have generally remained elusive. We show (n-3) PUFA specifically target the organization of lipid rafts more than nonrafts, which has relevance toward understanding how these fatty acids disrupt downstream intracellular signaling events or cell-cell communication. The data from this study have implications for several model systems. We present 2 examples for simplicity. A recent study demonstrated DHA treatment prevented dimerization and recruitment of the Toll-like Receptor 4 into lipid rafts (49). Our data suggest DHA could prevent recruitment of the protein into lipid rafts from nonrafts as a consequence of the disruption in the spatial distribution of rafts (which may be occurring on several length scales). As another example, it was proposed that suppression of MDA-MB-231 breast cancer migration with (n-3) PUFA was due to an inability of the chemokine receptor CXCR4 to cluster, but the mechanism was not investigated (50). We propose a disruption in lipid rafts could prevent CXCR4 clustering and one approach would be to measure CXCR4 clustering with FRET imaging on a nanometer scale.
In summary, data from EL4 and primary B cells suggest lipid rafts are far more sensitive to disruption in response to intervention with (n-3) PUFA than nonraft domains. Future studies will have to address how (n-3) PUFA acyl chains initiate a change in the spatial distribution of lipid rafts in the plasma membrane and how this affects protein clustering and ultimately cellular function.
Supplementary Material
Acknowledgments
B.D.R. designed studies, conducted experiments, wrote parts of the manuscript, and analyzed all data; A.F. conducted some FAST-DiI imaging studies and analysis; M.H. fed the mice and conducted chromatography experiments and flow cytometry; H.T. fed the mice and did lipid raft staining and analysis; A.R. analyzed diets; and S.R.S. designed the overall research, did some cell growth measurements, conducted some imaging and confirmed analysis of all the data, wrote parts of the manuscript, and has primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Supported by a Brody Shared Resources Equipment Grant (to S.R.S.) and a grant from the National Center for Complementary and Alternative Medicine at the NIH (R15AT006122) (to S.R.S.).
Supplemental Table 1 and Supplemental Figures 1–4 are available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of contents at jn.nutrition.org.
Abbreviations used: AA, arachidonic acid; C16-Bodipy, 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid; CD, control diet; FAST-DiI, 1,1′-dilinoleyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; FRET, Förster resonance energy transfer; HF, high-fat diet; LF, low-fat diet; MHC, major histocompatibility complex; OA, oleic acid.
Literature Cited
- 1.Fetterman JW, Jr, Zdanowicz MM. Therapeutic potential of n-3 polyunsaturated fatty acids in disease. Am J Health Syst Pharm. 2009;66:1169–79 [DOI] [PubMed] [Google Scholar]
- 2.Moghadasian MH. Advances in dietary enrichment with n-3 fatty acids. Crit Rev Food Sci Nutr. 2008;48:402–10 [DOI] [PubMed] [Google Scholar]
- 3.Kris-Etherton PM, Harris WS, Appel LJ. Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation. 2002;106:2747–57 [DOI] [PubMed] [Google Scholar]
- 4.Fedor D, Kelley DS. Prevention of insulin resistance by n-3 polyunsaturated fatty acids. Curr Opin Clin Nutr Metab Care. 2009;12:138–46 [DOI] [PubMed] [Google Scholar]
- 5.Kabir M, Skurnik G, Naour N, Pechtner V, Meugnier E, Rome S, Quignard-Boulange A, Vidal H, Slama G, et al. Treatment for 2 mo with n 3 polyunsaturated fatty acids reduces adiposity and some atherogenic factors but does not improve insulin sensitivity in women with type 2 diabetes: a randomized controlled study. Am J Clin Nutr. 2007;86:1670–9 [DOI] [PubMed] [Google Scholar]
- 6.Davidson MH, Stein EA, Bays HE, Maki KC, Doyle RT, Shalwitz RA, Ballantyne CM, Ginsberg HN. Efficacy and tolerability of adding prescription omega-3 fatty acids 4 g/d to simvastatin 40 mg/d in hypertriglyceridemic patients: an 8-week, randomized, double-blind, placebo-controlled study. Clin Ther. 2007;29:1354–67 [DOI] [PubMed] [Google Scholar]
- 7.Chapkin RS, Seo J, McMurray DN, Lupton JR. Mechanisms by which docosahexaenoic acid and related fatty acids reduce colon cancer risk and inflammatory disorders of the intestine. Chem Phys Lipids. 2008;153:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calder PC. Immunomodulation by omega-3 fatty acids. Prostaglandins Leukot Essent Fatty Acids. 2007;77:327–35 [DOI] [PubMed] [Google Scholar]
- 9.Galli C, Calder PC. Effects of fat and fatty acid intake on inflammatory and immune responses: a critical review. Ann Nutr Metab. 2009;55:123–39 [DOI] [PubMed] [Google Scholar]
- 10.Yaqoob P, Shaikh SR. The nutritional and clinical significance of lipid rafts. Curr Opin Clin Nutr Metab Care. 2010;13:156–66 [DOI] [PubMed] [Google Scholar]
- 11.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50 [DOI] [PubMed] [Google Scholar]
- 12.Shaikh SR, Rockett BD, Salameh M, Carraway K. Docosahexaenoic acid modifies the clustering and size of lipid rafts and the lateral organization and surface expression of MHC class I of EL4 cells. J Nutr. 2009;139:1632–9 [DOI] [PubMed] [Google Scholar]
- 13.Shaikh SR, Edidin MA. Membranes are not just rafts. Chem Phys Lipids. 2006;144:1–3 [DOI] [PubMed] [Google Scholar]
- 14.Wassall SR, Stillwell W. Docosahexaenoic acid domains: the ultimate non-raft membrane domain. Chem Phys Lipids. 2008;153:57–63 [DOI] [PubMed] [Google Scholar]
- 15.Shaikh SR, Cherezov V, Caffrey M, Soni SP, LoCascio D, Stillwell W, Wassall SR. Molecular organization of cholesterol in unsaturated phosphatidylethanolamines: X-ray diffraction and solid state 2H NMR reveal differences with phosphatidylcholines. J Am Chem Soc. 2006;128:5375–83 [DOI] [PubMed] [Google Scholar]
- 16.Shaikh SR, LoCascio DS, Soni SP, Wassall SR, Stillwell W. Oleic- and docosahexaenoic acid-containing phosphatidylethanolamines differentially phase separate from sphingomyelin. Biochim Biophys Acta. 2009;1788:2421–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schley PD, Brindley DN, Field CJ. (n-3) PUFA alter raft lipid composition and decrease epidermal growth factor receptor levels in lipid rafts of human breast cancer cells. J Nutr. 2007;137:548–53 [DOI] [PubMed] [Google Scholar]
- 18.Fan YY, McMurray DN, Ly LH, Chapkin RS. Dietary (n-3) polyunsaturated fatty acids remodel mouse T-cell lipid rafts. J Nutr. 2003;133:1913–20 [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Tan L, Wang C, Li N, Li Y, Xu G, Li J. Polyunsaturated eicosapentaenoic acid changes lipid composition in lipid rafts. Eur J Nutr. 2006;45:144–51 [DOI] [PubMed] [Google Scholar]
- 20.Lichtenberg D, Goñi FM, Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci. 2005;30:430–6 [DOI] [PubMed] [Google Scholar]
- 21.Heerklotz H. Triton promotes domain formation in lipid raft mixtures. Biophys J. 2002;83:2693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingwood D, Simons K. Detergent resistance as a tool in membrane research. Nat Protoc. 2007;2:2159–65 [DOI] [PubMed] [Google Scholar]
- 23.Rockett BD, Salameh M, Carraway K, Morrison K, Shaikh SR. N-3 PUFA improve fatty acid composition, prevent palmitate-induced apoptosis, and differentially modify B cell cytokine secretion in vitro and ex vivo. J Lipid Res. 2010;51:1284–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves PG. Components of the AIN-93 diets as improvements in the AIN-76A diet. J Nutr. 1997;127:S838–41 [DOI] [PubMed] [Google Scholar]
- 25.Shaikh SR, Edidin M. Immunosuppressive effects of polyunsaturated fatty acids on antigen presentation by human leukocyte antigen class I molecules. J Lipid Res. 2007;48:127–38 [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Mouton C, Lacalle RA, Mira E, Jimenez-Baranda S, Barber DF, Carrera AC, Martinez-A C, Manes S. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J Cell Biol. 2004;164:759–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seveau S, Eddy RJ, Maxfield FR, Pierini LM. Cytoskeleton-dependent membrane domain segregation during neutrophil polarization. Mol Biol Cell. 2001;12:3550–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowness P, Caplan S, Edidin M. MHC molecules lead many lives. workshop on MHC class I molecules at the interface between biology & medicine. EMBO Rep. 2009;10:30–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edidin M. Class I MHC molecules as probes of membrane patchiness: from biophysical measurements to modulation of immune responses. Immunol Res. 2010;47:265–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goebel J, Forrest K, Flynn D, Rao R, Roszman TL. Lipid rafts, major histocompatibility complex molecules, and immune regulation. Hum Immunol. 2002;63:813–20 [DOI] [PubMed] [Google Scholar]
- 31.Kenworthy AK, Petranova N, Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol Biol Cell. 2000;11:1645–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenworthy AK, Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J Cell Biol. 1998;142:69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P, Brand L. Resonance energy transfer: methods and applications. Anal Biochem. 1994;218:1–13 [DOI] [PubMed] [Google Scholar]
- 34.Bastiaens PI, Jovin TM. Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta I. Proc Natl Acad Sci USA. 1996;93:8407–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Image J. Available from: http://rsbweb.nih.gov/ij/
- 36.Stepensky D. FRETcalc plugin for calculation of FRET in non-continuous intracellular compartments. Biochem Biophys Res Commun. 2007;359:752–8 [DOI] [PubMed] [Google Scholar]
- 37.Thumser AE, Storch J. Characterization of a BODIPY-labeled fluorescent fatty acid analogue. binding to fatty acid-binding proteins, intracellular localization, and metabolism. Mol Cell Biochem. 2007;299:67–73 [DOI] [PubMed] [Google Scholar]
- 38.Stillwell W, Wassall SR. Docosahexaenoic acid: membrane properties of a unique fatty acid. Chem Phys Lipids. 2003;126:1–27 [DOI] [PubMed] [Google Scholar]
- 39.Verlengia R, Gorjao R, Kanunfre CC, Bordin S, de Lima TM, Martins EF, Newsholme P, Curi R. Effects of EPA and DHA on proliferation, cytokine production, and gene expression in Raji cells. Lipids. 2004;39:857–64 [DOI] [PubMed] [Google Scholar]
- 40.Mannini A, Kerstin N, Calorini L, Mugnai G, Ruggieri S. Dietary n-3 polyunsaturated fatty acids enhance metastatic dissemination of murine T lymphoma cells. Br J Nutr. 2009;102:958–61 [DOI] [PubMed] [Google Scholar]
- 41.Lyons AB. Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J Immunol Methods. 2000;243:147–54 [DOI] [PubMed] [Google Scholar]
- 42.Kim W, Fan YY, Barhoumi R, Smith R, McMurray DN, Chapkin RS. n-3 Polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J Immunol. 2008;181:6236–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zech T, Ejsing CS, Gaus K, de Wet B, Shevchenko A, Simons K, Harder T. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signaling. EMBO J. 2009;28:466–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers KR, Kikawa KD, Mouradian M, Hernandez K, McKinnon KM, Ahwah SM, Pardini RS. Docosahexaenoic acid alters epidermal growth factor receptor related signaling by disrupting its lipid raft association. Carcinogenesis. 2010;31:1523–30 [DOI] [PubMed] [Google Scholar]
- 45.Chen W, Jump DB, Esselman WJ, Busik JV. Inhibition of cytokine signaling in human retinal endothelial cells through modification of caveolae/lipid rafts by docosahexaenoic acid. Invest Ophthalmol Vis Sci. 2007;48:18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stulnig TM, Huber J, Leitinger N, Imre EM, Angelisova P, Nowotny P, Waldhausl W. Polyunsaturated eicosapentaenoic acid displaces proteins from membrane rafts by altering raft lipid composition. J Biol Chem. 2001;276:37335–40 [DOI] [PubMed] [Google Scholar]
- 47.Chapkin RS, Wang N, Fan Y, Lupton JR, Prior IA. Docosahexaenoic acid alters the size and distribution of cell surface microdomains. Biochim Biophys Acta. 2008;1778:466–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi J, Yang T, Kataoka S, Zhang Y, Diaz AJ, Cremer PS. GM1 clustering inhibits cholera toxin binding in supported phospholipid membranes. J Am Chem Soc. 2007;129:5954–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong SW, Kwon M, Choi AMK, Kim H, Nakahira K, Hwang DH. Fatty acids modulate toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species-dependent manner. J Biol Chem. 2009;284:27384–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altenburg JD, Siddiqui RA. Omega-3 polyunsaturated fatty acids down-modulate CXCR4 expression and function in MDA-MB-231 breast cancer cells. Mol Cancer Res. 2009;7:1013–20 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.