Abstract
Intracellular fibril formation by Ure2p produces the non-Mendelian genetic element [URE3] in S. cerevisiae, making Ure2p a prion protein. We show that solid state NMR spectra of full-length Ure2p fibrils, seeded with infectious prions from a specific [URE3] strain and labeled with uniformly 15N,13C-enriched Ile, include strong, sharp signals from Ile residues in the globular C-terminal domain (CTD), with both helical and non-helical 13C chemical shifts. Treatment with proteinase K (PK) eliminates these CTD signals, leaving only non-helical signals from the Gln- and Asn-rich N-terminal segment, which are also observed in solid state NMR spectra of Ile-labeled fibrils formed by residues 1-89 of Ure2p. Thus, the N-terminal segment, or “prion domain” (PD), forms the fibril core, while CTD units are located outside the core. We additionally show that, after PK treatment, Ile-labeled Ure2p fibrils formed without prion seeding exhibit a broader set of solid state NMR signals than do the prion-seeded fibrils, consistent with the idea that structural variations within the PD core account for prion strains. Measurements of 13C-13C magnetic dipole-dipole couplings among 13C-labeled Ile carbonyl sites in full-length Ure2p fibrils support an in-register parallel β-sheet structure for the PD core of Ure2p fibrils. Finally, we show that a model in which CTD units are attached rigidly to the parallel β-sheet core is consistent with steric constraints.
Keywords: amyloid structure, yeast prions, dipolar recoupling, magic-angle spinning
Introduction
Ure2p of S. cerevisiae is a 354-residue protein, consisting of a 245-residue C-terminal domain (CTD) with a globular fold, structurally homologous to glutathione S-transferase1,2, and a 110-residue, Asn- and Gln-rich N-terminal segment that is natively unfolded3-5. Under typical experimental conditions, the CTD dimerizes through multiple noncovalent interactions, principally hydrophobic in nature, on its flat face1,2,6. The normal biological function of Ure2p is to regulate genes involved in catabolism of certain nitrogen sources by binding to the transcription factor Gln3p. [URE3], a non-Mendelian genetic element of S. cerevisiae described in 1971 by Lacroute7, was shown in 1994 to be an autoinactivating infectious form of Ure2p8, making Ure2p the first non-mammalian prion protein to be identified. Aggregation of Ure2p into amyloid fibrils proved to be the basis of [URE3]9,10, as is likely the case for the mammalian prion diseases11-14. [URE3] is undetectable in wild yeast strains15, implying that [URE3] is most likely a prion “disease”, not an advantageous or functional condition.
Polypeptides from the N-terminal segment (e.g., Ure2p1-65, Ure2p1-89, and Ure2p10-3910,16,17, with Ure2pn-m representing residues n to m of Ure2p) form amyloid fibrils in vitro, protease treatment of full-length Ure2p amyloid yields only N-terminal fragments18, [URE3] propagates stably in cells that express only the N-terminal segment (but not in cells that express only the CTD)19, and protease-treated Ure2p amyloid induces [URE3] when transfected into S. cerevisiae20. Overexpression of N-terminal polypeptides (e.g., Ure2p1-65) within S. cerevisiae also induces [URE3]9. Therefore, the N-terminal segment (more specifically, the Asn- and Gln-rich portion thereof, residues 1-89) is the “prion domain” (PD) of Ure2p. Solid state nuclear magnetic resonance (NMR) measurements on Ure2p1-8916 and Ure2p10-3917 fibrils indicate that the cross-β structure within these fibrils, identified by electron diffraction21, has an in-register parallel organization, as also observed in full-length β-amyloid fibrils22, amylin fibrils23, and other yeast prion fibrils24-27. In-register, parallel β-sheet formation is apparently favored by intermolecular interactions among Gln and/or Asn sidechains, which are automatically aligned by the parallel structure so as to permit “polar zipper” interactions28-31 regardless of the order or spacing of Gln and Asn residues within the amino acid sequence17,25.
Recently, Loquet et al. have reported the first solid state NMR studies of full-length Ure2p fibrils, prepared with uniform 15N and 13C labeling32. Two-dimensional (2D) solid state NMR spectra of these fibrils clearly show strong, sharp signals attributable to the CTD, nearly identical to the signals observed in 2D spectra of microcrystals formed by the CTD alone. Thus, it appears that the CTD retains its globular fold in full-length Ure2p fibrils. This observation is consistent with experiments by Baxa et al., which showed that fibrils formed by fusions of the Ure2p PD with several globular partners (including barnase, carbonic anhydrase, glutathione S-transferase, and green fluorescent protein) retain the activity of the partner protein33 and that the calorimetric signature of the CTD unfolding is identical in soluble and fibrillized Ure2p3, as well as experiments by Bai et al., which showed that Ure2p retains glutathione peroxidase activity in the amyloid state34. An important implication of the solid state NMR data of Loquet et al. is that the CTD is not highly mobile in full-length Ure2p fibrils32, ruling out the possibility that CTD monomers are tethered to the fibril core by a disordered linker segment that is sufficiently long to permit large and rapid reorientational motions of the CTD.
Loquet et al. suggest that their results may support a model in which the core of biologically relevant Ure2p fibrils is constructed from CTD dimers, with the N-terminal segment located on the periphery (see Fig. S9 of ref. 32). Such a model is inconsistent with previous work described above, especially the infectivity of fibrils that contain only the PD, the induction of [URE3] by overexpression of the PD, and the stable propagation of [URE3] in cells that express only the PD9,19,20. An alternative interpretation of the solid state NMR results of Loquet et al. is that the PD forms a rigid, cross-β core that is contained within a helical shell of CTD units, which are immobilized by a combination of covalent linkage at their N-termini to the PD core, noncovalent CTD-CTD dimerization, and contacts among CTD dimers. Below, we present solid state NMR data to support this alternative interpretation. Data below also provide further evidence that the structural basis for distinct, self-propagating strains or variants of [URE3] lies in the PD core.
Results
Description of samples
The relatively low resolution of previously reported solid state NMR spectra of Ure2p1-89 fibrils is attributable in part to structural heterogeneity16, consistent with the production of a mixture of [URE3] prion variants on infection of yeast with such filaments20. In order to prepare samples with greater structural homogeneity, we seeded the growth of full-length Ure2p fibrils with prion aggregates extracted from S. cerevisiae cells that carried a specific variant of [URE3]. The slower kinetics of amyloid formation by full-length Ure2p compared with Ure2p1-8910 allows efficient seeding to occur under appropriate conditions (see Materials and Methods and Fig. S1). To eliminate NMR signals from outside the core of the Ure2p fibrils, the fibrils were treated with proteinase K (PK), a nonspecific protease that has been widely used to identify the stably structured cores of amyloid and prion fibrils35-37. As was shown previously18, such treatment generates protease-resistant fragments that correspond to the N-terminal region of Ure2p (Table 1 and Fig. S2). Full-length Ure2p fibrils seeded with S. cerevisiae extracts (EX-Ure2p) were compared with spontaneously formed full-length Ure2p fibrils (SP-Ure2p) and with fibrils formed by Ure2p1-89. For these samples, His-tagged Ure2p and Ure2p1-89 sequences were expressed in E. coli and isotopically labeled only at Ile residues, either with 15N and 13C at all sites (U-Ile) or with 13C only at the carbonyl site (1-Ile). Samples are therefore named U-Ile-EX-Ure2p, U-Ile-SP-Ure2p, PK-U-Ile-EX-Ure2p, PK-U-Ile-SP-Ure2p, U-Ile-SP-Ure2p1-89, and 1-Ile-SP-Ure2p. All solid state NMR measurements were performed on fully hydrated fibrils, which had never been dried or lyophilized. Details of sample preparation and experimental measurements are given in the Materials and Methods.
Table 1.
Ure2p fragments generated by proteinase K digestion of Ure2p fibrils, as determined by mass spectrometry. Fibrils were formed by recombinant Ure2p, either with seeding by Ure2p prions extracted from [URE3] S. cerevisiae or spontaneously as described in the Materials and Methods.
| Sample | Measured mass | Calculated mass | Relative abundance | residue numbers1 | |
|---|---|---|---|---|---|
| prion-seeded Ure2p fibrils | sample 1 | 8406.4 | 8405.7 | 100 | -8-67 |
| 8736.0 | 8735.1 | 60 | -8-70 | ||
| 8622.9 | 8622.9 | 65 | -10-66 | ||
| sample 2 | 8406.6 | 8405.7 | 100 | -8-67 | |
| 8735.8 | 8735.1 | 70 | -8-70 | ||
| 8622.0 | 8622.9 | 65 | -10-66 | ||
| sample 3 | 8406.4 | 8405.7 | 100 | -8-67 | |
| 8735.7 | 8735.1 | 70 | -8-70 | ||
| 8621.9 | 8622.9 | 60 | -10-66 | ||
| unseeded Ure2p fibrils | sample 1 | 8622.7 | 8622.9 | 100 | -10-66 |
| 8647.0 | 8646.0 | 40 | -11-65 | ||
| 9679.3 | 9678.9 | 30 | 2-88 | ||
| sample 2 | 8405.9 | 8405.7 | 100 | -8-67 | |
| 8622.6 | 8622.9 | 60 | -10-66 | ||
| 9164.4 | 9163.6 | 30 | -12-69 | ||
| sample 3 | 8623.0 | 8622.9 | 100 | -10-66 | |
| 9681.3 | 9681.9 | 40 | 1-87 | ||
| 9164.1 | 9163.6 | 30 | -12-69 |
Negative residue numbers refer to the MH6MYPRGN tag that precedes the full-length Ure2p sequence in our construct.
Ure2p contains 18 Ile residues, 14 of which are in the CTD. According to crystal structures of dimeric CTD1,2, nine Ile residues occur in helical segments (I130, I189, I212, I227, I308, I323, I327, I329, and I348), three occur in β-strand segments (I142, I169, and I178), and two occur in coil or turn segments (I241 and I325). The remaining four Ile residues occur in the PD (I21, I35, and I77) or in the segment between the PD and the CTD (I102). Since Ile residues in different secondary structure elements have distinctive 13C NMR chemical shifts38,39, solid state NMR spectra of Ile-labeled samples can provide useful structural information even without site-specific chemical shift assignments.
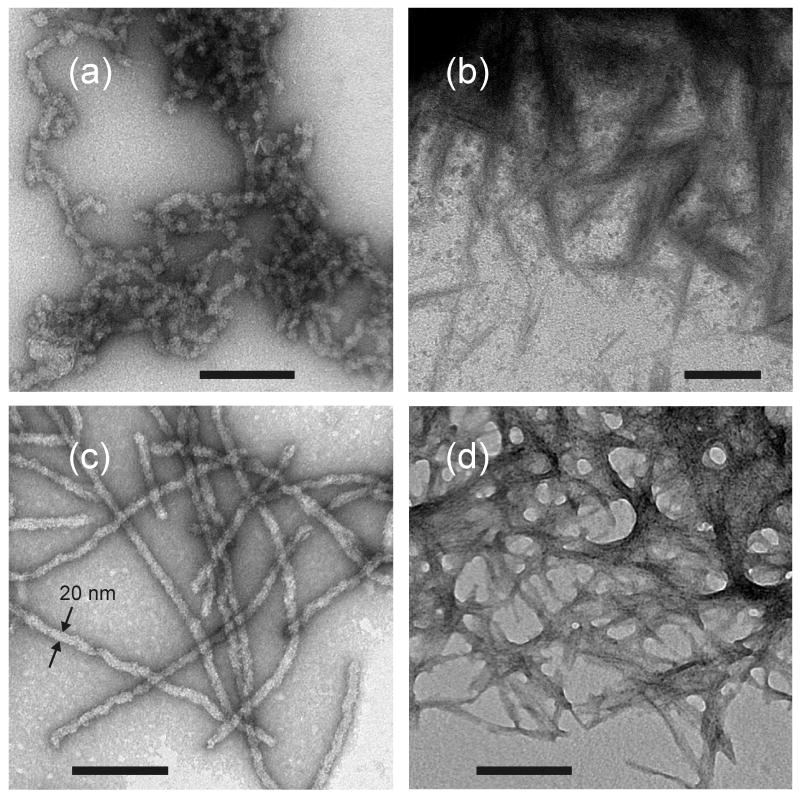
Fig. 1 shows transmission electron microscope (TEM) images of negatively stained fibril samples. Prion-seeded Ure2p fibrils (Fig. 1a) are typically less than 100 nm in length, while unseeded fibrils (Fig. 1b) have lengths approaching 1 μm. This difference arises from the fact that, in the case of seeded growth, the fibrils were sonicated after two days of incubation with prion seeds, then incubated for an additional two days (see Materials and Methods). Sonication generates a higher concentration of fibril ends, as required for efficient conversion of Ure2p to the fibrillar state in seeded growth. Sonication was not required in the production of unseeded fibrils because these fibrils were grown from a higher initial concentration of recombinant Ure2p. After PK treatment (Figs. 1c and 1d), the fibrils become thinner and tend to self-associate, making it difficult to assess the morphology of individual PK-treated fibrils in the TEM images
Figure 1.
TEM images of prion-seeded (a,b) and unseeded (c,d) Ure2p fibrils before (a,c) and after (b,d) proteinase K treatment. TEM grids are negatively stained with uranyl acetate. Scale bars are 200 nm.
Proteinase K treatment selectively removes the CTD from Ure2p fibrils
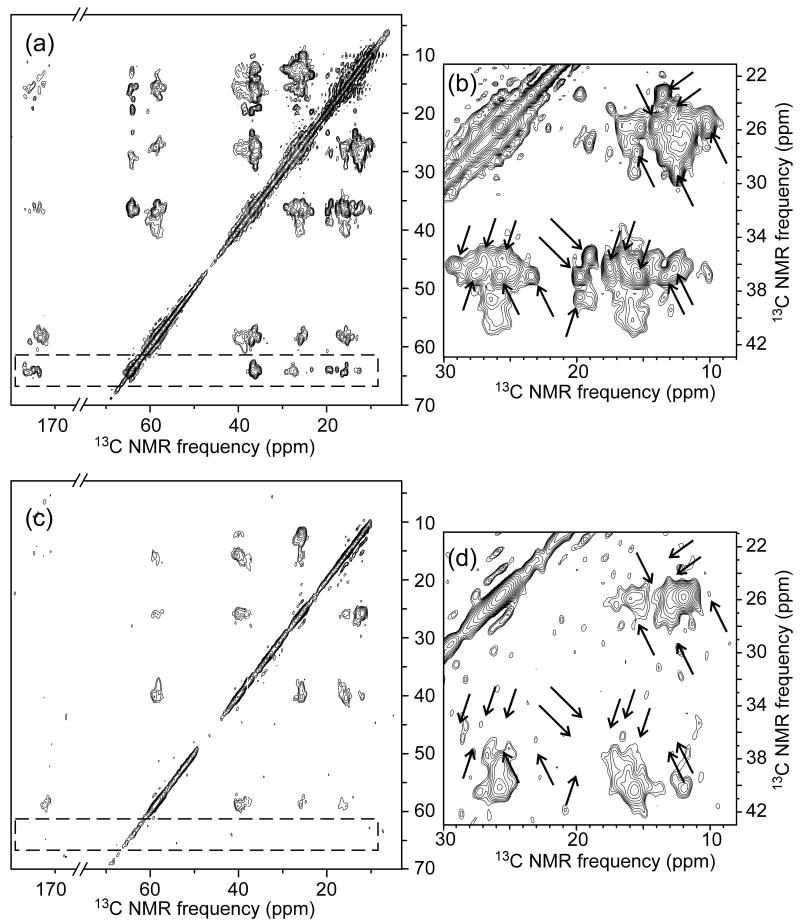
Fig. 2 shows the 2D 13C-13C NMR spectra of U-Ile-EX-Ure2p and PK-U-Ile-EX-Ure2p fibrils. 1D slices from these spectra are shown in Fig. S3 of supplementary data. For U-Ile-EX-Ure2p fibrils (Figs. 2a and 2b), crosspeak signals of Ile residues in helical segments of CTD are clearly separated from those in non-helical segments (of the CTD and PD), primarily due to the downfield shifts of helical Ile α-carbon signals (63-65 ppm) relative to non-helical Ile α-carbon signals (56-61 ppm). Consistent with the results of Loquet et al.32, signals from helical Ile sites in the CTD are strong and sharp (<1 ppm full-width-at-half-maximum). Sharp non-helical signals are also observed, which we attribute primarily to non-helical Ile residues in the CTD.
Figure 2.
2D 13C-13C NMR spectra of prion-seeded Ure2p fibrils before (a,b) and after (c,d) proteinase K treatment. Ile residues within Ure2p are uniformly 15N,13C-labeled. Spectra were obtained in a 14.1 T magnetic field. Contour levels increase by successive factors of 1.5 (a,c) or 1.3 (b,d). Dashed rectangle and arrows indicate Ile crosspeak signals that are eliminated by proteinase K treatment.
After PK treatment, nearly all sharp signals vanish from the 2D 13C-13C spectrum of PK-U-Ile-EX-Ure2p (Figs. 2c and 2d). Crosspeak signals from helical Ile residues vanish completely (dashed rectangular areas in Figs. 2a and 2c). Remaining signals come from residues 1-70 of Ure2p, as indicated by mass spectrometry of the PK-treated fibrils (Table 1). Thus, PK treatment selectively removes the CTD from Ure2p fibrils that were grown from biologically active prion seeds. This clearly shows that the fibril core is not comprised of CTD units.
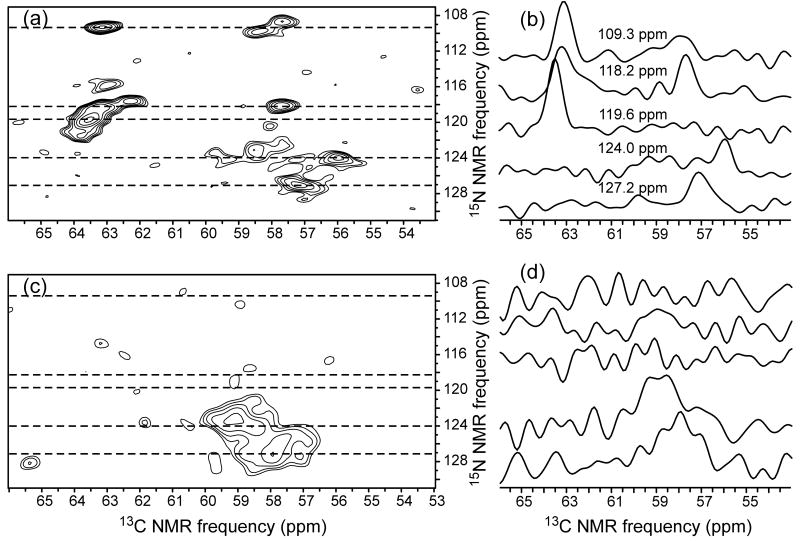
Fig. 3 shows 2D 15N-13C spectra of U-Ile-EX-Ure2p and PK-U-Ile-EX-Ure2p fibrils. These spectra also indicate that PK treatment selectively removes the sharp solid state NMR signals that arise from the globular CTD.
Figure 3.
2D 15N-13C spectra of prion-seeded Ure2p fibrils before (a) and after (c) proteinase K treatment (same samples as in Fig. 2). Contour levels increase by successive factors of 1.3. 1D slices at indicated 15N chemical shifts (dashed lines in the 2D spectra) are shown in (b,d).
Seeding with a specific [URE3] strain produces a more homogeneous Ure2p fibril core
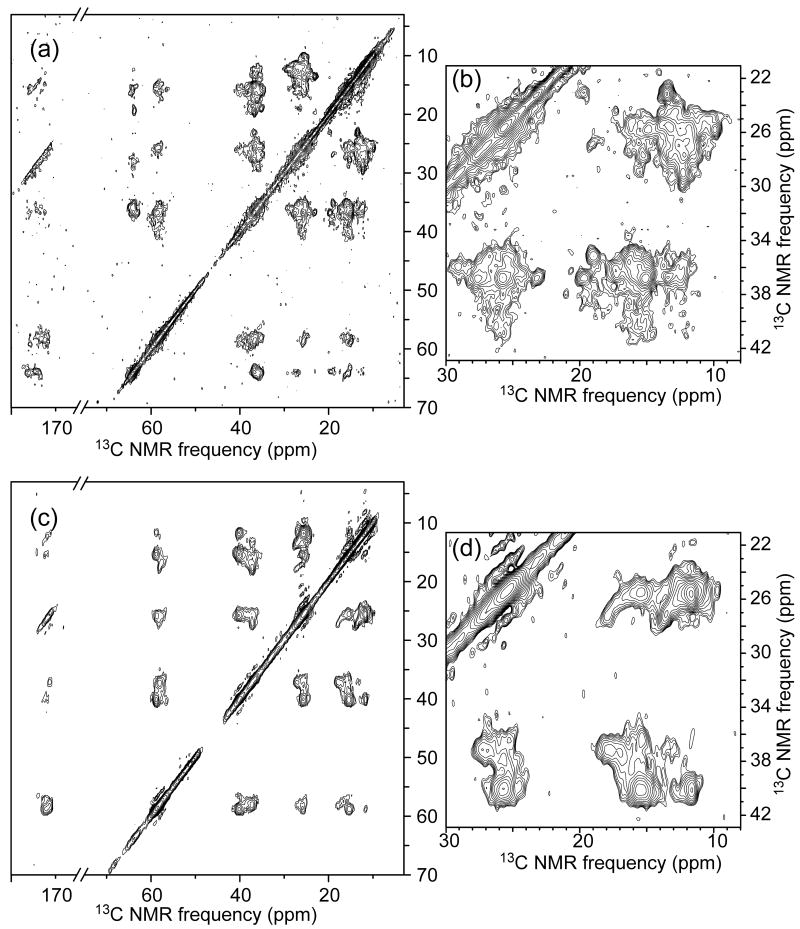
Fig. 4 shows 2D 13C-13C spectra of U-Ile-SP-Ure2p and PK-U-Ile-SP-Ure2p fibrils. The spectrum of U-Ile-SP-Ure2p fibrils (Figs. 4a and 4b) is similar to that of U-Ile-EX-Ure2p fibrils (Figs. 2a and 2b) because both spectra are dominated by signals from the CTD, which has the same structure in both samples. However, after PK treatment, the spectrum of PK-U-Ile-SP-Ure2p fibrils (Figs. 4c and 4d) differs significantly from that of PK-U-Ile-EX-Ure2p fibrils (Figs. 2c and 2d). In particular, the 2D 13C-13C spectrum of the prion-seeded sample shows fewer crosspeak signals (see especially Figs. 2d and 4d, as well as Fig. S3). This observation indicates that the PK-resistant core structure is more structurally homogeneous in the prion-seeded fibrils than in the unseeded fibrils, consistent with the idea that strain variations arise from structural variations in the fibril core. PK treatment also produced a somewhat different set of Ure2p fragments in the two cases, including a minor population of fragments that extend to residue 87 or 88 in the case of unseeded Ure2p fibrils (Table 1). Although I77 potentially contributes to solid state NMR signals of PK-U-Ile-SP-Ure2p fibrils, but not to those of PK-U-Ile-EX-Ure2p fibrils, this contribution should not be sufficient to explain the observed differences in crosspeak signals, as the longer PK fragments are a minor component (roughly 25% of the PK-treated sample, which means roughly 10% of the total NMR signal from Ile residues).
Figure 4.
2D 13C-13C NMR spectra of unseeded Ure2p fibrils before (a,b) and after (c,d) proteinase K treatment. Contour levels increase by successive factors of 1.5 (a,c) or 1.3 (b,d).
The 2D 13C-13C spectrum of U-Ile-SP-Ure2p1-89 fibrils (Fig. S4) is similar to spectra of PK-Ile-EX-Ure2p and PK-Ile-SP-Ure2p fibrils, as expected if the molecular structure in Ure2p1-89 fibrils is similar to the molecular structure in the core of full-length Ure2p fibrils. Structural homogeneity of U-Ile-SP-Ure2p1-89 fibrils may be intermediate between that of PK-Ile-EX-Ure2p fibrils and that of PK-Ile-SP-Ure2p fibrils.
The core of full-length Ure2p fibrils has an in-register parallel β-sheet structure
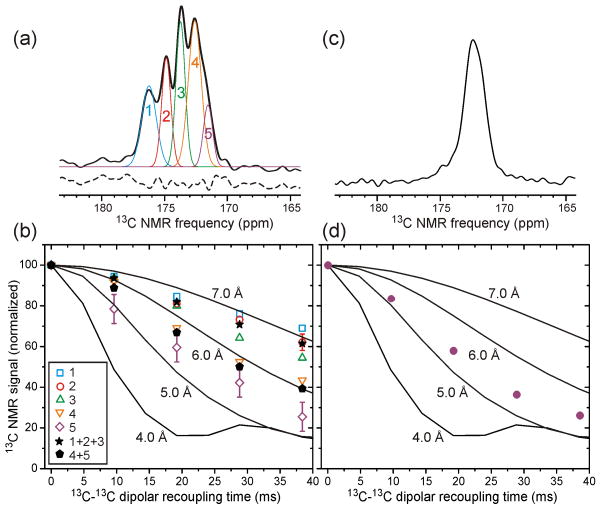
Fig. 5 shows measurements of nuclear magnetic dipole-dipole couplings among Ile carbonyl sites in 1-Ile-SP-Ure2p fibrils, using the PITHIRDS-CT technique40. We have previously used the same technique to characterize the β-sheet structures in other amyloid and prion fibrils22,24-26. When 13C labels form linear chains with 4.8 Å intermolecular spacings, as expected in an in-register parallel β-sheet, 13C-13C couplings produce a decay of 13C signals on the time scale of 30 ms. Larger spacings produce longer decay times, proportional to the inverse cube of the internuclear distance. In measurements on 1-Ile-SP-Ure2p fibrils, interpretation of the PITHIRDS-CT data is complicated by the fact that only a fraction of the Ile carbonyl 13C labels are in the PD and the fact that the CTD contains 13C pairs with relatively short distances. Examination of the CTD crystal structure shows that, among helical Ile residues, I327 and I329 have 4.5 Å nearest-neighbor 13C-13C distances, I323 has a 5.8 Å nearest-neighbor distance, and I212 and I308 have 6.5 Å nearest-neighbor distances. I325, in a coil segment, has a 5.8 Å nearest-neighbor distance (to I323). The remaining 8 Ile residues, including all CTD Ile residues in β-strands, have nearest-neighbor distances greater than 7.0 Å.
Figure 5.
(a) 13C NMR spectrum of unseeded Ure2p fibrils in which Ile residues are 13C-labeled only at carbonyl sites. Thin lines are a fit of the carbonyl lineshape with five Gaussian components. Dashed line is the residual after fitting, offset vertically for clarity. (b) Decay of 13C NMR signals due to 13C-13C magnetic dipole-dipole couplings for the five Gaussian components (open symbols), the sum of components 1-3 (filled stars), and the sum of components 4 and 5 (filled pentagons). Data were obtained with the PITHIRDS-CT dipolar recoupling technique40. Lines are numerical simulations of PITHIRDS-CT data for linear chains of 13C nuclei with the indicated spacings. (c) 13C NMR spectrum of the same sample after proteinase K treatment. (d) PITHIRDS-CT data (filled circles) of the same sample after proteinase K treatment.
Fig. 5a shows that the carbonyl 13C NMR signal from 1-Ile-SP-Ure2p fibrils can be deconvolved into five Gaussian components, at chemical shift values of 176.2, 174.9, 173.8, 172.6, and 171.5 ppm and with area ratios of 3.6:3.0:4.0:5.7:1.7 (in order of decreasing chemical shift, and normalized to a total area of 18 to match the 18 Ile residues in Ure2p). Carbonyl signals in the 2D 13C-13C spectrum of PK-U-Ile-SP-Ure2p fibrils (Fig. 4a) are entirely contained in the 170.8-173.0 ppm range. Based on both their positions and their areas, it is therefore reasonable to assign the two highest-field (lowest chemical shift) components of the carbonyl lineshape in Fig. 5a (i.e., peaks 4 and 5) to Ile sites in β-strands, including four sites in the N-terminal segment and three sites in the CTD. Fig. 5b shows that these components decay most rapidly in the PITHIRDS-CT data. The decay of the sum of peaks 4 and 5 lies between ideal simulations for linear chains of 13C nuclei with 5.0 Å and 6.0 Å spacings, as expected if three or four of the seven 13C-labeled sites that contribute to these peaks follow the 5.0 Å simulations and the remaining sites do not decay significantly on the 30 ms time scale. Given that the β-strand Ile sites in the CTD have no nearest-neighbor distances less than 7.0 Å, the experimentally observed decay of the sum of peaks 4 and 5 implies that the 13C-labeled sites in the PD (I21, I35, and I77, and possibly I102) have nearest-neighbor 13C-13C distances of approximately 5 Å. Since these sites are well separated in the Ure2p sequence, this result supports an in-register parallel β-sheet structure for the PD core in full-length Ure2p fibrils. As previously reported16, similar data for Ala, Leu, and Val residues in Ure2p1-89 fibrils also support an in-register parallel β-sheet structure.
As further support for our interpretation of the data in Figs. 5a and 5b, we performed PK digestion of the same 1-Ile-SP-Ure2p fibrils. The 13C NMR spectrum in Fig. 5c shows that the three low-field carbonyl signal components are selectively attenuated after PK digestion. PITHIRDS-CT data for the remaining carbonyl signals exhibit a rapid decay, as shown in Fig. 5d. The remaining deviation of the experimental PITHIRDS-CT data after PK digestion from the simulated 5.0 Å simulated curve can be attributed to a minor signal contribution from natural-abundance 13C at carbonyl sites, expected to be roughly 20% of the total carbonyl 13C signal after PK digestion.
CTD units are attached rigidly to the core of full-length Ure2p fibrils
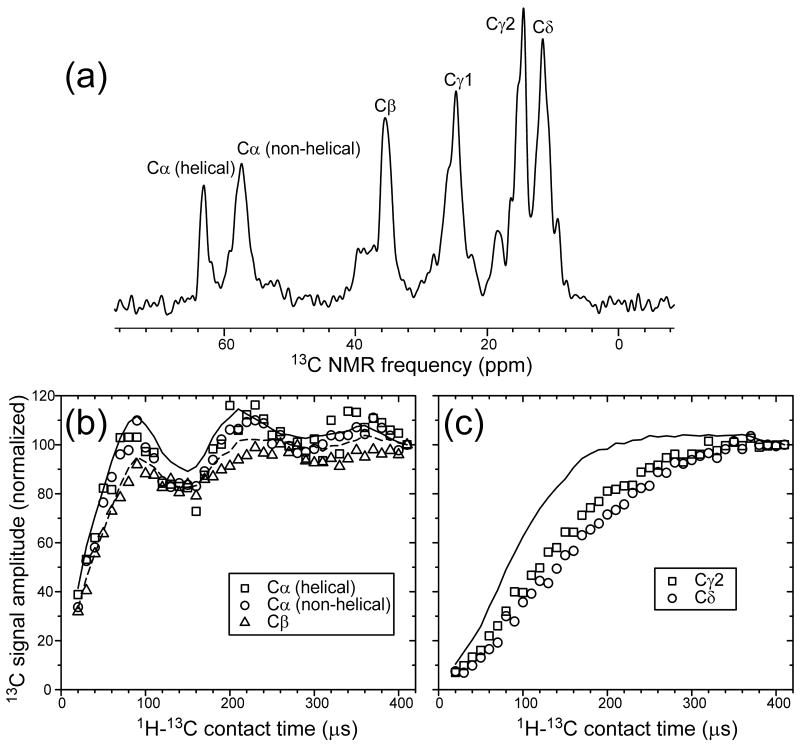
Fig. 6a shows the aliphatic region of the one-dimensional 13C NMR spectrum of U-Ile-EX-Ure2p fibrils. In this spectrum, as well as in the 2D spectra discussed above, signals from 13C nuclei were detected after transfer of spin polarization from 1H nuclei by Hartmann-Hahn 1H-13C cross-polarization (CP)41,42. The CP time scale in such measurements depends on the strength of 1H-13C dipole-dipole couplings, which are sensitive to motional averaging. Rapid, large-amplitude molecular motions can reduce the time-averaged couplings and increase the time scale for build-up of 13C polarization (and hence the build-up of 13C NMR signals) under CP.
Figure 6.
(a) Aliphatic region of the 1D 13C NMR spectrum of prion-seeded Ure2p fibrils in which Ile residues are uniformly 15N,13C-labeled, with assignments to Ile carbon sites. (b,c) 1H-13C cross-polarization build-up data for the indicated sites (open symbols). Solid and dashed lines in (b) are data for Cα and Cβ sites in uniformly 15N,13C-labeled L-valine powder, obtained under identical experimental conditions. Solid line in (c) is data for methyl sites in uniformly 15N,13C-labeled L-valine powder.
Fig. 6 compares experimental CP build-up measurements on U-Ile-EX-Ure2p fibrils with the same measurements on uniformly 15N,13C-labeled L-valine powder, in which large-amplitude molecular motions (other than methyl group rotation) are absent. The build-up curves for Ile α-carbon and β-carbon signals from U-Ile-EX-Ure2p fibrils are remarkably similar to the corresponding curves for L-valine powder (Fig. 6a), indicating that motional averaging of dipole-dipole couplings in the CTD backbone is minimal. Build-up of Ile methyl carbon signals (γ2-carbon and δ-carbon) is slower than build-up of L-valine methyl carbon signals (Fig. 6b), but this observation can be attributed to Ile sidechain motions in the U-Ile-EX-Ure2p fibrils. Thus, CTD units of full-length Ure2p fibrils do not undergo large-amplitude reorientational motions on the sub-millisecond time scale. The amplitude of diffusional or librational motions must be on the order of ±10° or less to be consistent with data in Fig. 6a.
Discussion
Interpretation of solid state NMR data for Ure2p fibrils
The initial solid state NMR studies of selectively 13C-labeled Ure2p1-89 fibrils by Baxa et al.16 showed that Leu, Val, and Ala residues have 13C chemical shifts and intermolecular 13C-13C dipole-dipole couplings indicative of an in-register parallel β-sheet structure. 2D 15N-13C and 13C-13C spectra of uniformly 15N,13C-labeled Ure2p1-89 fibrils did not show many sharp, well-resolved crosspeak signals, especially when compared with 2D spectra of uniformly 15N,13C-labeled HET-s prion domain fibrils obtained under identical experimental conditions16. The comparatively poor resolution observed in 2D spectra of uniformly 15N,13C-labeled Ure2p1-89 fibrils was attributed to structural heterogeneity, which may include both polymorphisms that account for distinct [URE3] strains and conformational disorder within fibrils with a single morphology. Quantitative analysis of the integrated intensities of crosspeak regions in 2D 13C-13C spectra suggested that the immobilized core of Ure2p1-89 fibrils contains only a subset of the amino acid sequence16.
Solid state NMR data for full-length Ure2p fibrils and Ure2p1-93 fibrils described subsequently by Loquet et al.32 are not inconsistent with the data of Baxa et al. Although 2D spectra of uniformly-labeled, full-length Ure2p fibrils do show many sharp crosspeak signals, these signals arise primarily from the globular CTD, as demonstrated by Loquet et al. through comparisons of spectra of full-length Ure2p fibrils with spectra of microcrystalline CTD32. 2D 13C-13C spectra of uniformly-labeled Ure2p1-93 fibrils reported by Loquet et al. (see Fig. S3 of ref. 32) are qualitatively similar to spectra of uniformly-labeled Ure2p1-89 fibrils reported by Baxa et al. (see Fig. 4 of ref. 16), with minor differences attributable to differences in the amino acid sequences, sample preparation conditions, and NMR measurement conditions (e.g., radio-frequency pulse sequences and magnetic field strengths). We see no major differences in the data that would necessitate a revision of the conclusions reached by Baxa et al. Loquet et al. also report that the resolution of 1D and 2D solid state 13C NMR spectra of full-length Ure2p fibrils is irreversibly impaired by heating to 50° C or higher32, but of course such heat treatment was not employed in the solid state NMR experiments of Baxa et al. or in the experiments described above.
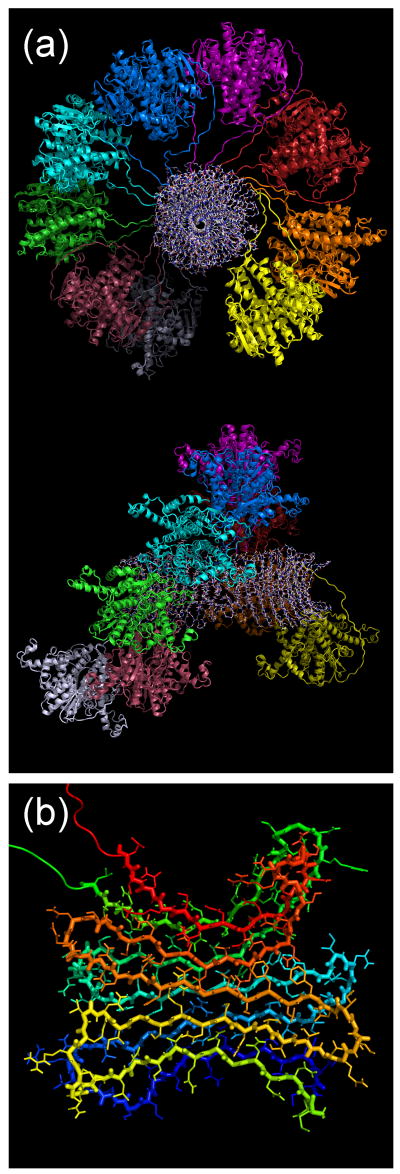
Loquet et al. report that their solid state NMR data are consistent with a model for full-length Ure2p fibrils in which the globular CTD (rather than the N-terminal PD) forms the central fibril core. Such a model, if correct, would imply that measurements on Ure2p1-89 and Ure2p1-93 fibrils are irrelevant to the properties of full-length Ure2p fibrils and irrelevant to the [URE3] phenomenon. Data presented above argue against such a model. While 2D spectra of full-length Ure2p fibrils that are uniformly 15N- and 13C-labeled at Ile residues show both sharp and broad crosspeak signals (Figs. 2a, 2b, 3a, 4a, 4b, S3a, and S3c), the sharp signals from the CTD are removed by PK treatment. The PK-treated fibrils retain only the PD (Table 1), and have only non-helical Ile chemical shifts. Moreover, measurements of intermolecular 13C-13C dipole-dipole couplings in full-length Ure2p fibrils that are 13C-labeled at carbonyl sites of Ile residues (Fig. 5) reveal couplings for β-strand sites in the PD that are consistent with the in-register parallel β-sheets identified in Ure2p1-89 and Ure2p10-39 fibrils16,17. All solid state NMR data, including those of Baxa et al., those of Loquet et al., and those presented above, support the model for full-length Ure2p fibrils shown in Fig. 7, in which the PD forms a cross-β core comprised of in-register parallel β-sheets and the CTD forms an outer helical shell. CTD units are immobilized by covalent attachment to the PD core at their N-termini, by their inherent dimerization, and by contacts between CTD dimers, but are susceptible to PK digestion.
Figure 7.
(a) Structural model for full-length Ure2p fibrils, generated as described in the text. A fibril section comprised of nine Ure2p dimers is shown, viewed down the fibril axis (top) and from the side (bottom). CTD dimers are shown in a cartoon representation, with each dimer in a distinct color. Residues 1-81 are shown in an all-atom representation. The fibril diameter is 20 nm, in good agreement with Fig 1c. (b) PD segments of one dimer, viewed down the fibril axis, representing a possible molecular structure for the cross-β fibril core.
Comparison of Figs. 2c and 2d with Figs. 4c and 4d shows that recombinant Ure2p fibrils that were seeded with [URE3] prions of a specific strain have a more homogeneous PD core structure than fibrils that form spontaneously (see also Fig. S3). This result provides evidence that distinct yeast prion strains arise from structural variations within the cross-β core. This result also suggests that full structure determination for yeast prion fibrils may be facilitated by seeded fibril growth, using infectious prion extracts.
Relation of solid state NMR data to previous studies of Ure2p
Our interpretation of solid state NMR data for Ure2p fibrils is also fully consistent with previous structural, biochemical, and genetic studies. Brachmann et al.20 have shown that protease-treated Ure2p amyloid, which contains only N-terminal fragments18, efficiently infects S. cerevisiae with the [URE3] prion. Masison et al. have shown that overexpression of N-terminal polypeptides within S. cerevisiae also induces [URE3]9 and that [URE3] propagates stably in cells that express only the PD19. Fibrils formed by fusions of the Ure2p PD with several globular partners retain the activity of the partner protein33; in addition, these fibrils (which lack the Ure2p CTD) induce [URE3] when transfected into S. cerevisiae20. Scanning transmission electron microscopy of full-length Ure2p fibrils and fibrils formed by Ure2p PD fusion proteins indicates mass-per-length values consistent with a cross-β structure with one protein molecule per β-sheet repeat spacing18, as in Fig. 7. High-resolution TEM images of full-length Ure2p fibrils and Ure2p PD fusion protein fibrils are also consistent with an inner PD core surrounded by globular C-terminal domains18,43.
The alternative model for Ure2p fibril structure discussed above, in which the fibril core is comprised of CTD units and does not have the cross-β motif of an amyloid, was originally suggested by Melki and coworkers based on observations that the secondary structure content of full-length Ure2p estimated from deconvolution of the (rather featureless) amide I band in infrared spectroscopy is unchanged upon fibril formation, and that the catalytic domain of Ure2p retains its substrate binding site upon fibril formation44. These observations are not conclusive, as both the approximate constancy of secondary structure and the retention of substrate binding are expected if the CTD (which accounts for about 70% of the infrared signal) remains folded while only portions of the PD adopt β-strand conformations in the fibril core. Absence of the characteristic 4.7-4.8 Å scattering peak in x-ray diffraction data has also been presented as evidence against a cross-β structure in full-length Ure2p fibrils, but such a scattering peak, arising from only a small fraction of the total protein mass, would be readily obscured in the fiber diffraction data reported by Bousset et al.45. Indeed, the 4.7-4.8 Å scattering peak has been detected in electron diffraction measurements, both for full-length Ure2p fibrils and for fibrils formed by N-terminal Ure2p peptides21. Thus, data presented as support for a non-cross-β structure do not establish the existence of such a structure. On the other hand, a structure for full-length Ure2p fibrils in which the PD forms a cross-β core is consistent with all data discussed above, as well as with additional data from Melki and coworkers, including their observations that the CTD alone can form oligomers or amorphous aggregates, but that these aggregates do not interact with fibrillar full-length Ure2p46, that GdnHCl denaturation disrupts the CTD but does not disassemble full-length Ure2p fibrils44,46, that proteolytic cleavage sites in the PD become protected in full-length Ure2p fibrils47, and that only the PD has increased protection from hydrogen/deuterium exchange upon fibril formation48.
Model for Ure2p prion fibril structure
The structural model for a Ure2p fibril in Fig. 7a was constructed computationally from nine copies of the Ure2p dimer, using simulated annealing and energy minimization within the XPLOR-NIH program49. The Ure2p dimer crystal structure from PDB 1G6Y2 was used as the starting point. N-terminal residues (1-97 and 1-108 for the two inequivalent molecules in the dimer) were added with PyMOL (http://pymol.org), initially as fully extended chains. Artificial backbone dihedral angle restraints were applied to force residues 3-12, 20-29, 37-46, 54-63, and 71-80 to form β-strands. Artificial interatomic distance restraints were applied to force the PD to adopt a five-stranded β-serpentine conformation50 and to force the β-strands to align intermolecularly as in-register parallel β-sheets, as shown in Fig. 7b. To make the computations feasible, all sidechain atoms were removed from residues 109-354 during simulated annealing and energy minimization. Residues 98-354 of each dimer were treated as a rigid unit to preserve the dimeric, globular CTD structure.
The purpose of the model in Fig. 7 is simply to show that it is geometrically feasible for Ure2p to form amyloid fibrils with a cross-β core comprised of the PD and with CTD dimers forming a helical outer shell. Such a structure is not prohibited by steric clashes among CTD dimers. The structure is rather tightly packed, suggesting that CTD dimers would be immobilized as observed experimentally. The helical twist required to accomodate the bulky CTD dimers while simultaneously preserving the 4.7-4.8 Å intermolecular spacing along the fibril axis is not too severe to allow β-sheet formation in the PD core. Obviously, this model is far from being uniquely determined by experimental data. In particular: (i) The orientations of CTD dimers relative to the fibril axis are not constrained by experimental data; (ii) The helical ordering of CTD dimers is a consequence of our modeling procedure and may be less perfect in real fibrils; (iii) Our choices of β-strand segments and contacts between β-sheets in the PD is arbitrary. Further experiments are required before any high-resolution model for Ure2p fibrils can be considered fully accurate. Nonetheless, the model in Fig. 7 is consistent with all experimental data known to us. We believe this model to be qualitatively correct.
Materials and Methods
Protein expression and purification
Expression of Ile-labeled Ure2p and Ure2p1-89 was performed according to published protocols24,51. The expression plasmids pKT41-1 and pKT5518 that code for proteins with sequences MH6MYPRGN-Ure2p1–354 and MH6-Ure2p1-89 respectively were used. The E. coli strain BL21-Codon Plus (DE3)-RIPL (Stratagene, La Jolla, CA) transformed with an expression plasmid was grown in Defined Amino Acid Medium (DAM) supplemented with amino acid mix (0.1 g/l each amino acid) at 37 °C to an optical density at 600 nm (OD600) of 0.8. Cells were harvested by centrifugation and resuspended in the same volume of pre-warmed DAM with the same amino acid composition, except that unlabeled Ile was substituted with 0.2 g/l of isotopically labeled Ile. The culture was returned to shaking at 37°C for 15 min before plasmid-based protein expression was induced by the addition of 1 mM IPTG. After 3.5 hr of induced protein expression, the cells were harvested and stored at -80°C. Unlabeled Ure2p was produced in rich medium (LB). When cell density reached OD600 of 0.8, protein expression was induced with 1 mM IPTG for 3.5 hours.
For purification of full-length Ure2p, the cells were resuspended in 50 mM sodium phosphate, 300 mM NaCl, pH 8.0, containing protease inhibitors (Complete EDTA-free, Roche Applied Science), and lysed by high pressure. To enhance solubilization, 0.2% Triton X-100 was added to the suspension. Insoluble material was removed by centrifugation (50,000 × g, 45 min). Ure2p was recovered using a nickel-nitrilotriacetic acid (Ni-NTA) Superflow column (Qiagen). The protein was bound to the column in 50 mM sodium phosphate, 300 mM NaCl, 40 mM imidazole, pH 8.0, then washed extensively with the same buffer and eluted with 40 mM Tris-HCl, 200 mM NaCl, 250 mM imidazole, 5% glycerol, pH 8.0. Immediately after elution, the protein solution was applied to PD-10 desalting columns (GE Healthcare), exchanging the buffer to assembly buffer A (40 mM Tris-HCl, 200 mM NaCl, pH 8.0). The concentration of Ure2p was measured at 280 nm using an extinction coefficient of 48,200 M-1 cm-16. All purification steps were done at 4° C to prevent self-assembly of Ure2p. Only freshly purified protein was used for fibril growth (see below).
For purification of Ure2p1-89, the cells were lysed by suspension in lysis buffer L (7 M guanidine, 50 mM sodium phosphate, 200 mM NaCl, 40 mM imidazole, pH 8.0; about 25 ml of lysis buffer per liter of culture) followed by 1 hr incubation at room temperature. The lysates were cleared by centrifugation (70,000 × g, 1 hr). The cleared lysate was applied to a Ni-NTA Superflow column, equilibrated with buffer L. After several washing steps with buffer L, the protein was eluted with 8 M urea, 40 mM Tris-HCl, 200 mM NaCl, 250 mM imidazole, pH 8.0. Purified protein was dialyzed against buffer A to initiate fibril formation.
Fibril formation
Recombinant Ure2p and Ure2p1-89 rapidly form filaments in buffer A at concentrations higher than 0.5 mg/ml; these conditions were used to obtain spontaneously formed (SP) Ure2p and Ure2p1-89 fibrils. Purified proteins (2 mg/ml in buffer A) were incubated for 3 days at room temperature without rotation. The fibrils were harvested by centrifugation (50,000 × g, 30 min), washed several times with H2O and collected for solid state NMR analysis.
To prepare full-length Ure2p fibrils by seeding with prion extracts (EX-Ure2p), we looked for conditions where fibril growth in the presence of seeds is much more rapid than spontaneous fibril formation. First, buffer A was used because it stabilizes the structure of soluble, functional Ure2p6, preventing Ure2p misfolding and aggregation. Second, different concentrations of Ure2p were tested for spontaneous fibril assembly, using Thioflavin T fluorescence to monitor fibril formation as previously described52 (Fig. S1); 0.2 mg/ml Ure2p in buffer A was chosen for further experiments, since only minor spontaneous fibril growth was detected. Partial purification of Ure2p prion seeds from yeast cells was performed as described by Tanaka and Weissman53. Briefly, strain BY241 (MATa ura3 leu2 trp1 PDAL5-ADE2 kar1 [URE3]) was grown in liquid YPD medium (2% glucose, 2% peptone, 1% yeast extract). At OD600 of 2.0, yeast cells were harvested, washed in buffer Y (25 mM Tris–HCl, 150 mM NaCl, 1 mM dithiothreitol, 5% glycerol, pH 7.4 and complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)) and lysed by glass beads in the same buffer. Cell debris was removed by centrifugation at 8000 × g for 10 min. Prion aggregates were sedimented by high-speed centrifugation (100,000 × g, 1 hr). The pellet was resuspended with 1 M lithium acetate, incubated on ice for 30 min with gentle agitation, and further sedimented at 100,000 × g for 30 min. Finally, the pellet was resuspended in buffer A and sonicated two times for 15 s each at 40 W using a Branson Sonifier 250 (Branson Ultrasonics Corp., Danbury, CT). Ure2p concentration in the preparation of prion aggregates was estimated by standard Western blotting using anti-Ure2p antibodies and 2% (w/w) seeds were added to recombinant Ure2p in the seeding reaction, which was performed at room temperature without rotation for several days. This amount of seeds was sufficient for efficient amyloid conversion of Ure2p, as was shown in control experiments by ThT fluorescence (Fig. S1) and acquired sodium dodecyl sulphate (SDS) resistance (Fig. S5). To enhance the rate and efficiency of conversion for Ile-labeled Ure2p samples, the seeded solution was subjected to a brief sonication after two days of incubation, when about half of the recombinant Ure2p was converted to the fibrillar state (estimated by centrifugation and SDS-PAGE analysis). After additional incubation for two days, the fibrils were harvested by centrifugation (50,000 × g, 30 min), washed several times with H2O, and collected for solid state NMR experiments.
Proteinase K digestion and mass spectrometry
PK digestion of Ure2p filaments was performed in buffer A at 37 °C as described by Baxa et al.18. PK concentration and treatment time were optimized to avoid overdigestion (i.e., to preserve as much material as possible for NMR measurements), while simultaneously ensuring complete digestion of sensitive regions (Fig. S2). The final conditions were as follows: 2 ml of Ile-labeled Ure2p fibril solution at 15 mg/ml in buffer A was treated with 0.01 mg/ml PK for 4 hr at 37 °C. Efficiency of digestion was confirmed by SDS-PAGE and by electron microscopy (Fig. 1). The insoluble material was collected by centrifugation at 20,000 × g for 30 min at 4° C and washed five times by resuspension in H2O and subsequent centrifugation. The pellet after the last washing step was used in solid state NMR experiments on PK-Ure2p fibrils. Liquid chromatography-mass spectrometry (LC-MS) was performed on an HP1100 LC-MSD system (Agilent Technologies, Palo Alto, CA) as previously described18.
Electron microscopy and solid state NMR
TEM images in Fig. 1 were obtained with an FEI Morgagni microscope, operating at 80 keV. Fibril solutions were adsorbed to glow-discharged carbon films over lacey carbon supports on the TEM grids, after dilution of the fibril solution with deionized water to achieve an appropriate coverage. Grids were rinsed twice with deionized water before staining with 2% uranyl acetate solution, blotting, and drying in air.
Solid state NMR experiments were performed at 150.7 MHz and 100.4 MHz 13C NMR frequencies (14.1 T and 9.4 T fields), using Varian InfinityPlus spectrometers and Varian magic-angle spinning (MAS) probes with 3.2 mm diameter rotors. 2D 13C-13C spectra in Figs. 2 and 4 were obtained at 14.1 T with 12.00 kHz MAS, using 2.67 ms finite-pulse radio-frequency-driven recoupling (fpRFDR) mixing periods54,55. 13C π pulses in the fpRFDR sequence were 20.0 μs, with the carrier frequency at 38 ppm. Maximum t1 periods were 3.8-7.5 ms. 2D 13C-13C spectra were obtained with 98,000-221,000 scans and 1.5 s recycle delays. The maximum t1 period and number of scans varied due to variations in the NMR linewidths and sample quantities (10-20 mg of pelleted fibrils, including the mass of the buffer). 2D 15N-13C spectra in Fig. 3 were also obtained at 14.1 T with 12.00 kHz MAS, using 4 ms 15N-13C CP mixing periods, maximum t1 values of 5.8-11.0 ms, and 154,000-197,000 scans. Proton decoupling fields in 2D measurements were 105 kHz during the t1, t2, and mixing periods, with two-pulse phase modulation (TPPM)56 during t1 and t2. Samples were cooled to approximately 15° C (determined from the 1H NMR frequency of water in the samples57 under experimentally relevant MAS and 1H irradiation conditions) to avoid degradation during long experiments. CP build-up data in Fig. 6 were obtained at 14.1 T with 12.00 kHz MAS, using constant 1H and 13C radio-frequency field amplitudes during the CP period.
PITHIRDS-CT data in Fig. 5 were obtained at 9.4 T with 20.00 kHz MAS, using experimental conditions described previously40. The 13C-13C recoupling period was 38.4 ms. 1H decoupling fields were 110 kHz, with TPPM during signal detection. Each data point in Fig. 5b was obtained with 10944 scans and a 2.0 s recycle delay.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health. We thank Eric Anderson for mass spectrometry measurements, Frank Shewmaker for assistance with electron microscopy, and Charles Schwieters for assistance with molecular modeling.
Footnotes
Supplementary Data: Supplementary data for this article can be found online at XXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Umland TC, Taylor KL, Rhee S, Wickner RB, Davies DR. The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p. Proc Natl Acad Sci U S A. 2001;98:1459–1464. doi: 10.1073/pnas.041607898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bousset L, Belrhali H, Janin J, Melki R, Morera S. Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae. Structure. 2001;9:39–46. doi: 10.1016/s0969-2126(00)00553-0. [DOI] [PubMed] [Google Scholar]
- 3.Baxa U, Ross PD, Wickner RB, Steven AC. The N-terminal prion domain of Ure2p converts from an unfolded to a thermally resistant conformation upon filament formation. J Mol Biol. 2004;339:259–264. doi: 10.1016/j.jmb.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 4.Pierce MM, Baxa U, Steven AC, Bax A, Wickner RB. Is the prion domain of soluble Ure2p unstructured? Biochemistry. 2005;44:321–328. doi: 10.1021/bi047964d. [DOI] [PubMed] [Google Scholar]
- 5.Shewmaker F, Mull L, Nakayashiki T, Masison DC, Wickner RB. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics. 2007;176:1557–1565. doi: 10.1534/genetics.107.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrett S, Freeman SJ, Butler PJG, Fersht AR. Equilibrium folding properties of the yeast prion protein determinant Ure2. J Mol Biol. 1999;290:331–345. doi: 10.1006/jmbi.1999.2872. [DOI] [PubMed] [Google Scholar]
- 7.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner RB. [Ure3] as an altered Ure2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 9.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 10.Taylor KL, Cheng NQ, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 11.Legname G, Baskakov IV, Nguyen HOB, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 2010;119:177–187. doi: 10.1007/s00401-009-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JI, Cali I, Surewicz K, Kong QZ, Raymond GJ, Atarashi R, Race B, Qing LT, Gambetti P, Caughey B, Surewicz WK. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010;285:14083–14087. doi: 10.1074/jbc.C110.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, Tycko R. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 17.Chan JCC, Oyler NA, Yau WM, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baxa U, Taylor KL, Wall JS, Simon MN, Cheng NQ, Wickner RB, Steven AC. Architecture of Ure2p prion filaments: The N-terminal domains form a central core fiber. J Biol Chem. 2003;278:43717–43727. doi: 10.1074/jbc.M306004200. [DOI] [PubMed] [Google Scholar]
- 19.Masison DC, Maddelein ML, Wickner RB. The prion model for [URE3] of yeast: Spontaneous generation and requirements for propagation. Proc Natl Acad Sci U S A. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxa U, Cheng NQ, Winkler DC, Chiu TK, Davies DR, Sharma D, Inouye H, Kirschner DA, Wickner RB, Steven AC. Filaments of the Ure2p prion protein have a cross-β core structure. J Struct Biol. 2005;150:170–179. doi: 10.1016/j.jsb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 26.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci U S A. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid β-peptide of amyloid plaques. Proc Natl Acad Sci U S A. 2002;99:5596–5600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 31.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loquet A, Bousset L, Gardiennet C, Sourigues Y, Wasmer C, Habenstein B, Schutz A, Meier BH, Melki R, Bockmann A. Prion fibrils of Ure2p assembled under physiological conditions contain highly ordered, natively folded modules. J Mol Biol. 2009;394:108–118. doi: 10.1016/j.jmb.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci U S A. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem. 2004;279:50025–50030. doi: 10.1074/jbc.M406612200. [DOI] [PubMed] [Google Scholar]
- 35.de Laureto PP, Taddei N, Frare E, Capanni C, Costantini S, Zurdo J, Chiti F, Dobson CM, Fontana A. Protein aggregation and amyloid fibril formation by an SH3 domain probed by limited proteolysis. J Mol Biol. 2003;334:129–141. doi: 10.1016/j.jmb.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci U S A. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinley MP, Meyer RK, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner SB. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Amyloid fibril formation by Aβ16-22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 39.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 40.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann SR, Hahn EL. Nuclear double resonance in the rotating frame. Phys Rev. 1962;128:2042–2053. [Google Scholar]
- 42.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 43.Ranson N, Stromer T, Bousset L, Melki R, Serpell LC. Insights into the architecture of the Ure2p yeast protein assemblies from helical twisted fibrils. Protein Sci. 2006;15:2481–2487. doi: 10.1110/ps.062215206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bousset L, Thomson NH, Radford SE, Melki R. The yeast prion Ure2p retains its native α-helical conformation upon assembly into protein fibrils in vitro. EMBO J. 2002;21:2903–2911. doi: 10.1093/emboj/cdf303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bousset L, Briki F, Doucet J, Melki R. The native-like conformation of Ure2p in fibrils assembled under physiologically relevant conditions switches to an amyloid-like conformation upon heat treatment of the fibrils. J Struct Biol. 2003;141:132–142. doi: 10.1016/s1047-8477(02)00606-8. [DOI] [PubMed] [Google Scholar]
- 46.Thual C, Bousset L, Komar AA, Walter S, Buchner J, Cullin C, Melki R. Stability, folding, dimerization, and assembly properties of the yeast prion Ure2p. Biochemistry. 2001;40:1764–1773. doi: 10.1021/bi001916l. [DOI] [PubMed] [Google Scholar]
- 47.Bousset L, Redeker V, Decottignies P, Dubois S, Le Marechal P, Melki R. Structural characterization of the fibrillar form of the yeast Saccharomyces cerevisiae prion Ure2p. Biochemistry. 2004;43:5022–5032. doi: 10.1021/bi049828e. [DOI] [PubMed] [Google Scholar]
- 48.Redeker V, Halgand F, Le Caer JP, Bousset L, Laprevote O, Melki R. Hydrogen/deuterium exchange mass spectrometric analysis of conformational changes accompanying the assembly of the yeast prion Ure2p into protein fibrils. J Mol Biol. 2007;369:1113–1125. doi: 10.1016/j.jmb.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 49.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 50.Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: The parallel superpleated β-structure. Proc Natl Acad Sci U S A. 2004;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanco FJ, Hess S, Pannell LK, Rizzo NW, Tycko R. Solid state NMR data support a helix-loop-helix structural model for the N-terminal half of HIV-1 Rev in fibrillar form. J Mol Biol. 2001;313:845–859. doi: 10.1006/jmbi.2001.5067. [DOI] [PubMed] [Google Scholar]
- 52.Kryndushkin DS, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008;27:2725–2735. doi: 10.1038/emboj.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka M, Weissman JS. An efficient protein transformation protocol for introducing prions into yeast. Method Enzymol. 2006;412:185–200. doi: 10.1016/S0076-6879(06)12012-1. [DOI] [PubMed] [Google Scholar]
- 54.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- 55.Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG. Homonuclear radio frequency-driven recoupling in rotating solids. J Chem Phys. 1998;108:9463–9479. [Google Scholar]
- 56.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 57.Dvinskikh SV, Castro V, Sandstrom D. Heating caused by radiofrequency irradiation and sample rotation in 13C magic angle spinning NMR studies of lipid membranes. Magn Reson Chem. 2004;42:875–881. doi: 10.1002/mrc.1477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.