Abstract
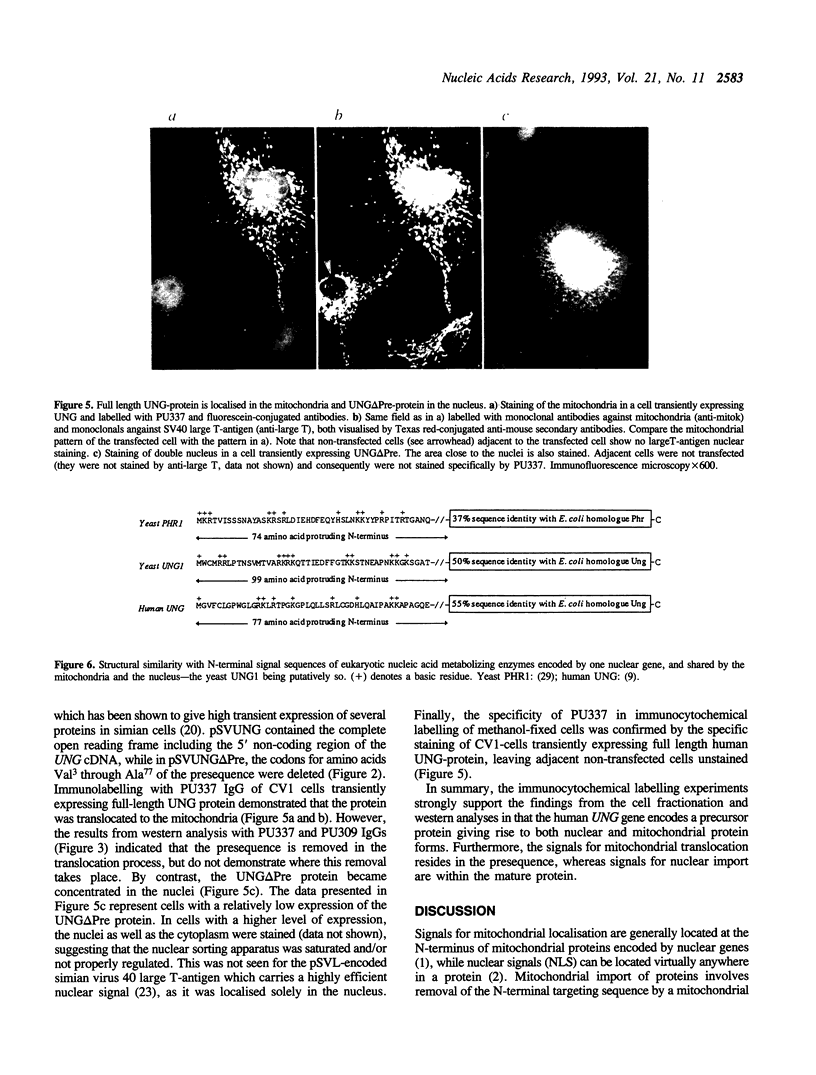
Recent cloning of a cDNA (UNG15) encoding human uracil-DNA glycosylase (UDG), indicated that the gene product of M(r) = 33,800 contains an N-terminal sequence of 77 amino acids not present in the presumed mature form of M(r) = 25,800. This led to the hypothesis that the N-terminal sequence might be involved in intracellular targeting. To examine this hypothesis, we analysed UDG from nuclei, mitochondria and cytosol by western blotting and high resolution gel filtration. An antibody that recognises a sequence in the mature form of the UNG protein detected all three forms, indicating that they are products of the same gene. The nuclear and mitochondrial form had an apparent M(r) = 27,500 and the cytosolic form an apparent M(r) = 38,000 by western blotting. Gel filtration gave essentially similar estimates. An antibody with specificity towards the presequence recognised the cytosolic form of M(r) = 38,000 only, indicating that the difference in size is due to the presequence. Immunofluorescence studies of HeLa cells clearly demonstrated that the major part of the UDG activity was localised in the nuclei. Transfection experiments with plasmids carrying full-length UNG15 cDNA or a truncated form of UNG15 encoding the presumed mature UNG protein demonstrated that the UNG presequence mediated sorting to the mitochondria, whereas UNG lacking the presequence was translocated to the nuclei. We conclude that the same gene encodes nuclear and mitochondrial uracil-DNA glycosylase and that the signals for mitochondrial translocation resides in the presequence, whereas signals for nuclear import are within the mature protein.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aasland R., Olsen L. C., Spurr N. K., Krokan H. E., Helland D. E. Chromosomal assignment of human uracil-DNA glycosylase to chromosome 12. Genomics. 1990 May;7(1):139–141. doi: 10.1016/0888-7543(90)90532-y. [DOI] [PubMed] [Google Scholar]
- Anderson C. T., Friedberg E. C. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res. 1980 Feb 25;8(4):875–888. [PMC free article] [PubMed] [Google Scholar]
- Bakke O., Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990 Nov 16;63(4):707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Braun H. P., Emmermann M., Kruft V., Schmitz U. K. The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J. 1992 Sep;11(9):3219–3227. doi: 10.1002/j.1460-2075.1992.tb05399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Verly W. G. Intracellular localization of rat-liver uracil-DNA glycosylase. Purification and properties of the chromatin enzyme. Eur J Biochem. 1983 Aug 15;134(3):415–420. doi: 10.1111/j.1432-1033.1983.tb07583.x. [DOI] [PubMed] [Google Scholar]
- Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol. 1992 Apr;12(4):1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domena J. D., Mosbaugh D. W. Purification of nuclear and mitochondrial uracil-DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry. 1985 Dec 3;24(25):7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- Ellis S. R., Hopper A. K., Martin N. C. Amino-terminal extension generated from an upstream AUG codon increases the efficiency of mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferases. Mol Cell Biol. 1989 Apr;9(4):1611–1620. doi: 10.1128/mcb.9.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M. N. Nuclear protein localization. Biochim Biophys Acta. 1991 Mar 7;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Gupta P. K., Sirover M. A. Stimulation of the nuclear uracil DNA glycosylase in proliferating human fibroblasts. Cancer Res. 1981 Aug;41(8):3133–3136. [PubMed] [Google Scholar]
- Hartl F. U., Neupert W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science. 1990 Feb 23;247(4945):930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Henkel T., Zabel U., van Zee K., Müller J. M., Fanning E., Baeuerle P. A. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-kappa B subunit. Cell. 1992 Mar 20;68(6):1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- Jameson B. A., Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988 Mar;4(1):181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Krokan H., Bjorklid E., Prydz H. DNA synthesis in isolated HeLa cell nuclei. Optimalization of the system and characterization of the product. Biochemistry. 1975 Sep 23;14(19):4227–4232. doi: 10.1021/bi00690a012. [DOI] [PubMed] [Google Scholar]
- Krokan H., Wittwer C. U. Uracil DNa-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981 Jun 11;9(11):2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M., Hopper A. K., Martin N. C. N2,N2-dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J Cell Biol. 1989 Oct;109(4 Pt 1):1411–1419. doi: 10.1083/jcb.109.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Siegler K., Mauro D. J., Seal G., Wurzer J., deRiel J. K., Sirover M. A. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S. J., Caradonna S. Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim Biophys Acta. 1991 Feb 16;1088(2):197–207. doi: 10.1016/0167-4781(91)90055-q. [DOI] [PubMed] [Google Scholar]
- Méjean V., Rives I., Claverys J. P. Nucleotide sequence of the Streptococcus pneumoniae ung gene encoding uracil-DNA glycosylase. Nucleic Acids Res. 1990 Nov 25;18(22):6693–6693. doi: 10.1093/nar/18.22.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Krokan H. E., Helland D. E. Human uracil-DNA glycosylase complements E. coli ung mutants. Nucleic Acids Res. 1991 Aug 25;19(16):4473–4478. doi: 10.1093/nar/19.16.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Rydberg B., Spurr N., Karran P. cDNA cloning and chromosomal assignment of the human O6-methylguanine-DNA methyltransferase. cDNA expression in Escherichia coli and gene expression in human cells. J Biol Chem. 1990 Jun 5;265(16):9563–9569. [PubMed] [Google Scholar]
- Sadler I., Chiang A., Kurihara T., Rothblatt J., Way J., Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J Cell Biol. 1989 Dec;109(6 Pt 1):2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 1985 Nov 25;13(22):8231–8246. doi: 10.1093/nar/13.22.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G., Olsen L. C., Helland D., Aasland R., Krokan H. E. Cell cycle regulation and in vitro hybrid arrest analysis of the major human uracil-DNA glycosylase. Nucleic Acids Res. 1991 Oct 11;19(19):5131–5137. doi: 10.1093/nar/19.19.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilin N. V., Aprelikova O. N. Uracil-DNA glycosylases and DNA uracil repair. Int Rev Cytol. 1989;114:125–179. doi: 10.1016/s0074-7696(08)60860-8. [DOI] [PubMed] [Google Scholar]
- Wittwer C. U., Bauw G., Krokan H. E. Purification and determination of the NH2-terminal amino acid sequence of uracil-DNA glycosylase from human placenta. Biochemistry. 1989 Jan 24;28(2):780–784. doi: 10.1021/bi00428a055. [DOI] [PubMed] [Google Scholar]
- Wittwer C. U., Krokan H. Uracil-DNA glycosylase in HeLa S3 cells: interconvertibility of 50 and 20 kDa forms and similarity of the nuclear and mitochondrial form of the enzyme. Biochim Biophys Acta. 1985 Dec 20;832(3):308–318. doi: 10.1016/0167-4838(85)90264-x. [DOI] [PubMed] [Google Scholar]
- Yasui A., Yajima H., Kobayashi T., Eker A. P., Oikawa A. Mitochondrial DNA repair by photolyase. Mutat Res. 1992 Mar;273(2):231–236. doi: 10.1016/0921-8777(92)90084-g. [DOI] [PubMed] [Google Scholar]





