Abstract
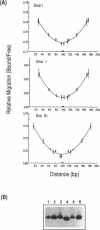
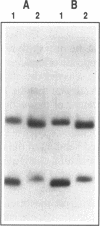
The SmaI and XmaI endonucleases are imperfect isoschizomers that recognize the sequence CCCGGG. SmaI cleaves between the internal CpG to produce blunt end scissions whereas XmaI cleaves between the external cytosines to produce a four base, five prime overhang. Each of the endonucleases forms stable, specific complexes with DNA in the absence of magnesium. Circular permutation analyses of the protein-DNA complexes revealed that each of the endonucleases induces bending of the DNA. Phase sensitive detection analyses verified the existence of the SmaI and XmaI induced bends. Furthermore, bending of the helix axis by the endonucleases appeared to be directed in opposite orientations. The orientation of the SmaI induced bend appeared to be towards the major groove and is reminiscent of the direction of the bend induced by EcoRV which similarly induces blunt end scissions. Conversely, XmaI appeared to bend the DNA towards the minor groove.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiken C. R., Fisher E. W., Gumport R. I. The specific binding, bending, and unwinding of DNA by RsrI endonuclease, an isoschizomer of EcoRI endonuclease. J Biol Chem. 1991 Oct 5;266(28):19063–19069. [PubMed] [Google Scholar]
- Banks K. M., Hare D. R., Reid B. R. Three-dimensional solution structure of a DNA duplex containing the BclI restriction sequence: two-dimensional NMR studies, distance geometry calculations, and refinement by back-calculation of the NOESY spectrum. Biochemistry. 1989 Aug 22;28(17):6996–7010. doi: 10.1021/bi00443a033. [DOI] [PubMed] [Google Scholar]
- Butkus V., Petrauskiene L., Maneliene Z., Klimasauskas S., Laucys V., Janulaitis A. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 1987 Sep 11;15(17):7091–7102. doi: 10.1093/nar/15.17.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Lipanov A. A., Fedoroff OYu, Kim S. G., Kintanar A., Reid B. R. Sequence effects on local DNA topology. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9087–9091. doi: 10.1073/pnas.88.20.9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M. I., Miner J. N., Yoshinaga S. K., Yamamoto K. R. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990 Sep 14;249(4974):1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- Feuerstein B. G., Pattabiraman N., Marton L. J. Molecular mechanics of the interactions of spermine with DNA: DNA bending as a result of ligand binding. Nucleic Acids Res. 1990 Mar 11;18(5):1271–1282. doi: 10.1093/nar/18.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D. E., Parent L. A., Sharp P. A. Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11779–11783. doi: 10.1073/pnas.89.24.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Heitman J. How the EcoRI endonuclease recognizes and cleaves DNA. Bioessays. 1992 Jul;14(7):445–454. doi: 10.1002/bies.950140704. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Rooney T. F., Austin R. H. Evidence for kinks in DNA folding in the nucleosome. Nature. 1987 Aug 6;328(6130):554–557. doi: 10.1038/328554a0. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Bertuccioli C., Takada R., Wang J., Yamamoto T., Roeder R. G. Transcription factor TFIID induces DNA bending upon binding to the TATA element. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1060–1064. doi: 10.1073/pnas.89.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991 Jul 26;66(2):317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Steponaviciene D., Maneliene Z., Petrusyte M., Butkus V., Janulaitis A. M.Smal is an N4-methylcytosine specific DNA-methylase. Nucleic Acids Res. 1990 Nov 25;18(22):6607–6609. doi: 10.1093/nar/18.22.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986 Sep 19;233(4770):1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Koudelka G. B. Bending of synthetic bacteriophage 434 operators by bacteriophage 434 proteins. Nucleic Acids Res. 1991 Aug 11;19(15):4115–4119. doi: 10.1093/nar/19.15.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. R., Hosur R. V., Roy K. B., Miles H. T., Govil G. Resonance assignment of the 500-MHz proton NMR spectrum of self-complementary dodecanucleotide d-GGATCCGGATCC: altered conformations at BamHI cleavage sites. Biochemistry. 1985 Dec 17;24(26):7703–7711. doi: 10.1021/bi00347a030. [DOI] [PubMed] [Google Scholar]
- Lahm A., Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991 Dec 5;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Site-specific methylation: effect on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2045–2071. doi: 10.1093/nar/19.suppl.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J., Espinosa M. The RepA repressor can act as a transcriptional activator by inducing DNA bends. EMBO J. 1991 Jun;10(6):1375–1382. doi: 10.1002/j.1460-2075.1991.tb07657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Thielking V., Selent U., Köhler E., Landgraf A., Wolfes H., Alves J., Pingoud A. Mg2+ confers DNA binding specificity to the EcoRV restriction endonuclease. Biochemistry. 1992 Apr 21;31(15):3727–3732. doi: 10.1021/bi00130a001. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M. D., Thresher R. J., Conrad M., Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991 Feb 19;30(7):2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Travers A., Klug A. Nucleoprotein complexes. DNA wrapping and writhing. 1987 May 28-Jun 3Nature. 327(6120):280–281. doi: 10.1038/327280a0. [DOI] [PubMed] [Google Scholar]
- Withers B. E., Ambroso L. A., Dunbar J. C. Structure and evolution of the XcyI restriction-modification system. Nucleic Acids Res. 1992 Dec 11;20(23):6267–6273. doi: 10.1093/nar/20.23.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- van der Vliet P. C., Verrijzer C. P. Bending of DNA by transcription factors. Bioessays. 1993 Jan;15(1):25–32. doi: 10.1002/bies.950150105. [DOI] [PubMed] [Google Scholar]