Abstract
In the cerebral cortex, local circuits consist of tens of thousands of neurons, each of which makes thousands of synaptic connections. Perhaps the biggest impediment to understanding these networks is that we have no wiring diagrams of their interconnections. Even if we had a partial or complete wiring diagram, however, understanding the network would also require information about each neuron's function. Here we show that the relationship between structure and function can be studied in the cortex with a combination of in vivo physiology and network anatomy. We used two-photon calcium imaging to characterize a functional property—the preferred stimulus orientation—of a group of neurons in the mouse primary visual cortex. We then used large-scale electron microscopy (EM) of serial thin sections to trace a portion of these neurons’ local network. Consistent with a prediction from recent physiological experiments, inhibitory interneurons received convergent anatomical input from nearby excitatory neurons with a broad range of preferred orientations, although weak biases could not be rejected.
Past studies of the synaptic connections in the cortex have focused on a few cells at a time, so that today we have only partial information about the structure of highly interconnected cortical networks. Until recently, anatomists relied on the sparse labelling of individual neurons to trace the extensive axonal and dendritic arbours of cortical neurons. Synaptic connectivity was originally inferred from the overlap of axonal and dendritic arbours1, an approach that remains fruitful today2,3. Combined physiological and anatomical studies of cortical slices in vitro have extended this analysis to include rules of pair-wise connectivity between different cell types and cortical layers4–6. It has been argued that the cortical network might be random beyond these simple statistical rules7, but recent studies suggest that there could be higher-order patterns of connections in cortical networks, such as mutually interconnected triplets of cells8, and subnetworks of neurons that are highly connected within a group, but not between groups9,10. Although these hypothesized patterns are based on in vitro physiological data, they have never been demonstrated anatomically, nor have they been related to information processing as measured from neural response properties in vivo.
Electron microscopy (EM) is an ideal tool for characterizing the highly interconnected structure of cortical networks. From visualizing quantal release by synaptic vesicles11 to generating a complete wiring diagram of a model organism12, it has provided definitive data for examining the relationship between structure and function in the nervous system. Electrophysiology combined with light microscopy and serial-section EM has allowed the inspection of structure-function relationships of single cells within neural circuits, such as the hippocampus13, retina14,15, thalamus16, and cortex17–20. A crucial difference from light microscopy is that serial-section EM can be used to follow the three-dimensional contours of neuronal membranes, so that any given axon or dendrite can be traced over hundreds of micrometres without selective, sparse staining of individual neurons. This feature of serial-section EM has been used to examine small volumes of cortical tissue, typically numbering in the thousands of cubic micrometres, in which portions of multiple dendrites and axons were reconstructed to examine synaptic relationships amongst them21,22, cf. 23.
Here, we exploited recent improvements in computer speed and storage capacity to perform serial-section EM of a volume that encompasses millions of cubic micrometres, sufficient to contain large portions of the dendritic and axonal arbours of more than 1,000 cells. With this data set, we could attempt a sampling—targeted to a subset of functionally imaged cells—of the dense interconnections found in a cortical network. In particular, we tested a prediction from recent physiological work24–27, cf. 28,29: that inhibitory interneurons in the mouse primary visual cortex receive dense, convergent input from nearby excitatory (pyramidal) neurons with widely varying preferred stimulus orientations (Figure 1a).
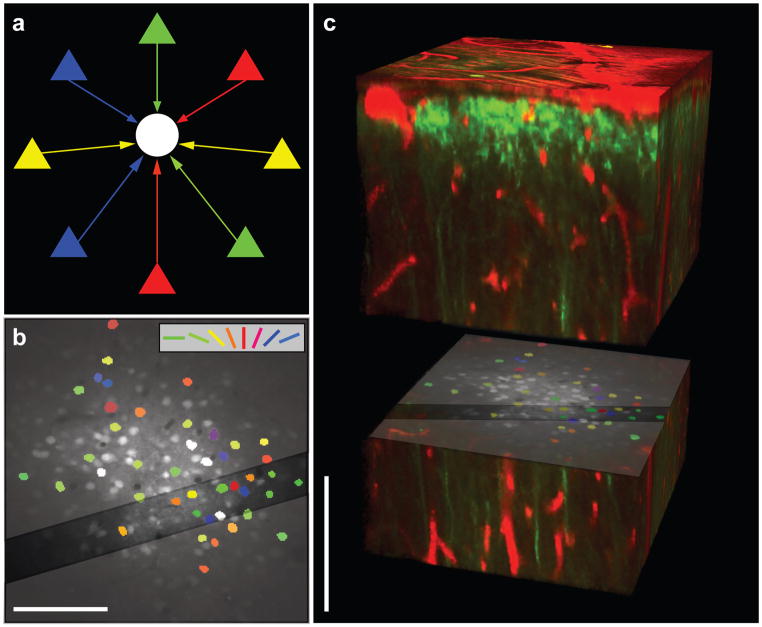
Figure 1. Functional characterization of neurons before anatomical reconstruction.
a, Schematic representation of diverse input to inhibitory interneurons. Excitatory pyramidal cells (triangles) with varied preferred orientations (different colours) provide input (coloured arrows) to an inhibitory interneuron (white circle). b, Cell-based visual orientation preference map in the mouse visual cortex from in vivo two-photon calcium imaging. Visually responsive cells are coloured according to their estimated preferred orientation, with broadly tuned cells (OSI ≤ 0.2) coloured white. Dark diagonal band: region targeted for acquisition of EM sections, cut orthogonal to image plane. c, In vivo two-photon fluorescence image of the 3-D anatomical volume (red: blood vessels or astrocytes, green: OGB or YFP) separated to expose the functionally imaged plane. Scale bars, 100 μm.
Functional imaging and large-scale EM
We used in vivo two-photon calcium imaging to determine the preferences for stimulus orientation of a cohort of cells in layer 2/3 of mouse primary visual cortex30. After loading cells with the fluorescent calcium indicator Oregon Green BAPTA-1 AM (OGB)31, we recorded neuronal activity as reflected by increases of intracellular calcium from a single plane of cells 186 μm beneath the brain surface. Black and white bars of varying orientations and directions were presented to the anesthetized animal, and the cellular responses were used to generate a map of orientation preference (Fig. 1b and Supplementary Fig. 1; see Methods). We concluded the in vivo experiment by collecting a stack of images from a volume surrounding the calcium-imaged plane (Fig. 1c; red: vessels, green: neurons). We then immediately perfused the animal and prepared the corresponding volume of visual cortex for serial-section transmission electron microscopy (TEM) (see Methods).
After finding the calcium-imaged region of cortex (see Methods), we cut a series of 1,215 thin sections (40–45 nm), oriented radially, at right angles to the functionally imaged plane (Fig. 1). Each section was wide enough (450 μm) to encompass the imaged plane, and tall enough (350 μm) to include cortical layers 1, 2/3 and upper 4 (Fig. 2a–b, Supplementary Movie 1). Although cortical axons can travel for millimetres, the proximal axonal collaterals of pyramidal cells arbourize more locally7, such that a portion of their local connectivity could be sampled within the span of the EM volume. Consistent with previous work we found that in order to reliably trace the finest neural processes and to identify chemical synapses (Fig. 2f, Supplementary Movie 2), we needed <50 nm section thickness and <5 nm lateral resolution32.
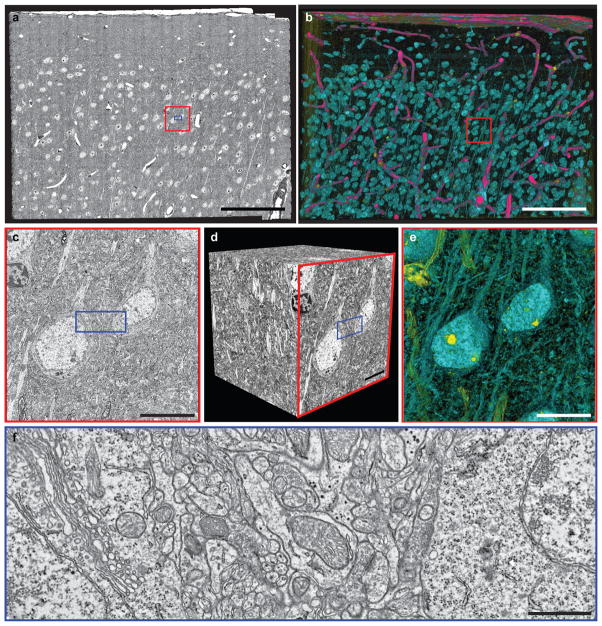
Figure 2. Large-scale electron microscopy.
a, Electron micrograph of an entire 120,000 x 80,000 pixel thin section, showing the pial surface (top) and cortical layers 1 through upper 4 (bottom). c, f: Zoomed-in view of (c) two functionally characterized cells (red outline in a), and (f) the neuropil between them (blue outlines in a, c, and d), illustrating the density of axons and dendrites coursing between cell bodies. d, A cube of the EM volume with c as one face. b, e: 3–D renderings of the EM volume (b, Supplementary Movie 3) through the entire series and (e) through 50 sections of the cube in d (red outline in b, Supplementary Movie 4). Pink represents electron transparent regions (e.g. blood vessels), yellow represents regions that are electron dense (e.g. nucleoli, oligodendrocyte nuclei, and myelin), and aqua denotes regions with pixel values in between (e.g. nuclei, cell bodies and dendrites). Scale bars, a–b: 100 μm, c–e: 10 μm, f: 1 μm.
We built a custom TEM Camera Array (TEMCA) that used four high-speed CCD cameras to efficiently acquire EM images of the required size and resolution (Supplementary Fig. 2, see Methods). The TEMCA achieves an order of magnitude increase in throughput over most commercially available TEM imaging systems. This system enabled us to collect the thousands of individual images required to mosaic each of the serial sections in approximately 20 min (Fig. 2a), so that we could image the entire series over the course of several months.
The 3.2 million camera images in our data set were converted into a seamless three-dimensional image volume by globally registering and aligning the EM volume after automated stitching of adjacent images into sections (see Methods). The raw image data was approximately 36 terabytes (TB), with the final stitched and registered EM data set encompassing approximately ten million megapixels (10 TB), spanning ~450 x 350 x 52 μm and containing approximately 1500 cell bodies (Fig. 2b).
We next re-located in the EM volume the cells whose function had been characterized by in vivo calcium imaging. The large extent of the EM dataset permitted registration of the fluorescently labelled vasculature and neurons between two-photon and EM imaging (Fig. 3a–c). We corresponded blood vessels of successively finer calibre, followed by the cell bodies, in order to identify individual neurons of known orientation selectivity within the EM volume (Fig. 3d, Supplementary Fig. 3; see Methods). The EM volume intersected the cell bodies of fourteen visually responsive neurons (Fig. 1 and Supplementary Fig. 1). Thirteen of the cells were selective for stimulus orientation. The fourteenth neuron was visually responsive, but non-selective for orientation. It had a non-pyramidal neuronal morphology, received asymmetric contacts onto the cell body, and made symmetric synapses onto its postsynaptic targets (Supplementary Fig. 4), suggesting it was an inhibitory, GABAergic interneuron33. Both the precision of physical registration and the alignment of appropriate functional properties with neuronal morphology (orientation selective excitatory cells and an unselective inhibitory neuron) demonstrate that we successfully combined micrometre-scale in vivo functional imaging and nanometre-scale EM ultrastructure.
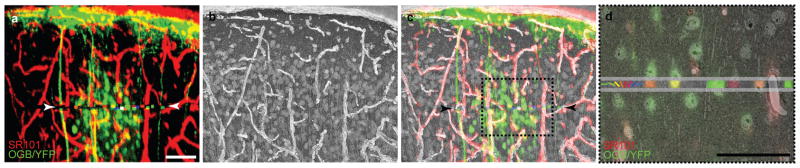
Figure 3. Correspondence between in vivo fluorescence anatomy and electron microscopy.
a, Maximum-intensity projection of the in vivo fluorescence anatomy corresponding to the EM volume. Red: blood vessels, green: cells. Colors between arrowheads correspond to orientation preference of neurons in the imaged plane, as in d. b, Projection through 37 EM sections evenly spaced in the series. c, Merge of EM and fluorescence projections. d, Zoomed-in view of the region outlined in c, showing the overlay of in vivo and EM data in a single thin section. Horizontal grey lines delineate the functionally imaged plane. Within the functionally imaged plane, the cell bodies of six neurons of known orientation preference (from left to right: cells 13, 12, 11, 10, 9 and 8, coloured as in Fig. 1b) are well registered with their EM ultrastructure. Outside the functionally imaged plane, blood vessels and astrocytes in the EM are well registered with the red SR101 staining. Scale bars, 50 μm.
EM circuit reconstruction and the resultant network graph
To analyze the circuit’s anatomical connectivity, we manually traced a wire-frame model of the dendritic and axonal arbours of each functionally characterized neuron, and noted the location of each synapse along the axons. For each synapse we then reconstructed the postsynaptic dendrite centripetally until we reached either the cell body or the boundary of the EM volume (see Methods). In this directed manner, we were able to identify all of the postsynaptic targets that could be found in the EM-imaged volume and to determine when multiple axons converged onto a common postsynaptic target, without unnecessary tracing of the target cells’ dendrites (Fig. 4 and Supplementary Fig. 5).
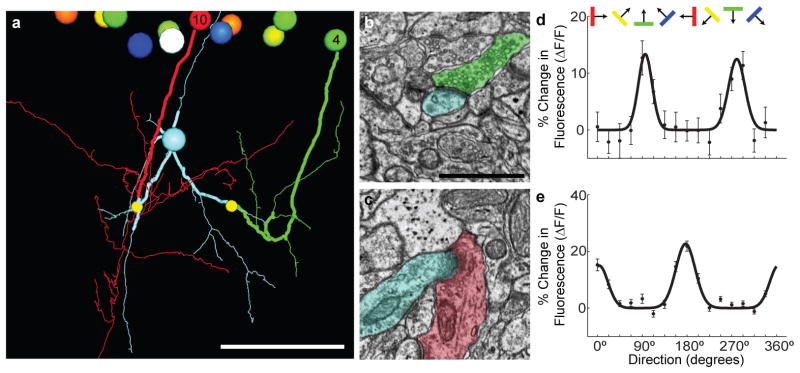
Figure 4. Convergent synaptic input onto inhibitory interneurons.
a, 3-D rendering of axonal contacts onto a postsynaptic neuron. Large balls at the top represent cell bodies of excitatory cells within the functionally imaged plane. Axons of a horizontally tuned neuron (cell 4; green) and a vertically tuned neuron (cell 10; red) descend and make synapses (small yellow balls) onto dendrites of an inhibitory interneuron (cyan). The axonal and dendritic segments leading to the convergence were independently traced by a second person, blind to the original segmentation (thick tracing). Cell bodies and axons coloured by orientation preference, as in Fig. 1b. Scale bar, 50 μm. b–c, Electron micrographs showing the synapses onto the inhibitory neuron from (b) cell 4 and (c) cell 10 with corresponding colours overlaid. Scale bar, 1 μm. d–e, Orientation tuning curves derived from in vivo calcium imaging of the cell bodies of (d) cell 4 and (e) cell 10. Error bars are +/− SEM.
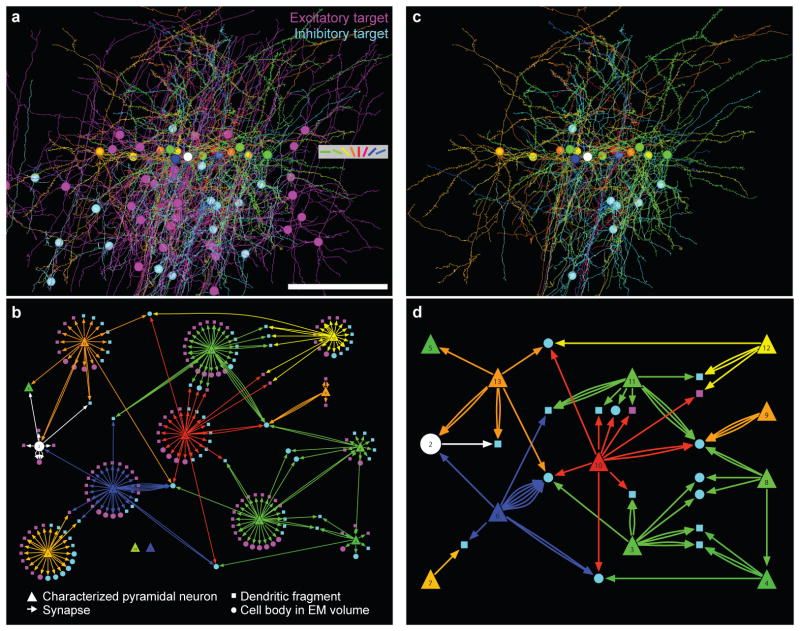
We categorized the postsynaptic targets as either excitatory or inhibitory based on morphology. Pyramidal cell dendrites were densely studded with spines, while inhibitory interneuron dendrites were sparsely spinous and receive more asymmetric (excitatory) input on their dendritic shafts33. Of 245 synapses originating from 10 functionally characterized pyramidal neurons that made synapses in the EM-imaged volume, 125 (51%) were onto inhibitory dendrites and 120 (49%) onto excitatory dendrites. Of the 185 distinct postsynaptic targets, 71 (38%) were inhibitory (Fig. 5, cyan) and 114 (62%) were excitatory (Fig. 5, magenta). The proportion of inhibitory cells in the target population was less than the proportion of synapses onto inhibitory targets because individual axons often made multiple synapses with inhibitory targets. We rendered the anatomical reconstructions of the functionally characterized cells and their postsynaptic targets as a graph (Fig. 5b) to examine the functional logic of the network.
Figure 5. From anatomy to connectivity graphs.
a, 3-D rendering of the dendrites, axons, and cell bodies of 14 neurons in the functionally imaged plane (coloured according to their orientation preference, as in Fig. 1b), and the dendrites and cell bodies of all their postsynaptic targets traced in the EM volume (magenta: excitatory targets; cyan: inhibitory targets; spines on postsynaptic targets not shown; Supplementary Movie 5). Scale bar, 100 μm. b, Directed network diagram of the functionally characterized cells and their targets, derived from a. Postsynaptic excitatory (magenta) and inhibitory (cyan) targets with cell bodies contained within the EM volume are drawn as circles. Other postsynaptic targets (dendritic fragments) are drawn as squares. c, 3-D rendering of the arbors and cell bodies of functionally characterized neurons, along with postsynaptic targets that either receive convergent input from multiple functionally characterized neurons, or were themselves functionally characterized (Supplementary Movie 5). d, A subset of the network graph showing only the connections in c, all independently verified.
Convergent excitatory input to inhibitory interneurons
In the graph of connectivity (Fig. 5b), we found multiple examples of pyramidal cells with diverse preferred stimulus orientations that provided convergent input to inhibitory neurons. We restricted subsequent analysis to a reduced and verified subnetwork that included convergent connections, as well as connections amongst the functionally imaged neurons (Fig. 5c–d, Supplementary Fig. 6–7; see Methods). Most strikingly, some inhibitory targets received input from three or four distinct cells with a range of preferred orientations (Fig. 5d, Supplementary Fig. 7a–d).
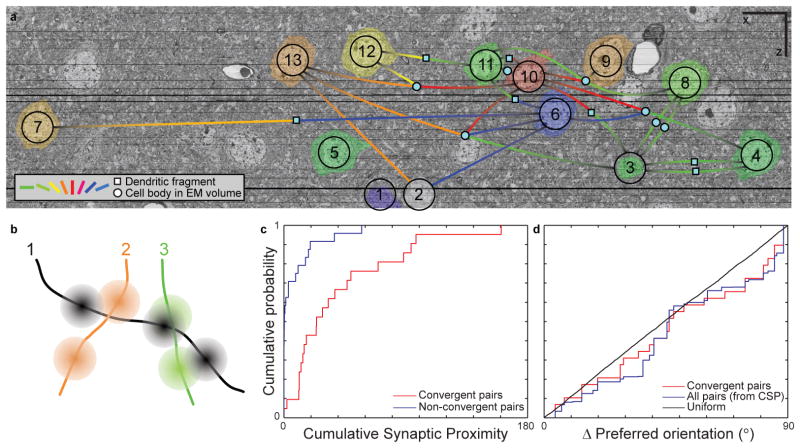
When we examined all of the convergences onto inhibitory targets, most were from pairs of cells that were close to each other (Fig. 5c, 6a), independent of the difference between their preferred orientations, which ranged from nearly orthogonal (Fig. 5d centre, Supplementary Fig. 7e–g) to nearly identical (Fig. 5d lower right, Supplementary Fig. 7m–o). The strongest predictor of whether two axons converged on a common target was found by examining how many of their synapses were nearby in space (Fig. 6b–c, cumulative synaptic proximity, p < 1.3 x 10-5, see Methods).
Figure 6. Convergent synaptic input onto inhibitory interneurons is predicted by proximity, not function.
a, A portion of the aligned and registered EM image series re-sliced parallel to the functionally imaged plane, through 1153 EM sections. Overlaid is a network graph of the convergences onto inhibitory interneurons. Visually responsive cell bodies are pseudo-coloured according to their preferred orientation (as in Fig. 1b),and numbered as in Figs. 4–5. Convergences onto inhibitory neuronal targets are represented by lines, corresponding to one or more synapses, leading either to solid cyan circles (targets traced to cell bodies in the EM volume) or squares (dendritic fragments). Cell 14 was partially contained in the EM volume and is not shown. Scale bars, 10 μm. b, Schematic of cumulative synaptic proximity (CSP). Line segments represent axons, with 3–D Gaussians centred at each synapse. A CSP was calculated for each pair of orientation-tuned neurons by summing all pair-wise overlaps of Gaussians from the two axons (σ = ~12 μm, see Methods). c, Pairs of axons whose synaptic boutons were in close proximity were more likely to converge onto a common target. The CSP of axon pairs participating in convergences (red) was significantly greater than for non-converging pairs (blue; p<1.3 x 10-5, two-sample Kolmogorov-Smirnov test, nconvergent pairs = 21, nnon-convergent pairs = 24). d, Convergences were not predicted by the difference in orientation preference between presynaptic cell pairs. The distribution of differences in orientation preference was not significantly different from a uniform distribution (p > 0.30, two-sample Kolmogorov-Smirnov test, nconvergences = 29) or a model distribution (see Methods) based on CSP (p > 0.68, two-sample Kolmogorov- Smirnov test, nconvergences = 29).
The functional properties of a pair of excitatory cells, specifically the difference in their preferred orientations, was not predictive of whether they converged onto an inhibitory target. Pairs that converged had a distribution indistinguishable from the uniform distribution (Fig. 6d, red line, p > 0.30) and from a distribution in which all possible presynaptic pairs were considered, weighted by their proximity (Fig. 6d, blue line, p > 0.68; see Methods). In summary, the axonal geometry of two excitatory neurons was a good predictor of whether they converged upon a nearby inhibitory interneuron, while the visual physiology of the excitatory neurons was not.
Discussion
The ability to study populations of neurons with a combination of network anatomy and in vivo physiology creates new opportunities for examining how neuronal circuits process information. Here, we explored how both the geometry and the function of cortical neurons influence the patterns of connections between them. In the case of excitatory input to local inhibitory interneurons, geometry appeared to dominate over function7. This finding may provide an anatomical substrate for a prediction from recent physiological studies of mouse visual cortex, in which inhibitory neurons were found to be less selective than excitatory neurons24–27, cf. 28,29 (Fig. 1a). Inhibitory interneurons that pool excitatory input could be used to set the gain of orientation-selective pyramidal cells26,34,35; they might also be involved in modulation of brain state34 or in attention-dependent normalization of cortical activity36.
A few studies have found some subtypes of inhibitory neurons to be selective in the mouse28,29. We did not classify different subtypes of inhibitory neurons, and thus cannot rule out the possibility that some might receive more selective input. Furthermore, although our sample size was large enough to exclude a strong bias in the preferred orientation tunings of convergent cell pairs, a larger sampling is necessary to exclude weaker biases (see Methods).
Until it is possible to fully reconstruct large EM volumes22,37,38, analysis of network connectivity will be limited to a partial sampling of the underlying anatomy. Here we concentrated on reconstructing the axons of functionally characterized pyramidal cells and their postsynaptic targets. Within our sample, we found that 51% of synapses were onto inhibitory targets, despite the preponderance of excitatory neurons in the cortex39–41, and reports that 10–20% of the synapses made by pyramidal cells are onto inhibitory targets in cat17 and macaque42. Whether the higher percentage we observed is due to a species difference43, cf. 44, or to the fact that we sampled synapses from proximal portions of the pyramidal cell axonal arbours, it resulted in our ability to sample a large number of convergences onto inhibitory targets (Fig. 5d).
Although the volume we imaged using electron microscopy was comparatively large, it proved to be near the minimum required to perform an analysis relating cortical function to network anatomy. We collected the series of wide-field (350 × 450 μm) high-resolution EM images to encompass the axonal arbours of the functionally characterized neurons and the dendrites of their targets. Nonetheless, we were limited by the shortest dimension in our volume (52 μm), determined by the number of thin sections, so we could trace only 245 out of the thousands of synapses made by the functionally characterized neurons.
We anticipate that the size of serial EM volumes will increase substantially in the near future, due to increases in imaging throughput and series length made possible by automated techniques45,46. The time required to trace connectivity between neurons will likely remain a limiting factor, although semi-automated techniques have already achieved 10-fold increases in throughput over purely manual approaches22. For large-scale reconstruction, data quality is paramount. To trace unlabelled, fine-calibre axons, minimum section thickness, minimum section loss, and optimal tissue quality are essential47,48.
In the current study, we found a large number of convergent inputs onto inhibitory neurons principally because they were densely innervated by the excitatory axons we reconstructed. Probing the network anatomy of more sparsely interconnected (and possibly weakly biased49) excitatory neurons50, however, will require larger samples. Here, we sought to limit tissue damage from the infrared laser, so two-photon calcium imaging was confined to a single plane, or less than 1% of the cells in the volume (Supplementary Fig. 8a). Recent advances in calcium imaging47,48, however, should now allow physiology to be collected from many more cells in a volume while maintaining tissue quality.
It is fortunate that increases in the dimensions of an EM-imaged volume, and the number of physiologically characterized cells within it, produce combinatorial increases in the number of network motifs8 that can be analyzed in a single experiment (Supplementary Fig. 8b–d). In particular, if a population of neurons is sparsely sampled, the number of interconnections found between them increases as the square of the sampling density. With moderate gains in the number of functionally imaged cells, or in the volumes encompassed by EM reconstructions, insight into the functional logic of cortical networks should therefore increase at an accelerating pace.
Methods Summary
We performed two-photon imaging in the mouse visual cortex as described previously27,30 by recording calcium responses to visual stimuli consisting of drifting gratings in each of 16 directions. We then acquired an in vivo fluorescent anatomical volume after injecting the tail vein with SR101 (100 mM) to label vasculature. The animal was perfused transcardially (2% paraformaldehyde/2.5% glutaraldehyde) and the brain was processed for serial-section TEM. Serial thin (<50 nm) sections were cut, picked up on pioloform-coated slot grids, and then post-stained with uranyl acetate and lead citrate. 1,215 serial sections were imaged at 120 kV on a JEOL 1200 EX with a custom scintillator atop optical-quality leaded vacuum glass at the end of a custom-built vacuum chamber extension. Custom software controlled automated x–y stage motion and image acquisition with a 2x2 array of CCD cameras (Imperx IPX-11M5) and Zeiss lenses. Images suitable for circuit reconstruction were acquired at a net rate of 5–8 MPix/s. Camera images were aligned in 2–D by registering adjacent camera images and dewarping, followed by histogram equalization and stitching. Then adjacent sections were registered and 3-D deformations were equalized in aligning the EM volume. Axonal and dendritic arbours of the functionally characterized neurons were manually reconstructed using TrakEM2 and objects were classified using classical criteria33. Neurons or dendritic fragments receiving synapses from multiple functionally characterized cells were included in analysis of convergence. For each synapse participating in a convergence, a second individual (blind to the original reconstruction) traced the pre- and the post-synaptic processes, starting from the synapse. Segmentation that diverged between the two tracers was excluded from further analysis. Cumulative synaptic proximity (CSP) of pairs of axons was calculated by centring a 3-D Gaussian density function at each synaptic bouton and taking the sum of their dot products over all pairs of synapses.
Methods
Animal preparation and in vivo calcium imaging
We imaged the visual cortex of an adult mouse (male Thy1-YFP-H51), as described previously27,30, using a custom-built two-photon microscope with a 20X objective (Olympus, 0.95 NA) at a frame rate of 2 Hz. Randomized drifting gratings (100% contrast, 2 Hz) were presented for 4 sec, followed by 4 sec of uniform mean luminance on a LCD monitor, positioned 18 cm from the contralateral eye, spanning approximately 80o (azimuth) by 60o (elevation) of visual space. To determine orientation tuning, square-wave gratings (0.05 cycles per degree) were presented at 16 directions of motion. Cellular responses were measured with the calcium indicator Oregon Green BAPTA-1 AM (OGB; Invitrogen). We next injected the tail vein with SR101 (100 mM; Invitrogen) to fluorescently label vasculature and acquired an in vivo fluorescent anatomy volume of the region (600 × 600 × 250 μm) surrounding the calcium-imaged plane (Figure 1c).
EM material preparation
Following in vivo two-photon imaging the animal was perfused transcardially (2% paraformaldehyde/2.5% glutaraldehyde) and the brain was processed for serial-section TEM. Seven 300 μm thick coronal vibratome sections were cut. The vibratome sections were post-fixed and stained with 1% osmium tetroxide/1.5% potassium ferrocyanide followed by 1% uranyl acetate, dehydrated with a graded ethanol series, and embedded in resin (TAAB 812 Epon, Canemco). After locating the calcium-imaged region by matching vasculature between in vivo fluorescence and serial thick (1 μm) toluidine blue (EMS) sections cut from an adjacent vibratome section, 4,150 serial thin (<50 nm) sections were cut on an ultramicrotome (Leica UC6) using a 35 degree diamond knife (EMS-Diatome) and picked up on 1x2 mm dot-notch slot grids (Synaptek) that were coated with a pale gold pioloform (Ted Pella) support film, carbon coated, and glow-discharged. Following section pickup, grids were post-stained with uranyl acetate and lead citrate. Of the 4,150 serial sections, we targeted 1,215 serial sections for imaging that contained the greatest diversity of orientation selectivities and the fewest lost sections. Within this range, a total of 28 sections were lost: 2 during pickup, 16 during post-staining, 2 before imaging, and 8 during imaging.
Transmission Electron Microscope Camera Array (TEMCA)
A 120 kV TEM (JEOL 1200 EX) was placed atop scaffolding with a custom-built 1.2 m vacuum chamber extension beneath the microscope’s final projector lens, culminating in a 226 mm diameter scintillator (Grant Scientific) resting above optical-quality leaded vacuum glass (Computer Optics) (Supplementary Fig. 2). Custom software (LabView) controlled x–y stage motion and image acquisition from a 2×2 array52 of CCD cameras (Imperx IPX-11M5) coupled to lenses (Zeiss Makro-planar T* 50 mm F/2.0 ZF). Each acquired camera frame was cross-correlated, translated, and summed by the camera’s dedicated acquisition computer, which wrote data over a local area network to storage servers (InterPRO Microsystems). Magnification at the scope was 2000x, accelerating potential was 120 kV, and beam current was ~90 microamperes through a tungsten filament. Images suitable for circuit reconstruction were acquired at a net rate of 5–8 MPix/s.
EM image registration and correspondence
A detailed description of the computational methods for registration and alignment of the EM volume will appear elsewhere. Briefly, camera images were aligned in 2-D by registering adjacent camera images and dewarping, followed by histogram equalization and stitching. Then adjacent sections were registered and 3-D bending stresses were equalized to align the EM volume. The imaged volume will be publicly accessible at the Whole Brain Catalog (http://wholebraincatalog.org) and the UCSD Cell Centered Database53 (http://ccdb.ucsd.edu/, accession number 8448).
The aligned volume (sections 2,990-4,150) was imported into a customized version of TrakEM254 for visualization and segmentation. Although we targeted imaging to 1,215 serial-sections, the final stitched and registered data set contains 1,153 sections, since some fragmentary material, typically small pieces cut from an edge of the EM block face, was excluded. Volume renderings of the raw, un-segmented EM image data (Fig. 2b, e) were made in TrakEM2 using a false-colour lookup table. To find the correspondence between the cells imaged in vivo with those in the EM data set, we used successively finer scales of vasculature and then somata to re-locate the calcium-imaged neurons in the EM-imaged volume (Fig. 3 and Supplementary Fig. 3). Pixel size (4 nm/pix) was calibrated using a latex bead-coated replica grating (EMS). Section thicknesses of ~45 nm were estimated using the cylindrical diameters method55.
Tracing and independent verification
Axonal and dendritic arbours of the functionally characterized neurons were traced by manually placing a series of marker points down the midline of each process and rendering a wire-frame model of the arbours using TrakEM254. Infrequently, ambiguities arose from poor local data quality or missing sections. In this case, tracing of a process was halted. Fine axons generally could not be traced across three or more consecutive missing sections. Synapses were identified using classical criteria33. For each synapse on the axon of a functionally characterized cell, we traced the dendrites of the postsynaptic neuron either to the boundaries of the volume or centripetally back to the cell body. Neurons or dendritic fragments receiving synapses from multiple functionally characterized cells were considered convergence targets.
A second individual who had not previously reconstructed the pre- or post-synaptic process and was blind to previous tracing work, independently verified the anatomical connectivity underlying convergences and connections between functionally characterized cells. Verification started at each synapse (small yellow spheres, Fig. 4a and Supplementary Fig. 7) and the pre- and postsynaptic processes were each traced centripetally (thick tracing, Fig. 4a and Supplementary Fig. 7). When the initial reconstruction and the subsequent verification of the reconstruction diverged, that connection and all segmentation work distal from the point of the divergence was excluded from further analysis. The verification process revealed that our initial segmentation effort was highly reproducible, consistent with earlier tests looking at reproducibility of full axonal arbours (Supplementary Fig. 6). All postsynaptic dendritic processes were verified. Of 64 presynaptic axonal contacts participating in convergences, 55 were independently verified. Three experienced team members did the bulk of the tracing and verification over a period of three months. Apical dendrites and dendritic spines (Fig. 5a, c and Supplementary Figs. 3, 7) not participating in either the full (Fig. 5b) or independently verified (Fig. 5d) connectivity graphs were traced by a small team of students and were not included in the analysis.
Categorization of postsynaptic targets as inhibitory or excitatory
Dendrites postsynaptic to axons of physiologically characterized cells were unambiguously identified as belonging to inhibitory or excitatory neurons. Inhibitory dendrites were smooth and densely coated with asymmetric shaft synapses, whereas excitatory dendrites were spiny and had fewer shaft synapses (Supplementary Fig. 4a–b). One hundred thirty-three (54%) of the synapses were onto dendritic spines, and 112 (46%) were onto dendritic shafts. Only four (1.6%) of the shaft synapses were onto excitatory targets, and 12 (4.9%) of the spine synapses were onto inhibitory targets. Starting at 10 randomly selected synapses made by functionally characterized neurons, the postsynaptic dendrites’ percentage of synapses on spines was calculated by tracing along the dendrite and counting up to 10 synapses (including those on spines and the shaft) in either direction. The distribution of spine synapses and shaft synapses was non-overlapping between cell types42 (Supplementary Fig. 4b) and was consistent with the prior categorization of the target. In all cases (n = 8) where the postsynaptic dendrite was categorized as inhibitory and could be traced to a cell body (cyan circles in Figs. 5–6), asymmetric synapses were found contacting the soma (Supplementary Fig. 4c). Cell 2, which was visually responsive, but not selective for stimulus orientation (Supplementary Fig. 1), exhibited all of the aforementioned characteristics. Furthermore, its axon made symmetric contacts with both dendrites and somata (Supplementary Fig. 4d).
Data analysis
Calcium-imaging data analyses were performed with Matlab (Mathworks) and ImageJ (NIH) as described previously27,30. Only neurons with an estimated photonic noise floor < 3% ΔF/F were included. Neurons were considered visually responsive if the response to the best direction was >6% ΔF/F (morphologically identified pyramidal neurons) or >4%ΔF/F (non-pyramidal neurons). Orientation tuning curves for each neuron were generated from the average responses to 16 directions fit to the sum of two Gaussians (Supplementary Fig. 1) and the preferred orientation was estimated from this fit. Orientation selectivity was calculated as the magnitude of the vector average divided by the sum of all responses: ((ΣR(θi)sin(2θi))2 + (ΣR(θi)cos(2θi))2)1/2/ΣR(θi)56,57.
The cumulative synaptic proximity (CSP) for a pair of axons was calculated by centring a clipped three dimensional Gaussian density function at each synaptic bouton, calculating the dot product of these Gaussians for each pair of boutons (one bouton from each axon), and summing over all pairs (Fig. 6b). The dot products were normalized so that two superimposed synapses yielded a value of 1.0. The distribution of CSPs between convergent and non-convergent pairs was significantly different (two-sample Komogorov-Smirnov test, σ = ~12 μm). A model population of convergences was generated by allowing each possible pair of physiologically characterized cells to contribute a number of convergences to the model proportional to the pair’s CSP, with high CSP pairs contributing many convergences, and low CSP pairs contributing few. The distribution of relative preferred orientations from the model population (Fig. 6d, blue line) did not differ significantly from either the uniform distribution or the population of convergent pairs (two-sample Kolmogorov-Smirnov test). A separate set of model populations was generated in which the probability of convergence by a cell pair increased with the similarity of their orientation preferences. Drawing randomly from these model populations we found our sample size (Fig. 5d, nconvergences = 29) was sufficient to detect a statistically significant difference from the uniform distribution (two-sample Kolmogorov-Smirnov test, α = 0.05, power ~ 0.80) in strongly biased model populations, i.e., populations in which convergent pairs with orthogonal preferred orientation tunings were excluded. However, in hypothetical populations exhibiting weak biases larger sample sizes are necessary (data not shown).
Supplementary Material
Acknowledgments
We thank E. Raviola for discussions and technical advice on all aspects of EM; J. Stiles for support and advice for the alignment and stitching effort at PSC; J. Lichtman for invaluable discussions and, along with K. Blum and J. Sanes, support from the Center for Brain Science; A. Cardona for help with TrakeEM2, and many helpful modifications of it; and H. Pfister and S. Warfield for discussions and help with computational issues. We also thank S. Butterfield for programming, L. Benecchi and HMS EM Core Facility for technical support. For technical help with EM and its interpretation, we thank K. Harris, M. Bickford, M. Ellisman, K. Martin and T. Reese. We thank J. Assad, R. Born, J. Maunsell, J. Stiles, E. Raviola, and members of the Reid lab for critical reading of the manuscript. This work was supported by the Center for Brain Science at Harvard University, Microsoft Research, and the NIH though the NEI to RCR (EY10115 and EY18742) and through resources provided by the NRBSC (P41 RR06009), which is part of the Pittsburgh Supercomputing Center; and by fellowships from Harvard Center of Neurodegeneration and Repair to DB and the NEI to WCL (EY18532).
Footnotes
Authors' Contributions
DB, ES, SY, RCR designed and built the TEMCA system, and DB programmed it. AK and MA performed the in vivo imaging and AK analyzed it. DB processed the tissue and DB and WCL aligned the block with the in vivo imaging. WCL sectioned the series and DB, WCL, HK imaged it on the TEMCA. GH, AW stitched and aligned the images into a volume. DB, WCL, HK did most of the segmentation. DB, WCL, SY performed quantitative analysis on the tracing. DB, WCL and RCR designed the experiment and wrote the paper, with considerable help from the other authors.
Author Information
The imaged volume will be publicly accessible at the Whole Brain Catalog (http://wholebraincatalog.org) and the UCSD Cell Centered Database (http://ccdb.ucsd.edu; accession number MP8448).
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Ramón y, Cajal S. Textura del Sistema Nervioso del Hombre y de los Vertebrados. Moya; 1904. [Google Scholar]
- 2.Binzegger T, Douglas RJ, Martin KA. A quantitative map of the circuit of cat primary visual cortex. J Neurosci. 2004;24:8441–8453. doi: 10.1523/JNEUROSCI.1400-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stepanyants A, et al. Local potential connectivity in cat primary visual cortex. Cereb Cortex. 2008;18:13–28. doi: 10.1093/cercor/bhm027. [DOI] [PubMed] [Google Scholar]
- 4.Mason A, Nicoll A, Stratford K. Synaptic transmission between individual pyramidal neurons of the rat visual cortex in vitro. J Neurosci. 1991;11:72–84. doi: 10.1523/JNEUROSCI.11-01-00072.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markram H, Lubke J, Frotscher M, Roth A, Sakmann B. Physiology and anatomy of synaptic connections between thick tufted pyramidal neurones in the developing rat neocortex. J Physiol. 1997;500 (Pt 2):409–440. doi: 10.1113/jphysiol.1997.sp022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson AM, Bannister AP. Interlaminar connections in the neocortex. Cereb Cortex. 2003;13:5–14. doi: 10.1093/cercor/13.1.5. [DOI] [PubMed] [Google Scholar]
- 7.Braitenberg V, Schüz A. Cortex : statistics and geometry of neuronal connectivity. 2. Springer; Berlin: 1998. rev. [Google Scholar]
- 8.Song S, Sjostrom PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 2005;3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshimura Y, Callaway EM. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat Neurosci. 2005;8:1552–1559. doi: 10.1038/nn1565. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimura Y, Dantzker JL, Callaway EM. Excitatory cortical neurons form fine-scale functional networks. Nature. 2005;433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- 11.Heuser JE, et al. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J Cell Biol. 1979;81:275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans Royal Soc London B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 13.Sorra KE, Harris KM. Stability in synapse number and size at 2 hr after long-term potentiation in hippocampal area CA1. J Neurosci. 1998;18:658–671. doi: 10.1523/JNEUROSCI.18-02-00658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dacheux RF, Raviola E. The rod pathway in the rabbit retina: a depolarizing bipolar and amacrine cell. J Neurosci. 1986;6:331–345. doi: 10.1523/JNEUROSCI.06-02-00331.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterling P. Microcircuitry of the cat retina. Annu Rev Neurosci. 1983;6:149–185. doi: 10.1146/annurev.ne.06.030183.001053. [DOI] [PubMed] [Google Scholar]
- 16.Hamos JE, Van Horn SC, Raczkowski D, Sherman SM. Synaptic circuits involving an individual retinogeniculate axon in the cat. J Comp Neurol. 1987;259:165–192. doi: 10.1002/cne.902590202. [DOI] [PubMed] [Google Scholar]
- 17.Kisvarday ZF, et al. Synaptic targets of HRP-filled layer III pyramidal cells in the cat striate cortex. Exp Brain Res. 1986;64:541–552. doi: 10.1007/BF00340492. [DOI] [PubMed] [Google Scholar]
- 18.Anderson JC, Douglas RJ, Martin KA, Nelson JC. Map of the synapses formed with the dendrites of spiny stellate neurons of cat visual cortex. J Comp Neurol. 1994;341:25–38. doi: 10.1002/cne.903410104. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed B, Anderson JC, Martin KA, Nelson JC. Map of the synapses onto layer 4 basket cells of the primary visual cortex of the cat. J Comp Neurol. 1997;380:230–242. [PubMed] [Google Scholar]
- 20.Tamas G, Somogyi P, Buhl EH. Differentially interconnected networks of GABAergic interneurons in the visual cortex of the cat. J Neurosci. 1998;18:4255–4270. doi: 10.1523/JNEUROSCI.18-11-04255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepherd GM, Harris KM. Three-dimensional structure and composition of CA3-->CA1 axons in rat hippocampal slices: implications for presynaptic connectivity and compartmentalization. J Neurosci. 1998;18:8300–8310. doi: 10.1523/JNEUROSCI.18-20-08300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishchenko Y, et al. Ultrastructural analysis of hippocampal neuropil from the connectomics perspective. Neuron. 2010;67:1009–1020. doi: 10.1016/j.neuron.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson JR, et al. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7:e1000074. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu BH, et al. Visual receptive field structure of cortical inhibitory neurons revealed by two-photon imaging guided recording. J Neurosci. 2009;29:10520–10532. doi: 10.1523/JNEUROSCI.1915-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Runyan CA, et al. Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron. 2010;67:847–857. doi: 10.1016/j.neuron.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma WP, et al. Visual representations by cortical somatostatin inhibitory neurons--selective but with weak and delayed responses. J Neurosci. 2010;30:14371–14379. doi: 10.1523/JNEUROSCI.3248-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohki K, Chung S, Ch'ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 31.Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris KM, et al. Uniform Serial Sectioning for Transmission Electron Microscopy. J Neurosci. 2006;26:12101–12103. doi: 10.1523/JNEUROSCI.3994-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters A, Palay SL, Webster Hd. The fine structure of the nervous system : neurons and their supporting cells. 3. Oxford University Press; New York: 1991. [Google Scholar]
- 34.Alitto HJ, Dan Y. Function of inhibition in visual cortical processing. Curr Opin Neurobiol. 2010;20:340–346. doi: 10.1016/j.conb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu BH, et al. Intervening inhibition underlies simple-cell receptive field structure in visual cortex. Nat Neurosci. 2010;13:89–96. doi: 10.1038/nn.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds JH, Heeger DJ. The normalization model of attention. Neuron. 2009;61:168–185. doi: 10.1016/j.neuron.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helmstaedter M, Briggman KL, Denk W. 3D structural imaging of the brain with photons and electrons. Curr Opin Neurobiol. 2008;18:633–641. doi: 10.1016/j.conb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Turaga SC, et al. Convolutional networks can learn to generate affinity graphs for image segmentation. Neural Comput. 2009;22:511–538. doi: 10.1162/neco.2009.10-08-881. [DOI] [PubMed] [Google Scholar]
- 39.Peters A, Kara DA. The neuronal composition of area 17 of rat visual cortex. II. The nonpyramidal cells. J Comp Neurol. 1985;234:242–263. doi: 10.1002/cne.902340209. [DOI] [PubMed] [Google Scholar]
- 40.White EL, Keller A. Cortical circuits: synaptic organization of the cerebral cortex - structure, function and theory. Birkhauser; Boston: 1989. [Google Scholar]
- 41.Markram H, et al. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 42.McGuire BA, Gilbert CD, Rivlin PK, Wiesel TN. Targets of horizontal connections in macaque primary visual cortex. J Comp Neurol. 1991;305:370–392. doi: 10.1002/cne.903050303. [DOI] [PubMed] [Google Scholar]
- 43.Holmgren C, Harkany T, Svennenfors B, Zilberter Y. Pyramidal cell communication within local networks in layer 2/3 of rat neocortex. J Physiol. 2003;551:139–153. doi: 10.1113/jphysiol.2003.044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson AM, West DC, Wang Y, Bannister AP. Synaptic connections and small circuits involving excitatory and inhibitory neurons in layers 2–5 of adult rat and cat neocortex: triple intracellular recordings and biocytin labelling in vitro. Cereb Cortex. 2002;12:936–953. doi: 10.1093/cercor/12.9.936. [DOI] [PubMed] [Google Scholar]
- 45.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayworth K, Kasthuri N, Schalek R, Lichtman J. Automating the Collection of Ultrathin Serial Sections for Large Volume TEM Reconstructions. Microsc and Microanal. 2006:86–87. [Google Scholar]
- 47.Grewe BF, Helmchen F. Optical probing of neuronal ensemble activity. Curr Opin Neurobiol. 2009;19:520–529. doi: 10.1016/j.conb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia H, Rochefort NL, Chen X, Konnerth A. Dendritic organization of sensory input to cortical neurons in vivo. Nature. 2010;464:1307–1312. doi: 10.1038/nature08947. [DOI] [PubMed] [Google Scholar]
- 50.Ohki K, Reid RC. Specificity and randomness in the visual cortex. Curr Opin Neurobiol. 2007;17:401–407. doi: 10.1016/j.conb.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–45. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 52.Peltier S, et al. Design of a new 8k x 8k lens coupled detector for wide-field, high-resolution transmission electron microscopy. Microsc Microanal. 2005:610–611. [Google Scholar]
- 53.Martone ME, et al. A cell-centered database for electron tomographic data. J Struct Biol. 2002;138:145–155. doi: 10.1016/s1047-8477(02)00006-0. [DOI] [PubMed] [Google Scholar]
- 54.Cardona A, et al. An Integrated Micro- and Macroarchitectural Analysis of the Drosophila Brain by Computer-Assisted Serial Section Electron Microscopy. PLoS Biol. 2010;8:e1000502. doi: 10.1371/journal.pbio.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiala JC, Harris KM. Cylindrical diameters method for calibrating section thickness in serial electron microscopy. J Microsc. 2001;202:468–472. doi: 10.1046/j.1365-2818.2001.00926.x. [DOI] [PubMed] [Google Scholar]
- 56.Ringach DL, Shapley RM, Hawken MJ. Orientation selectivity in macaque V1: diversity and laminar dependence. J Neurosci. 2002;22:5639–5651. doi: 10.1523/JNEUROSCI.22-13-05639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worgotter F, Eysel UT. Quantitative determination of orientational and directional components in the response of visual cortical cells to moving stimuli. Biol Cybern. 1987;57:349–355. doi: 10.1007/BF00354980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.